Abstract
Integration of foreign DNA was observed in the Gram-positive human pathogen Streptococcus pneumoniae (pneumococcus) after transformation with DNA from a recombinant Escherichia coli bacteriophage λ carrying a pneumococcal insert. Segments of λ DNA replaced chromosomal sequences adjacent to the region homologous with the pneumococcal insert, whence the name insertion-deletion. Here we report that a pneumococcal insert was absolutely required for insertion-deletion formation, but could be as short as 153 bp; that the sizes of foreign DNA insertions (289–2,474 bp) and concomitant chromosomal deletions (45–1,485 bp) were not obviously correlated; that novel joints clustered preferentially within segments of high GC content; and that the crossovers in 29 independent novel joints were located 1 bp from the border or within short (3–10 nt long) stretches of identity (microhomology) between resident and foreign DNA. The data are consistent with a model in which the insert serving as a homologous recombination anchor favors interaction and subsequent illegitimate recombination events at microhomologies between foreign and resident sequences. The potential of homology- directed illegitimate recombination for genome evolution was illustrated by the trapping of functional heterologous genes.
Genetic transformation, which was discovered in the Gram-positive Streptococcus pneumoniae (pneumococcus) (1), is believed to play a central role in the biology of this human pathogen through its contribution to genetic plasticity (see ref. 2 for a review). Transformation with naked DNA allows intraspecies and interspecies gene transfer. As such exchanges involve homologous recombination, they are generally “conservative,” i.e., they do not result in the creation of novel sequences but simply in a redistribution of previously existing genes. However, a pre-existing gene also can be modified when the two interacting sequences are partly divergent. Homologous recombination then leads to the production of mosaic genes, as exemplified in the case of the pbp genes of S. pneumoniae that encode altered penicillin-binding proteins with decreased affinity for β-lactam antibiotics (see ref. 3 for a review).
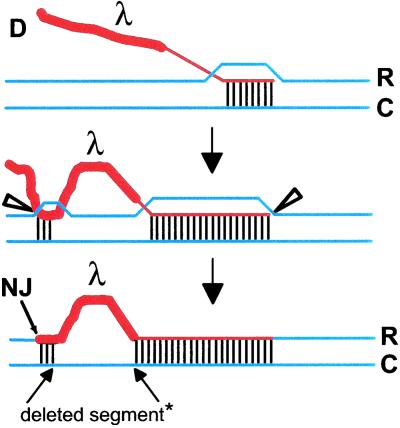
Besides these conservative facets of transformation, is there any potential for the creation of novel sequence combinations or genes? The observation that transformation with chimeric donor DNA is mutagenic (4) suggested that shuffling and reassembly of previously unrelated sequences could readily occur in S. pneumoniae. The chimeric DNA extracted from a recombinant Escherichia coli bacteriophage λ carrying a pneumococcal insert produced illegitimate recombinants at a frequency of about 0.5% that of homologous recombinants (4). Illegitimate recombinants resulted from simultaneous insertion of heterologous vector (i.e., λ) sequences and deletion of chromosomal sequences adjacent to the region homologous to the insert. These illegitimate events were therefore termed insertion-deletions (InsDels). As the pneumococcal insert in the chimeric donor seemed required for production of InsDels, the underlying mechanism was tentatively named homology-directed illegitimate recombination (5). Our working model for homology-directed illegitimate recombination postulates that pairing between homologous donor and recipient sequences (the homologous recombination anchor) favors transient pairing between foreign DNA adjacent to S. pneumoniae DNA in the donor (i.e., λ or any DNA) and resident sequences (Fig. 1). A key feature of the model is the presence of a short segment of sequence identity (hereafter termed microhomology) between the interacting heterologous donor and resident sequences. Resolution of the heteroduplex would result in integration of heterologous DNA and concomitant loss of resident sequences (Fig. 1).
Figure 1.
Model for the creation of NJs during transformation of S. pneumoniae. RecA-driven invasion of the homologous donor strand (D, in red) into the recipient duplex (in blue) produces a donor-recipient heteroduplex that favors transient pairing between heterologous donor DNA (λ or any DNA; thick red line) and the complementary strand (C) of the recipient chromosome at microhomologies (three vertical bars). Concomitant incision of the heterologous strand and the displaced chromosomal recipient strand (R) by a putative resolvase (open triangle) followed by ligation would create a NJ. DNA replication would then generate a wild-type and a mutant chromosome. No strand polarity is indicated as the polarity of heteroduplex extension for D-loop joint molecules is unclear (33). *, deleted in the R strand.
The aim of the present work was to characterize the trapping of foreign DNA with respect to the length and nature of the pneumococcal insert, the relationship between the sizes of insertions and accompanying deletions, and the structure of novel joints (NJs) formed in the process.
Materials and Methods
Bacterial Strains and Plasmids.
