Abstract
The active uptake of extracellular DNA and its genomic integration is termed natural transformation and constitutes a major horizontal gene-transfer mechanism in prokaryotes. Chromosomal DNA transferred within a species can be integrated effectively by homologous recombination, whereas foreign DNA with low or no sequence homology would rely on illegitimate recombination events, which are rare. By using the nptII+ gene (kanamycin resistance) as selectable marker, we found that the integration of foreign DNA into the genome of the Gram-negative Acinetobacter sp. BD413 during transformation indeed was at least 109-fold lower than that of homologous DNA. However, integration of foreign DNA increased at least 105-fold when it was linked on one side to a piece of DNA homologous to the recipient genome. Analysis of foreign DNA integration sites revealed short stretches of sequence identity (3–8 bp) between donor and recipient DNA, indicating illegitimate recombination events. These findings suggest that homologous DNA served as a recombinational anchor facilitating illegitimate recombination acting on the same molecule. Homologous stretches down to 183 nucleotides served as anchors. Transformation with heteroduplex DNA having different nucleotide sequence tags in the strands indicated that strands entered the cytoplasm 3′ to 5′ and that strands with either polarity were integrated by homologous recombination. The process led to the genomic integration of thousands of foreign nucleotides and often was accompanied by deletion of a roughly corresponding length of recipient DNA. Homology-facilitated illegitimate recombination would explain the introgression of DNA in prokaryotic genomes without the help of mobile genetic elements.
Transformation is considered a major horizontal gene-transfer mechanism contributing to genetic adaptation and evolution of prokaryotes (1–3). Among the transformable organisms, the two species Neisseria gonorrhoeae and Haemophilus influenzae take up preferentially DNA of their own species recognized by specific nucleotide sequences (4, 5), whereas other species such as Streptococcus pneumoniae, Bacillus subtilis, and Acinetobacter sp. do not discriminate between their own and foreign DNA (2). With sufficient nucleotide-sequence similarity between donor DNA and the recipient genome, integration can occur by homologous recombination leading to a replacement of alleles or the integration of heterologous sequences when bracketed by homologous regions. However, the efficiency of homologous recombination decreases strongly with decreasing nucleotide-sequence similarity (6, 7).
For the integration of foreign DNA with low or no sequence identity in bacteria, two basically different mechanisms have been identified. One relies on short specific nucleotide sequences recognized by cognate enzymes such as transposases or integrases that can cut and paste DNA at these sites. Nucleotide sequences bordered by such sites constitute genetic elements, examples being gene cassettes and integrons (8), transposons and insertion sequence elements (9), and conjugative transposons (10). The other mechanism is thought to depend on illegitimate (nonhomologous) recombination events that join DNA molecules at sites where they have no or only a few identical nucleotide pairs (11). Such events do not require RecA protein and often were identified as intramolecular recombination events following double-strand breaks. Examples are the formation of deletions in bacterial chromosomes (12) and plasmids (13, 14) and the formation of transducing phage after UV irradiation of lysogenic cells (15, 16). The integration of a linear piece of foreign DNA would require two such events.
Whether heterologous DNA taken up by competent cells could become part of their genome by illegitimate recombination is not known. To address this point we have chosen the naturally transformable environmental Gram-negative Acinetobacter sp. BD413 (17). Natural transformation of the strain is similar in several aspects to the transformation of other well studied species (2): (i) the level of DNA uptake competence is growth phase-dependent (18); (ii) as already mentioned, the cells bind and take up DNA from any source (19, 20); (iii) the active uptake initiates at an endonucleolytic cut within duplex DNA from where a single strand is translocated into the cytoplasm (20); and (iv) the genomic integration of homologous DNA is RecA-dependent (ref. 21 and J.d.V., unpublished data).
Here we show that Acinetobacter is transformed effectively by heterologous DNA fragments if they contain a single region providing homology to the recipient genome. This finding is reminiscent of the integration of phage λ DNA fragments into the S. pneumoniae genome during natural transformation when linked to chromosomal DNA (22). We have analyzed the integration of foreign DNA in Acinetobacter with respect to the sites of illegitimate recombination events and their nucleotide sequences, the length of the integrated fragment and the DNA loss from the recipient genome, the polarity of strand integration, and the effect of the length of the homologous region. The data show that homology-facilitated illegitimate recombination promotes foreign gene acquisition by genetic transformation, which would explain events of horizontal gene transfer between nonrelated organisms that were identified by genome sequencing and apparently are not associated with mobile genetic elements.
Materials and Methods
Strain Construction and Natural Transformation.
