Abstract
Differentiation of spermatids into spermatozoa is regulated via phosphorylated RNA-binding proteins that modulate the expression of stage-specific mRNAs. We demonstrate that the phosphoserine, -threonine or -tyrosine, interaction protein, Styx, complexes with a testicular RNA-binding protein and is essential for normal spermiogenesis. Ablation of Styx expression in mouse disrupts round and elongating spermatid development, resulting in a >1,000-fold decrease in spermatozoa production. Moreover, Styx−/− males are infertile because of structural head abnormalities in residual epididymal sperm. Immunoprecipitation of Styx with Crhsp-24, a phosphorylated RNA-binding protein implicated in translational repression of histone mRNAs, provides a strategy for regulating posttranscriptional gene expression.
The importance of RNA-binding proteins during spermatogenesis has been demonstrated in mice lacking the proteins Prbp (1) and TLS/FUS (2). In the absence of Prbp, males exhibit defects in nuclear remodeling and chromatin compaction attributable to the dysregulation of protamine expression (1). Likewise, TLS/FUS-deficient males manifest meiotic abnormalities through a mechanism involving either RNA or DNA binding (2). Interestingly, a spermatogenic arrest phenotype also has been seen in transgenic mice that prematurely translate protamine mRNA (3). Thus the timing of protein expression as dictated by RNA-binding proteins is thought to be central to the differentiation process; however, regulation of this activity is understood only partially (3, 4). In the case of TLS/FUS (5) and two other spermatogenic RNA-binding proteins, TB-RBP (6) and MSY2 (7), protein dephosphorylation diminishes their affinity for mRNA and thereby relieves translational repression of bound transcripts.
Reversible protein phosphorylation is a critical regulatory mechanism for cell metabolism and proliferation as well as differentiation. Specific kinases and phosphatases alter the phosphorylation state of individual proteins, whereas distinct noncatalytic domains facilitate protein-protein interactions via specific phosphorylated motifs (8, 9). Three unique protein modules (14-3-3, FHA, and WW domains) have been shown to bind discrete phosphoserine or -threonine sites, pS/T (8, 10), whereas Src homology 2 (SH2) and phosphotyrosine-interaction/binding (PI/PTB) domains recognize specific phosphotyrosine (pY) motifs in target proteins (11, 12). We previously have identified a pS/T or pY interaction protein, Styx, which possesses protein tyrosine phosphatase (PTP) structure but is inactivated catalytically by endogenous substitution of the essential PTP active-site cysteine, to glycine (13). Thus Styx and related “dead” PTP domains have been postulated to function as antagonists of endogenous phosphatase activity (14, 15); however, their physiological roles and effector mechanisms have not been established.
As a first attempt to demonstrate the biological significance of STYX-like domains, we selectively disrupted the prototype Styx gene in mouse and found it to be essential for normal spermatogenesis. Coimmunoprecipitation of Styx with a unique RNA-binding protein suggests that together they may regulate a translational checkpoint governing this process. These findings identify Styx as a candidate fertility gene in man and fundamentally establish STYX/dead-phosphatase domains as important components of biological systems.
Materials and Methods
Construction of Gene-Targeting Vector.
Plasmid pnlacF, encoding the lacZ gene, was a gift of R. Palmiter (University of Washington, Seattle). Plasmid pFlox was provided by K. Rajewsky (Cologne, Germany) and J. Marth (16). In short, a 3.1-kb NcoI/BamHI partial digest fragment of pnlacF was ligated to the 300-bp EcoRV/NcoI partial digest fragment of the Styx active site exon 7. The resulting Styx:lacZ fusion product was inserted into the NotI/SmaI site of pFlox, thereby replacing the entire coding sequence for the pFlox-HSVtk marker. To facilitate targeting of the Styx locus, a 2.3-kb HindIII/EcoRV partial digest fragment spanning Styx exons 5–6 was inserted into the SalI site of native pFlox. The resulting Styx-loxP product was subcloned into the pFlox SalI site upstream of Styx:lacZ sequence. Similarly, a 2.4-kb EcoRV/HindIII partial digest fragment containing Styx exons 7–8 was inserted into the BamHI/HindIII site downstream of the pFlox-PGKneor marker sequence. Finally, the active site glycine codon of exon 7 was mutated to encode a cysteine as described (13). The entire insert of the final targeting vector, pFlox-StyxlacZfloxPGC, was confirmed by sequencing.