Strain R800 (6) was used as a wild-type S. pneumoniae strain together with strains R304 as a donor of the streptomycin resistance marker str41, TCHA2, and TCHA3. Strain TCHA2 is a derivative of strain R800 harboring a 3′ deletion of hexA (from the PvuII site, at position 2264 with respect to the start, to the 3′ extremity of the gene; last 89 triplets deleted) and a 5′ truncated ermAM gene (up to the unique ScaI site; first 17 triplets missing). Strain TCHA3 is similar to strain TCHA2 except that the hexA gene is more severely truncated (from the EcoRV site to the 3′ extremity of the gene; last 278 triplets deleted). Construction of strains TCHA2 and TCHA3 and plasmids is described in additional Methods and Table 2, which are published as supporting information on the PNAS web site, www.pnas.org.
Plasmid pR313 (Table 2) is a ColE1 derivative carrying a 942-bp-long EcoRI–HindIII amiC–amiD fragment from the ami locus of S. pneumoniae (7). Plasmid pCHS101 is a ColE1 derivative carrying a NcoI–BamHI insert containing the Salmonella thyphimurium mutS gene (8) from plasmid pGW1812 (9) (Table 2).
DNA Manipulations.
Methods for preparing S. pneumoniae chromosomal DNA, obtaining plasmid DNA from E. coli or S. pneumoniae, and purifying DNA fragments have been described (10). Restriction enzymes and DNA modification enzymes were used as recommended by the manufacturer. The following primers were used in this study: MP105 (5′-cgcgccatggTTTTACCTGGGGCCACATG-3′; mutS, positions −65 to −37, and NcoI site), MP106 (5′-cgcggatccGCGCTATCGGGTCTGACACG-3′; mutS, 2583 to 2564, and BamHI site), MP112 (5′-GGGCTTCATCGCTGATAGTGCC-3′; mutS, 376 to 355), MP113 (5′-GCGGGTCGCGGCATCCA-3′; mutS, 828 to 812), MP114 (5′-GCGCGTTCCAGGAGGTCG-3′; mutS, 1231 to 1214), MP115 (5′-CCAGATGCGGCAACAGCAGA-3′; mutS, 1618 to 1599), MP116 (5′-CCAGACTGTTTTCCGTGGCATT-3′; mutS, 2068 to 2047), MP117 (5′-gcgccatggTCGAGATCAGCACGTTGCCA-3′; hexA, −114 to −95, and NcoI site). [Positions are given with respect to the start of each gene; lowercase letters indicate additional nucleotides containing restriction sites (underlined) introduced for cloning]. PCR was performed by using Hot Tub DNA polymerase (Amersham Pharmacia), primers (1 mM), and 25 cycles of amplification (30-s denaturation at 94°C, 1-min annealing at 55°C, and 4-min extension at 72°C) with a final 10-min extension at 72°C. PCR products amplified from chromosomal DNA were purified with the QIAquik PCR Purification Kit (Qiagen, Chatsworth, CA) and sequenced by using a CircumVent Thermal Cycle dideoxy DNA sequecing kit (New England Biolabs).
Growth and Transformation Media.
Cultures of E. coli and S. pneumoniae and methods for the preparation of competent cells and transformation have been described (10). Antibiotic concentrations used for the selection of transformants were: erythromycin (Ery), 0.2 mg ml−1; methotrexate (MTX), 3 × 10−6 M; streptomycin, 200 mg ml−1.
Results and Discussion
Trapping of a Foreign Gene During Transformation with Hybrid DNA.
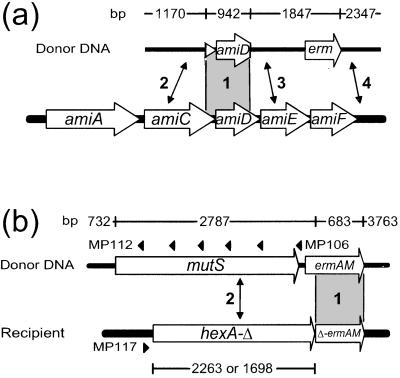
To evaluate the efficiency of InsDels for the trapping of foreign genes, linearized molecules from a nonreplicative recombinant plasmid, pR313, were used as donor in transformation of the wild-type S. pneumoniae strain R800. Plasmid pR313 carries a pneumococcal DNA insert from the ami operon overlapping the amiC and amiD genes and providing a 942-bp-long homologous anchor for recombination (region 1 in Fig. 2a). The ami insert is flanked by the Ery resistance gene, ermAM (Fig. 2a). Illegitimate recombination events occurring on either side of the insert were expected to lead to resistance to MTX, through inactivation of the ami operon (4), whereas only those in region 4 (Fig. 2a) would promote capture of the ermAM gene and result in Ery resistance.
Figure 2.
Experimental systems to investigate InsDel formation and the capture of foreign DNA. Lengths of donor segments are indicated above the map. (a) The R800 wild-type recipient was transformed by using as donor purified PstI-linearized plasmid pR313 DNA. (b) Purified HindIII-linearized plasmid pCHS101 DNA was used to transform the TCHA2 or TCHA3 recipients. Diagnostic oligonucleotides used as PCR primers (MP117 combined with MP112 to MP116 or MP106) for the analysis of recombinants are indicated by black triangles (numbering of MP113 to MP116, located from left to right between MP112 and MP106, was omitted for clarity). The homologous anchor provided by the amiC–amiD or the ermAM segments, present in both donor and recipient DNA in a and b respectively, is shown in gray (region 1). While NJs formed in regions 2, 3, or 4 in a lead to InsDels (scored as MTXR transformants), only those formed in region 4 promote capture of the ermAM gene (EryR transformants). (b) NJs formed to the left of ermAM (region 2) restore a functional ermAM gene. The truncated hexA coding region is 2,263 bp (49.1% identity with mutS) or 1,698 bp long (46.9% identity with mutS) in strain TCHA2 and TCHA3, respectively (Materials and Methods).