The recipient strain for natural transformation was Acinetobacter sp. BD413 trpE27 (DSM 588, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). The plasmids pKT210 (23), pMR7 (24), and pMR30 (Fig. 1) were introduced into Acinetobacter cells by electroporation (24). Selection was on LB media plates (25) containing chloramphenicol (25 μg/ml). Chromosomal DNA from Acinetobacter sp. BD4 trp+ (DSM 586) and plasmids were purified with the corresponding purification kits (Qiagen, Hilden, Germany). Natural transformation was carried out in aerated 1-ml cultures containing 2.5 × 108 recipient cells (24). Transforming DNA was included at 2.0 × 1010 nptII+ per ml, and nptII+ transformants were selected on LB agar containing 10 μg kanamycin/ml. Transformation by trpE+ DNA (100 ng/ml) was performed as described (26). Transformation frequencies are given as transformants per nptII+ or trpE+, respectively (genome size of Acinetobacter, 3.78 × 106 bp; ref. 27).
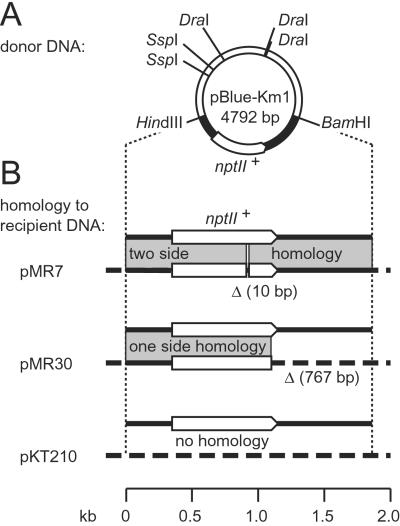
Figure 1.
The plasmids used for the analysis of recombination after natural transformation. The nptII is represented by arrows, shaded areas indicate regions of homology, and dashed lines indicate nonhomologous recipient DNA. (A) The donor DNA pBlue-Km1 carries the nptII+ gene on a 1.87-kb HindIII-BamHI fragment from Tn5. Restriction sites used for the linearization of the donor DNA are indicated. (B) The DNA corresponding to the HindIII-BamHI fragment of the donor DNA contains in pMR7 a 10-bp deletion and in pMR30 a substitution of the 51 C-terminal nucleotides of nptII plus the following 716 bp by eukaryotic tg4 terminator DNA. The vector plasmid of pMR7 and pMR30 was pKT210.
DNA Manipulations and Preparation of Heteroduplex DNA (HD) with Mismatches.
Cloning of DNA and sequencing was done by standard procedures (25). Site-directed mutagenesis was performed with the Quick Change kit (Stratagene) changing a GG downstream of nptII in pBlue-Km1 to TC in pBlue-Km2. The inserts of pBlue-Km1 and pBlue-Km2 [vector, pBluescript II SK(+)] were excised by HindIII and BamHI (Fig. 1) and ligated into the vector pBluescript II SK(−), yielding plasmids pBlue-Km3 and pBlue-Km4, respectively. Single-stranded plasmid DNA was purified from phage produced by Escherichia coli strain KK2186 (28) after infection with the helper phage R408 (Stratagene) and purified by gel electrophoresis. Linear duplex plasmid DNA was obtained by cleavage at the single XmnI site and purified by gel electrophoresis. Formation and purification of HD was carried out as described (29). Single-stranded pBlue-Km2 (-Km1) and double-stranded pBlue-Km1 (-Km2) yielded HD1 (or HD2), and double-stranded pBlue-Km3 and single-stranded pBlue-Km4 yielded HD3. HD was cut to completion by DraI, SspI, or HindIII as indicated.
Analysis of Transformants Obtained with HD.
The transformants were analyzed by colony PCR (≈105 cells per 10-μl PCR mixture) with a forward primer binding to nucleotides 7–26 of the nptII gene (5′-GAACAAGATGGATTGCACGC) and reverse primers specific for either the wild-type (5′-TTCGAACCCCAGAGTCCCGC) or mutant version of nptII (5′-TTCGAACCCCAGAGTCGAGC). The strand-discriminating nucleotides are in italics.
Results
Donor and Recipient DNA Molecules for Recombination Detection.
For the analysis of genetic recombination after natural transformation of Acinetobacter sp. BD413 trpE27, the donor DNA was a linear duplex and the recipient DNA a plasmid in the competent cells. The plasmid pBlue-Km1 treated with DraI and SspI provided the 3.72-kb donor DNA fragment carrying in its center nptII+ (conferring kanamycin resistance) as a selectable marker (Fig. 1A). Different plasmids in the recipient cells provided either no homology with the donor DNA or homology on only one or both sides of the selectable marker (Fig. 1B). The recipient plasmid pMR7 carried a defective nptII gene because of an internal 10-bp deletion, causing a frameshift mutation and deletion of an NcoI restriction site. pMR7 provided homologous sequences on both sides of the internal deletion (915 bp to the left, 945 bp to the right). In the plasmid pMR30, the last 51 nucleotides of the nptII+ gene and the adjacent Tn5-derived sequence downstream of nptII were replaced by heterologous DNA (the eukaryotic tg4 terminator; Fig. 1B), eliminating the kanamycin resistance. In this construct homology to the donor DNA was present only on the left side of the nptII truncation (1,096 bp). The vector pKT210 (23) without an insert was the recipient plasmid with no homology to the donor DNA. In the three recipient systems recombinative integration of nptII+ donor DNA can be detected by the formation of kanamycin-resistant transformants.