MTXR transformants were readily obtained by using linearized pR313 as donor (Table 1), indicating that a shorter recombination anchor (942 bp versus 2,695 bp in ref. 4) efficiently promoted the formation of InsDels. About 50% of InsDels resulted in capture of the ermAM gene as deduced from the EryR to MTXR ratio (Table 1). InsDels leading to trapping of another antibiotic resistance gene, cat, which confers resistance to chloramphenicol, was previously reported after transformation with hybrid DNA carrying part of the recA locus (10) or the comCDE locus (11), but the efficiency of the trapping process was not established. Here InsDel frequency using linear pR313 as donor was 1–2% of the frequency of transformation of a streptomycin resistance marker or of integration of the entire pR313 plasmid, using as donor-saturating amounts of chromosomal DNA or intact covalently closed circles, respectively (Table 1). The absolute frequency of ermAM gene capture in the transformed culture was about 10−4.
Table 1.
Trapping of foreign DNA during transformation with hybrid molecules
| Donor DNA type of event | R304 (chromosomal) point mutation transfer | pR313 ccc plasmid integration* | Linear pR313 InsDel |
|---|---|---|---|
| SmR ml−1 | 0.99 × 106 | — | — |
| MTXR ml−1 | — | 1.45 × 106 | 2.02 × 104 |
| EryR ml−1 | — | 1.31 × 106 | 8.6 × 103 |
| EryR/MTXR | — | 0.90 | 0.47 |
The most frequent event occurring with covalently closed circles (ccc) donors is integration of the entire plasmid (34). InsDel represent 5% of total events with ccc donors (unpublished observations).
The observation that 50% of InsDels lead to ermAM capture would be consistent with illegitimate recombination events occurring with equal probabilities on either side of the insert, the average size of integrated heterologous DNA segments being larger than 1,847 bp (the minimal size required for integration of an intact ermAM gene; Fig. 2a).
Selection of InsDels at the hexA Locus.
To establish whether partially divergent sequences could serve as a recombination anchor, a wild-type recipient was first transformed with linear DNA from pCHS101, a nonreplicative recombinant plasmid that carried the intact ermAM gene 174 bp downstream of the mutS gene of S. typhimurium (Fig. 2b). The mutS gene (2,559 bp) shares 48.9% identity with the S. pneumoniae mismatch repair hexA gene (2,532 bp) (12). No EryR transformants were obtained, indicating that hexA and mutS sequences could not interact to promote the capture of ermAM (data not shown). This finding confirmed the absolute requirement for a homologous recombination anchor. Taken together with our previous observation that homologous recombination in S. pneumoniae dropped by at least 6 orders of magnitude in a recA mutant (in fact below the limit of detection) (10), this result implies that production of InsDels strictly depends on RecA.
We then constructed an S. pneumoniae recipient strain in which the 3′ region of hexA was replaced by a truncated copy of ermAM to provide a 683-bp-long homologous recombination anchor (Fig. 2b; Materials and Methods). EryR transformants were readily obtained after transformation of this strain, TCHA2, with linearized pCHS101 DNA. EryR frequency was similar to that observed in Table 1, suggesting that inactivation of mismatch repair in the recipient strain TCHA2 had no effect on the rate of appearance of transformants (data not shown).
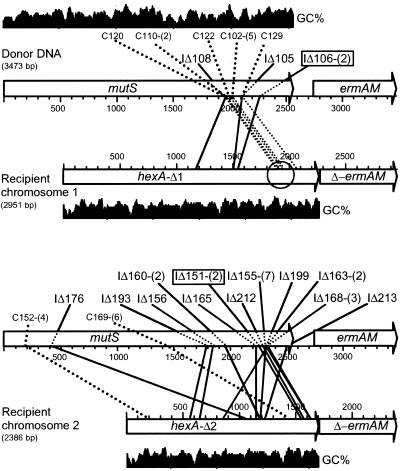
Detailed analysis of the molecular structure of hexA–mutS recombinants was carried out with a set of 30 clones by a combination of PCR and DNA sequencing. Briefly, a series of PCRs using a hexA-specific primer (MP117) combined with primers evenly distributed along the mutS gene (MP106, MP112–MP116; Fig. 2b) was first carried out to evaluate the position of the NJ. A preliminary assessment of the nature of recombinants was deduced from the sizes of PCR products (data not shown). For 26 clones (87%), PCR fragment lengths were consistent with predictions for in-frame full-sized fusions between hexA and mutS, potentially encoding full-sized chimeric proteins (hereafter termed interspecies recombinants; data not shown). Of 26 interspecies recombinants, 24 corresponded to crossovers located between MP115 and MP116 (Fig. 2b). PCR fragments were then sequenced as described in Materials and Methods by using the primer closest to each junction. DNA sequencing across the NJs for 10 (randomly chosen) of these 24 recombinants revealed that 9 of 10 crossovers were clustered within a 66-nt-long segment having the highest level of sequence identity (80.6%) between hexA (positions 1921–1986) and mutS (positions 1957–2023) (Fig. 3 Upper). This region contained the longest stretch of identity (19 consecutive nt) between the two sequences (circled in Fig. 3 Upper). These results strongly suggested that synapsis between homologous donor and recipient ermAM sequences favored “in register” pairing between mutS and hexA sequences, in the region of best match. Only four clones obtained in this experiment generated PCR products shorter than expected for in-frame full-sized fusions (data not shown) and were classified as InsDels (Fig. 3 Upper; see below).
Figure 3.
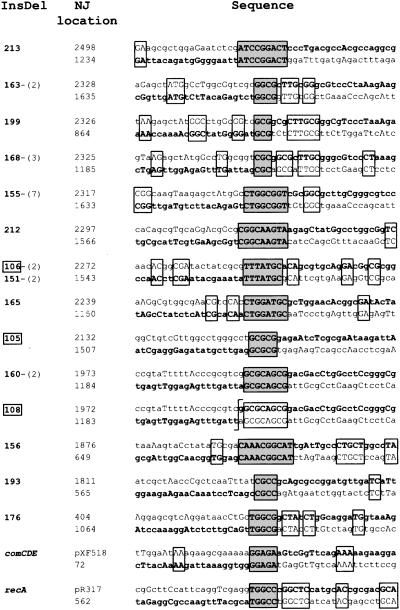
Distribution of NJs. Purified HindIII-linearized plasmid pCHS101 was used as donor in the transformation of strain TCHA2 (recipient chromosome 1, Upper) and TCHA3 (recipient chromosome 2, Lower) (Fig. 2b). NJ corresponding to in-frame hexA–mutS fusions that would direct synthesis of full-sized chimeric proteins are indicated by C, while InsDels are indicated by IΔ. Each recombinant is identified by a triple digit code. The number of independent isolations of the same InsDel is indicated between parenthesis. Identical InsDels independently isolated in the two recipients are boxed. GC% scale (window size = 40) varied between 32.5% and 80% for mutS and between 27.5 and 62.5% for hexA.
We hypothesized that deletion from the recipient chromosome of the region of best match between mutS and hexA would prevent the preferential formation of interspecies recombinants and increase the proportion of InsDels. Strain TCHA3, which lacks the last 278 triplets of hexA (Fig. 2b), was therefore constructed and used to isolate a new set of 62 EryR transformants. Analysis of the transformants by PCR revealed that as expected the proportion of InsDels now reached 82% (data not shown). Twenty three InsDels were then characterized by DNA sequencing (Fig. 3 Lower; see below).
Characteristics of InsDels Formed During Transformation with Hybrid DNA.
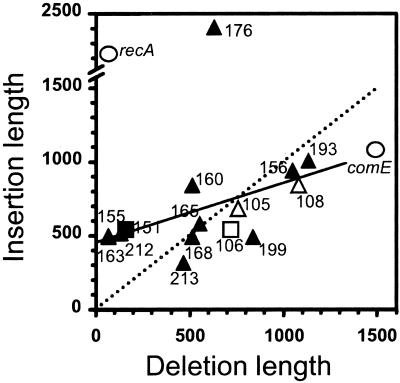
The shortest deleted segment observed previously was a 45-bp portion of the recA locus (10), and the longest was 1,485 bp of the comCDE locus (11). The 27 InsDels, including the four InsDels isolated in TCHA2 (Fig. 3), analyzed here fall within this range (64 to 1,134 bp; Fig. 4). It is worth pointing out that whereas there was no obvious constraint on the size of deleted segments, the trapping of cat required incorporation of ≈950 bp and ≈1,230 bp at the comC and the recA locus, respectively. At the hexA locus (Fig. 2b), reconstitution of a functional ermAM gene including the SD required integration of ≈70 bp and ≈140 bp to incorporate the ermAM promoter. Heterospecific segments integrated at the hexA locus varied in size between 289 bp and 2,474 bp (Fig. 4). The length of heterologous segments integrated at comCDE (1,074 bp) and recA (2,210 bp) also fell within this range.
Figure 4.
Correlation diagram of the length of inserted and deleted segments. Filled and open symbols indicate InsDels isolated in the TCHA3 and TCHA2 recipients, respectively. recA and comE InsDels are shown as open circles. Identical InsDel independently obtained in TCHA3 (IΔ151) and TCHA2 (IΔ106) recipients are indicated by a square. Note that the linear regression of points (excluding recA and comE InsDel) weighted differently to take into account multiple occurrences of the same NJ (solid line) differs from the dotted line. A fit to the latter (which corresponds to a slope of 1) would indicate a one-to-one correlation between the length of insertions and deletions.