Recombinative Integration Occurs with One-Side Homology.
Cells of Acinetobacter carrying pMR7, pMR30, or pKT210 and having equal competence were transformed with the pBlue-Km1 DNA fragment (Fig. 1 and Table 1). With two-side homology in the recipient cells (pMR7), nptII+ was restored with high efficiency (Table 1). The frequency of 10−4 was ≈100-fold lower than that obtained with the chromosomal trpE+ marker, which is caused by the relatively small fragment size of the nptII+ DNA compared with that of the chromosomal DNA (≈30 kb) as described (30). When the recipient cells (with pKT210) did not provide homology, the integration frequency dropped by at least 9 orders of magnitude and fell below the limit of detection (1.3 × 10−13 transformants per nptII+; Table 1) as previously observed (26). In this case, the target for illegitimate integration of nptII+ was the plasmid plus the whole bacterial chromosome. With one-side homology in the recipient cells (pMR30), numerous kanamycin-resistant transformants were obtained with a frequency of ≈0.01% of that with the two-side homologous pMR7. This frequency was at least 105-fold higher than in the absence of homology. The results show that a fully heterologous stretch of nucleotides (encoding the 16 C-terminal amino acids of the NptII protein) can be integrated effectively when neighbored on only one side by a region of homology to the recipient DNA. A recA mutation decreased the integration frequency of one-side homologous DNA by at least a factor of 10 (unpublished data).
Table 1.
Transformation frequencies of Acinetobacter cells providing homology on two, one, or no side of the selectable donor sequence*
| Homology type (plasmid) | Transformants per
|
||
|---|---|---|---|
|
nptll+
|
102trpE+† | ||
| Absolute | Relative | ||
| Two-side (pMR7) | 0.9 (± 0.1) × 10−4 | 1.0 | 1.0 (± 0.01) |
| One-side (pMR30) | 1.1 (± 0.03) × 10−8 | 1.2 × 10−4 | 1.8 (± 0.3) |
| None (pKT210) | ≤1.3 × 10−13 | ≤1.4 × 10−9 | 2.1 (± 0.3) |
Frequencies are given with standard deviation; n = 3.
The competence of the cells was determined by transformation with chromosomal trpE+ DNA isolated from Acinetobacter sp. BD4.
One-Side Homologous Recombination Can Integrate a Heterologous Gene.
At a distance of 20 bp downstream of nptII+, the donor DNA contained the blm+ gene of Tn5 conferring bleomycin resistance. Liquid culturing in the presence of bleomycin (50 μg/ml) of 20 randomly chosen kanamycin-resistant transformants obtained with one-side homology revealed that 18 had gained resistance to bleomycin. This result suggested that the integrated DNA in most transformants exceeded the 51 bp of the nptII+ terminus by at least the 381 bp of blm+.
Determination of the Illegitimate Recombination Sites.
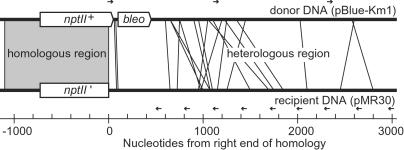
The recombination sites outside the homology in the 20 kanamycin-resistant transformants were localized by PCR (Fig. 2), and their nucleotide sequences were determined. The transformants (Fig. 2) had integrated 69–2,599 nucleotides of foreign DNA (average of 988 nucleotides) that substituted recipient DNA between 95 and 2,793 nucleotides (average of 1,120 nucleotides). Hot spots of illegitimate recombination were not apparent with respect to either the donor or recipient molecule (Fig. 2).
Figure 2.
Locations of illegitimate recombination sites of 20 transformants obtained with one-side homologous donor DNA. For each recombinant the fusion between donor and recipient DNA is shown by a line. The positions of forward and reverse primers complementary to the donor and recipient DNA used for the localizations are indicated by arrows.
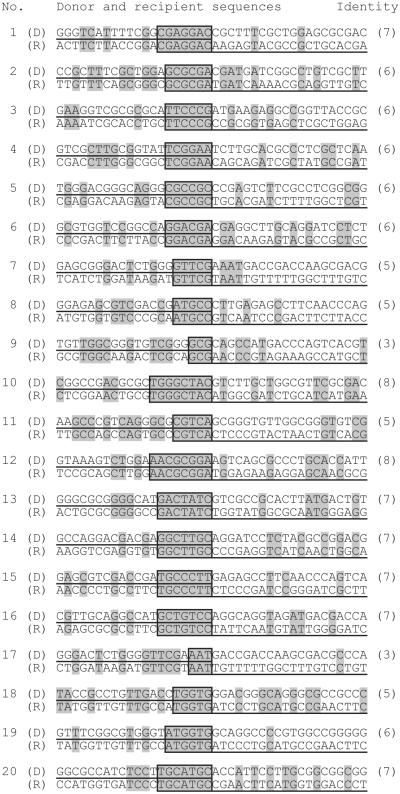
The sequences of the fusion sites between donor and recipient DNA revealed small regions of identity between both molecules (Fig. 3). These ranged from 3–8 nucleotides in length, resembling illegitimate recombination sites identified in other studies (13, 16, 31–34). Within the first 1 kb of the heterologous sequences, we found for the 995 possible hexanucleotides of the donor 281 matches in the recipient, a value close to that calculated for a random sequence with 50% GC content (242 matches). These calculations suggest that close contact between two unrelated sequences can lead to frequent matching of microhomologies. There was no consensus sequence discernible among the fusion points (Fig. 3).