As to the parameters governing the length of inserted and deleted material, no obvious correlation with the size of the homologous recombination anchor [which varied from 153 bp (comCDE) to 449 bp (recA), 683 bp (hexA), and 942 bp (ami)] was observed. The plot of the length of insertions versus the length of deletions for the set of InsDels available at the hexA locus ruled out the existence of a one-to-one correlation between the two lengths (Fig. 4). Large differences in the length of inserted segments could be observed for similarly sized deletions. For example, 636-bp and 721-bp-long deletions were associated with 2,412-bp and 544-bp-long insertions, respectively (IΔ176 and IΔ106; Fig. 4). In addition, the striking observation that the very same NJ was independently obtained twice with two different recipients (IΔ106 and IΔ121 in strain TCHA2, and IΔ151 and IΔ223 in strain TCHA3; Fig. 3), despite a large difference in the length of deleted segments (721 bp in strain TCHA2 versus 157 bp in strain TCHA3; squares in Fig. 4), suggested that factors intrinsic to both donor and recipient sequences governed the formation of NJs.
Structure of NJs: Distribution of Crossovers and GC Richness.
The structure of 27 NJs was established by sequencing across the junctions (Fig. 5). NJs appeared clearly nonrandom because more than 50% of them were clustered within two segments of mutS. Thirteen of 27 independent NJs were distributed within a 15-bp segment, from position 2317 to 2331 of mutS (see categories represented by IΔ163, IΔ199, IΔ168, and IΔ155 in Fig. 5). Three InsDels occurred between positions 1972 and 1979 (see categories IΔ160 and IΔ108 in Fig. 5). The GC content of these two mutS segments is 86.7% and 87.5%, respectively. A similar clustering also was observed at the recipient hexA locus. Fifteen of 27 independent NJs occurred within two segments of hexA, from position 1182 to 1189 (six cases represented by IΔ168, IΔ160, and IΔ108; Fig. 5) and from position 1633 to 1644 (nine cases represented by IΔ163 and IΔ155; Fig. 5). The GC content of these two hexA segments is 87.5% and 75%, respectively. Thus, GC richness of both donor and recipient strands appeared as a factor favoring the formation of NJs. Two NJs previously characterized at the comCDE and the recA locus also occurred in GC-rich segments (Fig. 5). On the other hand, GC richness was not observed for four NJs resulting from spontaneous deletion in a replicative plasmid in S. pneumoniae (13). However, unlike InsDels formed in transformation with chimeric molecules, plasmid deletions probably resulted from intramolecular events. In addition, they occurred with similar frequencies in recombination-deficient strains (13), suggesting that these plasmid deletions differed from InsDels in their mechanism of formation.
Figure 5.
Structure of NJs. The sequence of each InsDel is shown in bold and reads from left to right, starting on the bottom strand (hexA sequence) and shifting to the top strand (mutS sequence) within a stretch of sequence identity (boxed with gray shading; except for IΔ108). Additional microhomologies are also boxed. NJ location refers to the position (with respect to the start of mutS or hexA) of the first identical nucleotide in the crossover region. The number of independent occurrences of the same InsDel is indicated between parentheses after the triple-digit code. Boxed numbers correspond to InsDel isolated in strain TCHA2 (Fig. 3). For InsDel in the comCDE and recA chromosomal regions, the top strand corresponds to the sequence of the recombinant donor plasmid (pXF518 and pR317, respectively) and the bottom strand to comC or recA coding sequence.
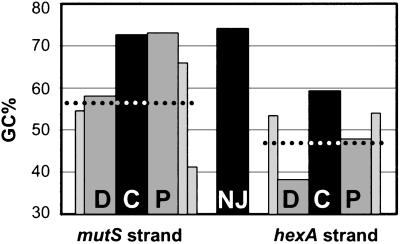
Calculation of the GC content of the entire set of sequenced NJs at the hexA locus revealed a value of 74.3% (Fig. 6), well above that for mutS (57.2%) or hexA (46.7%). Average GC% were also calculated for C, a 10-nt-long segment centered around NJs, and for 10-nt-long segments flanking C on each side. In the hexA recipient strand, the C segment was GC rich, whereas D (the segment distal to ermAM) had only 38.1% GC (Fig. 6). In the mutS strand, both C and P (the segment proximal to ermAM) were GC rich (72.6% and 73%). The segment next to P was also GC rich (65.9%), whereas the following one had only 41.1% GC (Fig. 6). This finding suggested that GC richness of a ≈30-nt-long segment in the donor strand might affect formation of InsDels.
Figure 6.
GC richness around NJs. Average GC% taking into account multiple occurrences of the same NJ were calculated for each entire stretch of sequence identity containing a NJ, a 10-nt-long segment centered around the NJ (C for central), and a 10-nt-long segment region immediately to the left (D) or to the right (P) of C, for the donor (mutS strand) and for the recipient strand (hexA strand). D and P are, respectively, distal and proximal to the ermAM donor-recipient heteroduplex joint (see Fig. 2b). The dotted lines indicate the average GC% of the entire mutS and hexA DNA sequences (57.2% and 46.7%, respectively). GC% values for additional 10-nt-long segments located immediately to the left of D and the right of P (two consecutive segments for P mutS) are also shown as thinner bars.