Figure 3.
The nucleotide sequence of illegitimate recombination sites in 20 transformants. Nucleotides identical in donor (D) and recipient (R) sequences are indicated by shading. The sequences present in the transformants are underlined. The core regions of identity (boxed) contain the presumptive fusion sites, and their nucleotide number is indicated in brackets.
Even Short Homologous Regions Facilitate Illegitimate Recombination.
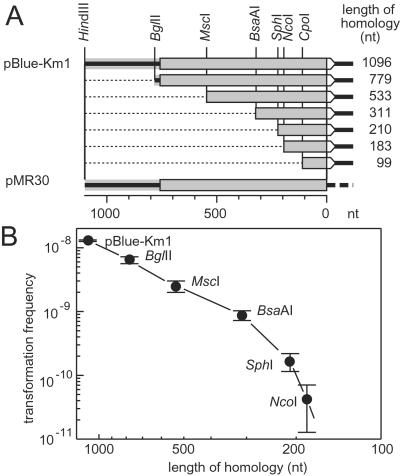
To determine the influence of the length of the homologous region on the frequency of illegitimate recombination events in its vicinity, a series of deletion derivatives of pBlue-Km1 was constructed (Fig. 4A). A decrease of the length of homology from 1,096 to 311 nucleotides decreased the transformation frequency 15-fold. With a further reduction to 183 homologous nucleotides, the transformation frequency dropped another 20-fold to a level that was still at least 500 times higher than that observed in the absence of homology. With 99 homologous nucleotides, it fell below the limit of detection (1.7 × 10−11 per donor molecule).
Figure 4.
Dependence of the transformation frequency with one-side homologous donor DNA on the length of the homologous region. The arrows represent the nptII gene and its deletion derivatives in the donor DNA, and the white arrowhead symbolizes the selective marker region. Shading indicates homology between donor and recipient (pMR30) DNA. (A) Deletion derivatives of pBlue-Km1 obtained by removal of nucleotides (dotted lines) between the HindIII site and the other indicated restriction sites. (B) Frequency of transformation (number of transformants per donor molecule) with pBlue-Km1 and the deletion derivatives. Data are given with SD (n = 3).
Heterologous 5′ and 3′ Ends Are Integrated.
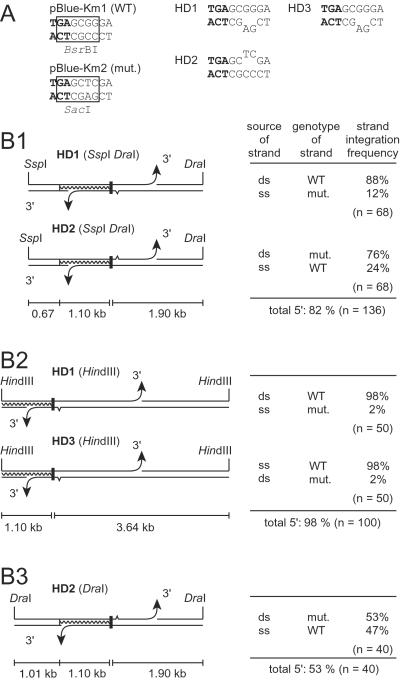
The polarity of strand uptake into the cytoplasm of Acinetobacter is unknown thus far. In duplex DNA molecules with one-side homology, the selective marker region (the 51 nucleotides of nptII+ missing in pMR30) is at the 5′ side of the homology on one strand and at the 3′ side on the other (Fig. 5B). To determine which of the donor strands was integrated into pMR30 we used HD in which the two strands differed by two adjacent mismatching bp, GG in the wild-type coding strand, and AG in the noncoding strand of the mutated DNA (Fig. 5A). These nucleotides served as genetic tags for the subsequent identification of the incorporated strand by PCR and restriction with BsrBI and SacI (Fig. 5A). The tag site was located 2 bp downstream of the stop codon of nptII in the heterologous DNA to provide high linkage with the marker and to prevent an influence of mismatch repair on its genomic establishment.
Figure 5.