Structure of NJs: Presence of Short Stretches of Identity at the Crossovers.
The point of crossover between donor and recipient sequences was unambiguous in only one case, IΔ108 (Fig. 5). In all other cases, the crossover occurred within short stretches (3–10 nt) of sequence identity between the interacting sequences (Fig. 5). Interestingly, the crossover point in IΔ108 was immediately adjacent to a stretch of eight identical nucleotides. Two NJs previously obtained at the comCDE and the recA locus occurred similarly within 5-nt-long segments of identity (Fig. 5). The presence of microhomoloies (4–6 bp) was also reported in four independent InsDels obtained at the comAB locus (14).
These data strongly suggested that microhomology plays a role in the formation of InsDels. Additional blocks of sequence identity were often present in the immediate vicinity of NJs. Remarkably, the highest incidence of additional microhomologies was observed for those InsDels corresponding to NJs that occurred within the shortest stretches of sequence identity (IΔ199 and IΔ168; Fig. 5). Therefore, neighborhood sequence identity appeared as another factor favoring the formation of NJs.
Are GC-Rich Sequences Recombination Hotspots in S. pneumoniae?
In E. coli, early steps of recombination are coordinated by RecA protein, RecBCD enzyme, and recombination hotspots called Chi (χ) sites. E. coli χ sites (χEc) are G-rich (5′-GCTGGTGG-3′) cis-acting regulatory sequences that modify the activities of the RecBCD enzyme and leads to loading of the DNA strand exchange protein RecA, onto the χ-containing DNA strand (see ref. 15 for a review). The RecA protein-single-stranded DNA filament then invades homologous double-stranded DNA to produce a D-loop structure (15). χ-like sites have been identified in two Gram-positive bacteria, Lactococcus lactis and Bacillus subtilis. Similarly to χEc, χLl and χBs modify the activities of RecBCD-like enzymes (namely RexAB and AddAB, respectively in L. lactis and B. subtilis), although it has not been shown that they constitute recombination hotspots in these species (16, 17). Interestingly, both χLl (5′-GCGCGTG-3′) and χBs (5′-AGCGG-3′) are also GC-rich sequences. It would be tempting to speculate that GC-rich sequences in the NJs correspond to a S. pneumoniae χ site (χSp). These putative χSp sites are unlikely to be recognized on donor DNA by S. pneumoniae RexAB because single-stranded segments enter the cells (see ref. 18 for a review) and χEc are recognized only from within double-stranded DNA by RecBCD enzyme (15). However, occurrence of double-strand breaks in the chromosome of competent cells, followed by RexAB processing up to a χSp site, and subsequent incorporation of a donor DNA segment to repair the break could account for the presence of χSp at the NJs. An alternative possibility still connecting the GC-rich regions to putative χSp sites would be that of a preferential binding of RecA to these sequences, as demonstrated for the E. coli RecA protein and χEc sites (19). If such a preferential binding were to result in an increased stability of RecA protein–single-stranded DNA complexes, this could increase the probability of interaction with recipient sequences. In addition, heterologous donor DNA complexed with RecA protein could be protected from DNases. It is known that extremities of donor fragments in transformation frequently are excluded from recombination, possibly because of degradation (20). In any case, if χSp sites are identified, it will be interesting to compare them with the sequences of NJs (Fig. 5).
A D-Loop Resolvase in Transformation?
A third possibility to account for the high GC content around NJs is suggested by the observation that the RuvC Holliday junction resolvase of E. coli cuts the DNA at preferred sites (cleavage occurs at the 3′ side of two thymine residues) (21). Assuming that a resolvase is required to process D loops formed during transformation into recombinants (Fig. 1), the high GC content around crossover points in InsDels could reflect some sequence specificity of the putative resolvase. Obviously, the sequences of the NJs do not fit the consensus for RuvC. This finding is not surprising because there is no RuvC orthologue in S. pneumoniae. However, a member of a newly defined family of Holliday junction resolvase typified by E. coli YqgF and suggested to be an alternative to RuvC particularly in low-GC Gram-positive bacteria (22) is encoded in the genome sequence of a virulent isolate of S. pneumoniae (The Institute for Genomic Research, http://www.tigr.org). It remains to be established whether S. pneumoniae YqgF processes D-loops during transformation and whether it exhibits any sequence preference for cutting.
In the context of the resolvase hypothesis, the NJs of IΔ108 and IΔ160 could represent alternative processing of the same donor-recipient heteroduplex (Fig. 5).
Local Sequence Identity in the Formation of NJs.
GC richness is not an absolute requirement for the formation of InsDels, as illustrated by IΔ106–IΔ151 (Fig. 5). The corresponding NJ occurred within a 7-nt-long stretch of identity that contained only two GC pairs. Let us consider the alternative possibility, that the first feature important for production of InsDels is the presence of microhomology. The role of the 3- to 10-bp-long microhomologies (Fig. 5) could be to allow transient pairing between heterospecific donor DNA and the recipient strand. Interestingly, NJs corresponding to interspecific hexA–mutS recombinants also occurred within 2- to 19-nt-long stretches of sequence identity (M.P. and J.-P.C., unpublished data), which could indicate a common mechanism for the production of InsDels and interspecies recombinants. With respect to the pairing hypothesis, it is relevant that the E. coli RecA protein has been shown to form synaptic complexes in vitro with less than one helical repeat of DNA (as few as eight bases of homology) (23).