Structures of HD molecules HD1, HD2, and HD3 and frequencies of genomic integration of the strands. (A) Nucleotide sequences including the stop codon of nptII (bold face) and the tag site of the donor plasmids. Restriction recognition sites are boxed in the wild-type (WT) and mutant (mut.) version of the plasmid. (B) Frequencies of genomic integration of strands with the selective marker region (black bar) on the 3′ or 5′ side of the homology (waved line). Restriction sites used for the formation of the transforming fragment are indicated. The arrowheads symbolize the 3′ ends of the marker-containing DNA fragment that would be released by a double strand cut in either the left or right flanking region of the marker. The sizes of the flanking regions are given. Mismatches are indicated by a tine in the mutant strand. The source (ds, double-stranded plasmid DNA; ss, single-stranded phage DNA), genotype (WT and mut.), and integration frequency are given for each strand. The numbers of transformants analyzed are indicated.
In transformants obtained with HD the nucleotide sequence at the tag site was analyzed by PCR with two primers specific for the two mismatches. In all cases PCR products were obtained with only one of them. The results were confirmed by restriction analysis with BsrBI and SacI (Fig. 5A). Transformants obtained with HD1 had acquired either strand, the majority of them with 5′ homology (88%; Fig. 5B1). The tag did not affect strand uptake and/or integration, because HD with the mutated sequence in the opposite strand (HD2) also gave predominantly transformants with 5′ homology (Fig. 5B1).
In HD1 the 5′-homologous strand was derived from the plasmid, whereas the 3′-homologous strand was the single-stranded phage DNA. For comparison we also constructed HD complementary to HD1 in which the 5′-homologous strand was the single-stranded phage DNA, and the 3′-homologous strand came from the plasmid (HD3, Fig. 5B2). HD1 and HD3 were cut with HindIII to remove the f1 replication origin upstream of nptII, which had opposite polarity in both HD and perhaps might influence integration frequencies. The HindIII cleavage moved the homologous region plus the marker region closer to the left end of the molecule (Fig. 5B). With both constructs the ratio of 5′/3′ ends integrated was identical, and an even stronger preponderance of the 5′-homologous integration was observed (98%; Fig. 5B2). We concluded that the proportion of integrated 5′- and 3′-homologous strands was independent of the DNA source but depended on the position of the selective marker region within the linear donor DNA fragment. This conclusion was supported by the results obtained with HD2 cleaved with DraI in which the selective marker region is located almost precisely in the middle of the DNA molecule (Fig. 5B3). This DNA gave almost equal proportions of 5′- and 3′-homologous strand integration (53 vs. 47%). The data in Fig. 5B suggest that the probability for initiation of strand uptake on each side of the marker is a function of the relative length of the DNA in the marker-flanking regions.
Discussion
Our studies on natural transformation of Acinetobacter show that illegitimate recombination events are enhanced by homologous recombination acting on the same DNA molecule. Although illegitimate recombination (leading to integration of a heterologous marker) was not detected in the absence of homology, its frequency was increased at least 105-fold when a 1-kb region of homology to the recipient DNA was present on the otherwise heterologous donor DNA (Table 1). We propose that the homologous sequence provides a recombinational anchor for a RecA-dependent strand transfer that links heterologous DNA to a recipient molecule and thereby creates favorable conditions for illegitimate interactions, probably by maintaining close proximity between the heterologous sequences. The frequencies of an illegitimate recombination event associated with a homologous integration was ≈10−4 (Table 1). The recombinational anchor function decreased with decreasing homology length but still was measurable at 183 bp (Fig. 4). In transformation of Acinetobacter with fully homologous DNA, a minimum length of 294 bp was observed previously (30).
Each strand of a fully homologous double-stranded donor DNA molecule has the same probability to enter the cytoplasm and be integrated into the chromosome. In the one-side homologous donor DNA, however, the two strands of the duplex differ in that the homology is located either 3′ or 5′ of the selected marker (Fig. 5). The fact that either strand was identified in transformants indicated that each can enter the cytoplasm and be integrated into the genome.
The proportion of integrated 5′- and 3′-homologous strands was influenced by the position of the marker within the linear donor DNA (Fig. 5B). Because the uptake of a single strand starts from a random double-strand cut (20), both strands of a linear duplex carrying a marker in its center would have the same chance to transfer the marker into the cytoplasm. In fact, with the selective marker region in the middle of the DNA, the transformation efficiency of each strand was close to 50% (Fig. 5B3). The reduction of the length of the DNA left to the marker from 2.1 to 1.8 and 1.1 kb reduced the fraction of integrated 3′-homologous strands from 47 to 18 and 2%, respectively (lower strands in Fig. 5B). This result indicates that the uptake into the cytoplasm occurs from 3′ to 5′, because a shorter length of DNA left to the marker gives a lower chance for the initial cut to occur 3′ of the selective marker region in the 3′-homologous strand and thus will reduce the fraction of transformants formed by this strand (Fig. 5B). The effect of shortening the flanking region left of the selective marker region is particularly strong in our construct, because the homologous anchor region is located on this side, and any truncation of it would decrease the integration efficiency (Fig. 4). The inferred 3′ to 5′ direction of strand uptake into the cytoplasm of Acinetobacter is identical to that found in H. influenzae (35) and S. pneumoniae (36).
In vitro joint molecule formation by RecA protein of E. coli does not require homology at a DNA end. Strand transfer can initiate at an internal homologous region and then proceed 5′ to 3′ with respect to the single strand originally bound by RecA (for a review see ref. 37). From studies on the natural transformation of S. pneumoniae, it was proposed also that strand exchange by RecA initiates at internal paranemic joints (not only from an end) and proceeds 5′ to 3′ (38). Because one-side homologous donor strands of both polarities transferred a heterologous marker into the recipient genome, there seems to be no specific requirement for a homologous 3′ or 5′ end. This conclusion would be in accord with the recent in vitro demonstration that 3′ strands are as reactive as 5′ strands (39). We propose that the homologous RecA-catalyzed strand transfer at the anchor region is the first step in the genomic integration of a flanking heterologous region at either end.
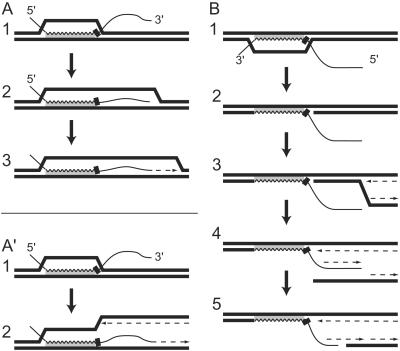
Because either strand can integrate in the homologous anchor region, the cells must have means to integrate 5′ and 3′ ends on the heterologous side. It is known that joint molecules formed as intermediates in homologous recombination lead to the priming of DNA replication when the 3′ end of the donor DNA is fully hybridized after branch migration (40). If the 3′ end is heterologous, priming may occur at transient contacts mediated by microhomologies. The concomitant primer extension would stabilize transient contacts and lead to the fusion of heterologous sequences to resident sequences (Fig. 6A). Alternatively, the fusion may occur at an approaching replication fork by using the 3′ end of the donor DNA at a microhomology as the primer for lagging strand synthesis, also resulting in primer extension (Fig. 6A′). In both models the establishment of the marker would require gene conversion, e.g., by replication. Fusion of heterologous DNA ends by an exonuclease-deficient derivative of the Klenow fragment of DNA polymerase I of E. coli has been demonstrated in vitro (41), and a promoting role of DNA polymerase I for illegitimate recombination has been established by in vivo experiments (42).
Figure 6.
Models for genomic integration of a heterologous 3′ (A A′) and 5′ end (B) of one-side homologous DNA. Homology between the donor (thin line) and recipient (bold lines) DNA is indicated by shading. In the donor DNA the homologous (waved) and selective marker (black bar) regions are indicated. The dotted arrows indicate newly synthesized DNA. For details see Discussion.
The integration of heterologous 5′ ends could start with the covalent joining of donor and recipient DNA within the anchor region (Fig. 6B 1 and 2). During the approach of a replication fork the unpaired part of the donor DNA could be converted into a double-strand by acting as a template for lagging strand synthesis (Fig. 6B 3 and 4). This synthesis would lead to the structure of a double-strand break in the lagging strand duplex with a 3′-protruding single-stranded region extending from the priming site of the last Okazaki fragment. Double-strand break repair taking advantage of microhomologies then could fuse duplex donor and recipient DNA, e.g., by a splice mechanism (Fig. 6B5). In E. coli, joining of DNA ends in the absence of homology occurs frequently at short direct repeats of 1 to ≈6 nucleotides (13). Such fusions are mediated by DNA polymerases, and the nucleotides between the direct repeats are deleted (43, 44). Exposure of matching sequences by the formation of single strands (Fig. 6B4) is promoted by nucleases and helicases (14, 45–48). The replication fork assembly at recombination intermediates such as in Fig. 6B2 is required for normal growth of E. coli and was demonstrated in vitro (49, 50).
Although in our experiments the selected marker was located at the very beginning of the heterologous donor DNA, there were frequently thousands of heterologous nucleotides incorporated beyond this point (Fig. 2). This can be explained by the fact that RecA can drive strand transfer over hundreds of bases 5′ to 3′ into heterologous regions (51). Alternatively, RuvAB-promoted branch migration proceeding in both directions can bypass even longer heterologous regions than the RecA-promoted reaction (52, 53). Both branch migration mechanisms produce plectonemically interwound strands in which the two strands probably would be roughly of the same length. This view would be consistent with the observation that many of the heterologous inserts substituted similar lengths of the recipient sequences.
Anchor sequence-facilitated illegitimate recombination can produce insertions, deletions, and substitutions next to the anchor. In our experiments the length of substitutions (and deletions) was limited to ≈2.9 kb, because longer exchanged tracks would have removed the replication origin from the recipient plasmid DNA. It should be noted that new one-side homologous substrates for further transfers are generated by new fusions in each illegitimate recombination event.