In this model, the GC richness of the donor strand over the last 30 nt and the resident strand in the region of the future NJ (Fig. 6) would only contribute to an increase in the stability of the joints. The consequence of an increased stability would be a stalling of the junction that would in turn increase the probability of resolution of recombination intermediates by the putative D-loop resolvase (Fig. 1 and above).
Alternatively replication could be initiated at the D loop (15), with DNA synthesis being primed at the heterologous 3′ end. The mechanism would be analogous to that proposed for the formation of spontaneous deletions by strand slippage during replication (24). We do not favor this hypothesis because IΔ108 contains a mismatched nucleotide (Fig. 5), whereas a perfect match at the 3′ terminus would probably be favored for elongation by the replication machinery.
Whatever the model, regions that harbor a lower than average GC% (i.e. the segment next to the central GC-rich region on the P side, in the donor strand, and segment D in the recipient strand; Fig. 6) may also contribute to the preferential location of NJ. These regions could possibly slow down the sliding of strands during the search for homology, thus contributing to a transient sequestering of the joint in the GC-rich segment.
Transformation, DNA Shuffling, and Capture of Foreign DNA in Nature.
Are InsDels produced only under laboratory conditions, when using artificially constructed chimeric molecules as donors in transformation, or could they occur in nature? Two features seem to be required in the donor DNA for the production of InsDels: a homologous anchor that can be as short as 153 bp (11) and a sequence arrangement not present in the recipient strain. This situation is encountered for incoming DNA fragments overlapping a region that is polymorphic in the chromosome of S. pneumoniae (e.g., the capsule locus, cap; see ref. 25 for a review). Homologous sequences flanking the polymorphic region on either side (i.e., dexB or aliA sequences for the cap locus) (25) can provide a homologous recombination anchor and favor the formation of InsDels in the flanking heterologous cap region. Evidence has been obtained that two S. pneumoniae isolates can differ by up to 10% of the chromosome (26). Therefore this scenario can lead to shuffling of DNA sequences at several places in the S. pneumoniae chromosome.
Our results demonstrate that chimeric donor DNA can lead to trapping of foreign genes through formation of InsDels during transformation of S. pneumoniae (Table 1). Could such a trapping occur in nature? Inasmuch as formation of InsDels requires only a short homologous recombination anchor, we suggested previously that any piece of homologous DNA, including a mobile genetic element (e.g., an insertion sequence, IS) present both in the incoming DNA and the recipient chromosome, can serve as a recombination anchor (5). Taking into account the high incidence of IS in the chromosome of S. pneumoniae (27, 28), and the wide interspecies distribution of these elements, IS can promote the trapping of DNA from distantly related species by generating InsDels through homology-directed illegitimate recombination. Such a mechanism would be of great potential in terms of genome evolution because, depending on the location of the NJ, it could lead to the production of chimeras and/or to the acquisition of completely heterologous genes. Similar mechanisms potentially leading to the capture of foreign DNA through natural transformation are likely to be widespread as suggested by the observations of de Vries and Wackernagel (35) using one-side homologous substrates in the Gram-negative Acinetobacter sp.
Since our initial observation of homology-directed illegitimate recombination in S. pneumoniae (4), related phenomena have been reported, for example in E. coli (29) and yeast (30, 31). NJs also were frequently found to be associated with microhomologies (29, 31). Another example strikingly similar to the formation of InsDels in S. pneumoniae is that of the parahomologous targeting described in Drosophila cells (32). Taken together, these observations suggest that homology-directed illegitimate recombination has been conserved during evolution and could therefore be intrinsic to the recombination-repair machinery.
Supplementary Material
Acknowledgments
We are grateful to David Lane for critical reading of the manuscript. We thank Graham Walker for providing us with plasmid pGW1812 and Stéphane Duhamel for his participation in some of the experiments. This work was supported in part by a Contrat de Recherche Externe (No. 920102) from the Institut National de la Santé et de la Recherche Médicale and by a Subvention (No. RECH/9407531) from the Région Midi Pyrénées. M.P. was the recipient of a travel award from the P. Philippe Foundation.
Abbreviations
- InsDel
insertion-deletion
- NJ
novel joint
- Ery
erythromycin
- MTX
methotrexate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Griffith F. J Hyg. 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claverys J P, Prudhomme M, Mortier-Barrière I, Martin B. Mol Microbiol. 2000;35:251–259. doi: 10.1046/j.1365-2958.2000.01718.x. [DOI] [PubMed] [Google Scholar]
- 3.Hakenbeck R, Grebe T, Zähner D, Stock J B. Mol Microbiol. 1999;33:673–678. doi: 10.1046/j.1365-2958.1999.01521.x. [DOI] [PubMed] [Google Scholar]
- 4.Claverys J P, Lefèvre J C, Sicard A M. Proc Natl Acad Sci USA. 1980;77:3534–3538. doi: 10.1073/pnas.77.6.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mortier-Barrière I, Humbert O, Martin B, Prudhomme M, Claverys J P. Microb Drug Res. 1997;3:233–242. doi: 10.1089/mdr.1997.3.233. [DOI] [PubMed] [Google Scholar]
- 6.Martin B, Prats H, Claverys J P. Gene. 1985;34:293–303. doi: 10.1016/0378-1119(85)90138-6. [DOI] [PubMed] [Google Scholar]
- 7.Alloing G, Trombe M C, Claverys J P. Mol Microbiol. 1990;4:633–644. doi: 10.1111/j.1365-2958.1990.tb00632.x. [DOI] [PubMed] [Google Scholar]
- 8.Haber L T, Pang P P, Sobell D I, Mankovich J A, Walker G C. J Bacteriol. 1988;170:197–202. doi: 10.1128/jb.170.1.197-202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang P P, Lundberg A S, Walker G C. J Bacteriol. 1985;163:1007–1015. doi: 10.1128/jb.163.3.1007-1015.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin B, García P, Castanié M P, Claverys J P. Mol Microbiol. 1995;15:367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 11.Alloing G, Martin B, Granadel C, Claverys J P. Mol Microbiol. 1998;29:75–84. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 12.Priebe S, Hadi S, Greenberg B, Lacks S A. J Bacteriol. 1988;170:190–196. doi: 10.1128/jb.170.1.190-196.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballester S, Lopez P, Espinosa M, Alonso J C, Lacks S A. J Bacteriol. 1989;171:2271–2277. doi: 10.1128/jb.171.5.2271-2277.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M S, Dougherty B A, Madeo A C, Morrison D A. Appl Environ Microbiol. 1999;65:1883–1890. doi: 10.1128/aem.65.5.1883-1890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 16.El Karoui M, Ehrlich D, Gruss A. Proc Natl Acad Sci USA. 1998;95:626–631. doi: 10.1073/pnas.95.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chédin F, Ehrlich S D, Kowalczykowski S C. J Mol Biol. 2000;298:7–20. doi: 10.1006/jmbi.2000.3556. [DOI] [PubMed] [Google Scholar]
- 18.Lacks S A. In: Microbial Interactions, Receptors, and Recognition. Reissig J L, editor. London: Chapman & Hall; 1977. pp. 179–232. [Google Scholar]
- 19.Tracy R B, Kowalczykowski S C. Genes Dev. 1997;10:1890–1903. doi: 10.1101/gad.10.15.1890. [DOI] [PubMed] [Google Scholar]
- 20.Lataste H, Claverys J P, Sicard A M. Mol Gen Genet. 1981;183:199–201. doi: 10.1007/BF00270163. [DOI] [PubMed] [Google Scholar]
- 21.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, Makarova K S, Koonin E V. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh P, Camerini-Otero C S, Camerini-Otero R D. Proc Natl Acad Sci USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albertini A M, Hofer M, Calos M P, Miller J H. Cell. 1982;29:319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- 25.Garcia E, Llull D, Munoz R, Mollerach M, Lopez R. Res Microbiol. 2000;151:429–435. doi: 10.1016/s0923-2508(00)00173-x. [DOI] [PubMed] [Google Scholar]
- 26.Hakenbeck R, Balmelle N, Weber B, Gardes C, Keck W, de Saizieu A. Infect Immun. 2001;69:2477–2486. doi: 10.1128/IAI.69.4.2477-2486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oggioni M R, Claverys J P. Microbiology. 1999;145:2647–2653. doi: 10.1099/00221287-145-10-2647. [DOI] [PubMed] [Google Scholar]
- 28.Tettelin H, Nelson K E, Paulsen I T, Eisen J A, Read T D, Peterson S, Heidelberg J, DeBoy R T, Haft D H, Dodson R J, et al. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 29.Kusano K, Sakagami K, Yokochi T, Naito T, Tokinaga Y, Ueda E, Kobayashi I. J Bacteriol. 1997;179:5380–5390. doi: 10.1128/jb.179.17.5380-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunes S, Botstein D, Fox M S. Genetics. 1990;124:67–80. doi: 10.1093/genetics/124.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mézard C, Pompon D, Nicolas A. Cell. 1992;70:659–670. doi: 10.1016/0092-8674(92)90434-e. [DOI] [PubMed] [Google Scholar]
- 32.Cherbas L, Cherbas P. Genetics. 1997;145:349–358. doi: 10.1093/genetics/145.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasta F, Sicard M A. Proc Natl Acad Sci USA. 1999;96:2943–2948. doi: 10.1073/pnas.96.6.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasseghi H, Claverys J P, Sicard A M. In: Transformation 1980. Polsinelli M, Mazza G, editors. Oxford: Cotswold; 1981. pp. 137–154. [Google Scholar]
- 35.de Vries J, Wackernagal W. Proc Natl Acad Sci USA. 2002;99:2094–2099. doi: 10.1073/pnas.042263399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.