Anchor sequences of the size of transfer RNA loci were active in our experiments (Fig. 4). Such loci are conserved and have been considered as integration sites for distinct DNA segments in different strains and species (54–57). Introgression of genes and operons (58, 59) and the evolution of gene clusters by horizontal gene transfer (60) could occur with the help of anchor sequences. A similar mechanism as described here was observed in S. pneumoniae naturally transformed by one-side homologous substrates [see accompanying paper by Prudhomme et al. (61)]. These observations on transformable bacteria display a remarkable similarity to various instances where illegitimate recombination events were found to be associated with homologous recombination. These cases include bacteria that are not naturally transformable (62, 63) and various eukaryotic cells such as yeast (64), insect cells (65), mammalian cells (66–68), and plant cells (69). Integration of heterologous DNA adjacent to homologous recombination regions may contribute substantially to genome evolution in which gene acquisition and gene loss play key roles (70, 71).
Acknowledgments
We thank Petra Meier for the preparation of HD and Era Cassuto for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie.
Abbreviation
- HD
heteroduplex DNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lorenz M G, Wackernagel W. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubnau D. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Arber W. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- 4.Danner D B, Deich R A, Sisco K L, Smith H O. Gene. 1980;11:311–318. doi: 10.1016/0378-1119(80)90071-2. [DOI] [PubMed] [Google Scholar]
- 5.Goodman S D, Scocca J J. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zawadzki P, Roberts M S, Cohan F M. Genetics. 1995;140:917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strätz M, Mau M, Timmis K N. Mol Microbiol. 1996;22:207–215. doi: 10.1046/j.1365-2958.1996.00099.x. [DOI] [PubMed] [Google Scholar]
- 8.Recchia G D, Hall R M. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 9.Berg D E, Howe M M. Mobile DNA. Washington, DC: Am. Soc. Microbiol.; 1989. [Google Scholar]
- 10.Salyers A A, Shoemaker N B, Li L Y, Stevens A M. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich S D. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 799–824. [Google Scholar]
- 12.Brake A J, Fowler A V, Zabin I, Kania J, Müller-Hill B. Proc Natl Acad Sci USA. 1978;75:4824–4827. doi: 10.1073/pnas.75.10.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albertini A M, Hofer M, Calos M P, Miller J H. Cell. 1982;29:319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- 14.Allgood N D, Silhavy T J. Genetics. 1991;127:671–680. doi: 10.1093/genetics/127.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franklin N C. In: The Bacteriophage Lambda. Hershey A D, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1971. pp. 175–194. [Google Scholar]
- 16.Shimizu H, Yamaguchi H, Ashizawa Y, Kohno Y, Asami M, Kato J, Ikeda H. J Mol Biol. 1997;266:297–305. doi: 10.1006/jmbi.1996.0794. [DOI] [PubMed] [Google Scholar]
- 17.Juni E. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmen R, Vosman B, Kok R, van der Zee J R, Hellingwerf K J. Mol Microbiol. 1992;6:1747–1754. doi: 10.1111/j.1365-2958.1992.tb01347.x. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz M G, Reipschläger K, Wackernagel W. Arch Microbiol. 1992;157:355–360. doi: 10.1007/BF00248681. [DOI] [PubMed] [Google Scholar]
- 20.Palmen R, Vosman B, Buijsman P, Breek C K D, Hellingwerf K J. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 21.Gregg-Jolly L A, Ornston L N. Mol Microbiol. 1994;12:985–992. doi: 10.1111/j.1365-2958.1994.tb01086.x. [DOI] [PubMed] [Google Scholar]
- 22.Claverys J P, Lefevre J C, Sicard A M. Proc Natl Acad Sci USA. 1980;77:3534–3538. doi: 10.1073/pnas.77.6.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 24.de Vries J, Wackernagel W. Mol Gen Genet. 1998;257:606–613. doi: 10.1007/s004380050688. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.de Vries J, Meier P, Wackernagel W. FEMS Microbiol Lett. 2001;195:211–215. doi: 10.1111/j.1574-6968.2001.tb10523.x. [DOI] [PubMed] [Google Scholar]
- 27.Gralton E M, Campbell A L, Neidle E L. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 28.Zagursky R J, Berman M L. Gene. 1984;27:183–191. doi: 10.1016/0378-1119(84)90139-2. [DOI] [PubMed] [Google Scholar]
- 29.Westmoreland J, Porter G, Radman M, Resnick M A. Genetics. 1997;145:29–38. doi: 10.1093/genetics/145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmen R, Hellingwerf K J. Gene. 1997;192:179–190. doi: 10.1016/s0378-1119(97)00042-5. [DOI] [PubMed] [Google Scholar]
- 31.Jones I M, Primrose S B, Ehrlich S D. Mol Gen Genet. 1982;188:486–489. doi: 10.1007/BF00330053. [DOI] [PubMed] [Google Scholar]
- 32.Marvo S L, King S R, Jaskunas S R. Proc Natl Acad Sci USA. 1983;80:2452–2456. doi: 10.1073/pnas.80.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumagai M, Ikeda H. Mol Gen Genet. 1991;230:60–64. doi: 10.1007/BF00290651. [DOI] [PubMed] [Google Scholar]
- 34.Meima R, Haijema B J, Dijkstra H, Haan G-J, Venema G, Bron S. J Bacteriol. 1997;179:1219–1229. doi: 10.1128/jb.179.4.1219-1229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barany F, Kahn M E, Smith H O. Proc Natl Acad Sci USA. 1983;80:7274–7278. doi: 10.1073/pnas.80.23.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mejean V, Claverys J P. Mol Gen Genet. 1988;213:444–448. doi: 10.1007/BF00339614. [DOI] [PubMed] [Google Scholar]
- 37.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasta F, Sicard M A. Proc Natl Acad Sci USA. 1999;96:2943–2948. doi: 10.1073/pnas.96.6.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIlwraith M J, West S C. J Mol Biol. 2001;305:23–31. doi: 10.1006/jmbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 40.Kuzminov A, Stahl F W. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King J S, Fairley C F, Morgan W F. J Biol Chem. 1996;271:20450–20457. doi: 10.1074/jbc.271.34.20450. [DOI] [PubMed] [Google Scholar]
- 42.Stodolsky M. Genetics. 1973;73:65–66. [PubMed] [Google Scholar]
- 43.King J S, Valcarcel E R, Rufer J T, Phillips J W, Morgan W F. Nucleic Acids Res. 1993;21:1055–1059. doi: 10.1093/nar/21.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King J, Fairley C, Morgan W. Nucleic Acids Res. 1998;26:1749–1754. doi: 10.1093/nar/26.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shanado Y, Kato J, Ikeda H. J Bacteriol. 1997;179:4239–4245. doi: 10.1128/jb.179.13.4239-4245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamashita T, Hanada K, Iwasaki M, Yamaguchi H, Ikeda H. J Bacteriol. 1999;181:4549–4553. doi: 10.1128/jb.181.15.4549-4553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanada K, Iwasaki M, Ihashi S, Ikeda H. Proc Natl Acad Sci USA. 2000;97:5989–5994. doi: 10.1073/pnas.100101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Marians K J. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Xu L, Sandler S J, Marians K J. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bianchi M E, Radding C M. Cell. 1983;35:511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- 52.Iype L E, Wood E A, Inman R B, Cox M M. J Biol Chem. 1994;269:24967–24978. [PubMed] [Google Scholar]
- 53.Adams D E, West S C. J Mol Biol. 1996;263:582–596. doi: 10.1006/jmbi.1996.0600. [DOI] [PubMed] [Google Scholar]
- 54.Lawrence J G, Ochman H. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanc-Potard A B, Groisman E A. EMBO J. 1997;16:5376–5585. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 57.Hensel M, Nikolaus T, Egelseer C. Mol Microbiol. 1999;31:489–498. doi: 10.1046/j.1365-2958.1999.01190.x. [DOI] [PubMed] [Google Scholar]
- 58.Lawrence J G, Roth J R. Genetics. 1996;142:11–24. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kroll J S, Wilks K E, Farrant J L, Langford P R. Proc Natl Acad Sci USA. 1998;95:12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence J G, Roth J R. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prudhomme M, Libante V, Claverys J-P. Proc Natl Acad Sci USA. 2002;99:2100–2105. doi: 10.1073/pnas.032262999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kusano K, Sakagami K, Yokochi T, Naito T, Tokinaga Y, Ueda E, Kobayashi I. J Bacteriol. 1997;179:5380–5390. doi: 10.1128/jb.179.17.5380-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuteja D, Dua M, Khanna R, Dhingra N, Khanna M, Kaur H, Saxena D M, Lal R. Plasmid. 2000;43:1–11. doi: 10.1006/plas.1999.1426. [DOI] [PubMed] [Google Scholar]
- 64.Kunes S, Botstein D, Fox M S. Genetics. 1990;124:67–80. doi: 10.1093/genetics/124.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cherbas L, Cherbas P. Genetics. 1997;145:349–358. doi: 10.1093/genetics/145.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berinstein N, Pennell N, Ottaway C A, Shulman M J. Mol Cell Biol. 1992;12:360–367. doi: 10.1128/mcb.12.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakagami K, Tokinaga Y, Yoshikura H, Kobayashi I. Proc Natl Acad Sci USA. 1994;91:8527–8531. doi: 10.1073/pnas.91.18.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang Y K, Park J S, Lee C S, Yeom Y I, Chung A S, Lee K K. J Biol Chem. 1999;274:36585–36591. doi: 10.1074/jbc.274.51.36585. [DOI] [PubMed] [Google Scholar]
- 69.Puchta H, Dujon B, Hohn B. Proc Natl Acad Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe H, Mori H, Itoh T, Gojobori T. J Mol Evol. 1997;44, Suppl 1:57–64. doi: 10.1007/pl00000052. [DOI] [PubMed] [Google Scholar]
- 71.Koonin E V, Galperin M Y. Curr Opin Genet Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]