Gene Targeting, Mouse Breeding, and Genomic Screening.
Early passage mouse R1 embryonic stem (ES) cells (17) were provided by A. Nagy, R. Nagy, W. Abramow-Newerly, J. Rossant, and J. Roder (Mount Sinai Hospital, Toronto). Before transfection, pFlox-StyxlacZfloxPGC was digested with NotI, and the entire 13.3-kb insert was isolated from vector DNA by agarose gel electrophoresis. Targeting of the Styx gene in ES cells was performed with the isolated StyxlacZfloxPGC insert as described (18) by using mouse embryonic fibroblast feeder cells and recombinant leukemia inhibitory factor (ESGRO, Life Technologies, Grand Island, NY) to inhibit ES cell differentiation. Individual ES cell colonies were replica-plated, and colonies that incorporated the targeting construct were identified by β-galactosidase (β-gal) activity of fixed cells by using 5-bromo-4-chloro-3-indolyl β-D-galactoside as substrate (19). Correct targeting of the StyxlacZfloxPGC insert into ES cells and identification of germ line transmission in subsequent transgenic animals was confirmed by Southern blot and PCR, respectively. PCR screening of Styx alleles was performed with the Expand long template PCR system (Roche Molecular Biochemicals) and primers I5-7f (5′-CGTTTTTTCCCTATGGTAAGTATCGG-3′), E7r (5′-ACTTCTAGAGATACCTGCATTCCCATGGAC-3′), and lacZr (5′-GCCAGGGTTTTCCCAGTCACGACG-3′). Reverse transcription–PCR of Styx:lacZ transcripts was performed with the same primers to confirm fusion protein production by using the mRNA capture kit and Titan one-tube reverse transcription–PCR System (Roche Molecular Biochemicals) by the manufacturer's specifications. Chimeric mice were created at the University of Michigan Transgenics Core, and mouse breeding was carried out within the Unit for Lab Animal Medicine under the guidelines of the University Committee on Use and Care of Animals.
A vector construct for bacteriophage Cre protein expression in mammalian cells, pMC-Cre-Hygro, was obtained from J. Marth and K. Rajewsky (above). To facilitate loxP recombination at targeted alleles, targeted ES sublines were cultured and transfected with pMC-Cre-Hygro as described above in the absence of antibiotic selection. Screening for Cre-mediated recombination of the StyxlacZfloxPGC allele to create the StyxloxC allele was performed as described above with the PCR primers I5–7f and E7r. Genotyping of mice was performed with DNA recovered from tail biopsies performed by standard methods.
Tissue Preparation, Morphology, and Histology.
Before dissection, tissues were prepared for histology by whole animal cardiac perfusion with PBS and paraformaldehyde. Whole tissues or sections were fixed routinely in freshly prepared 4% paraformaldehyde for 1 h at 4°C. For preparation of testis, either the tunica albuginea was removed or the tissue was sectioned after initial fixation. Staining for β-gal activity was carried out as described (19) by using 5-bromo-4-chloro-3-indolyl β-D-galactoside as a substrate. Histological preparations were performed at the University of Michigan Cell Morphology Core essentially as described (20, 21). In short, after initial fixation and staining, samples were postfixed in paraformaldehyde 4–24 h at 4°C before processing for embedding in glycol methacrylate. Cryosections were stained with hematoxylin and eosin or periodic acid Schiff reagent before morphological evaluation under light microscopy. Epididymal sperm were harvested, and morphology was assessed as described (22).
Northern and Western Blot Analysis.
Total RNA and protein were recovered from freshly dissected whole testis by using TRIzol reagent (Life Technologies) according to the manufacturer's protocol. Poly(A) RNA was purified from total RNA by using PolyATtract (Promega) and subsequently blotted onto nitrocellulose for probing with Styx cDNA as described (13). Proteins recovered from TRIzol preparations were resolved by SDS/PAGE and transferred onto nitrocellulose. For detection of Styx protein, membranes were blocked with 5% nonfat dry milk and probed with affinity-purified polyclonal antisera raised against full-length recombinant mouse Styx as described (M.J.W. and J.E.D., unpublished data). Antibodies against full-length Crhsp-24 were a gift of J. A. Williams (University of Michigan, Ann Arbor, MI). All other primary antibodies were obtained from Santa Cruz Biotechnology and used under the manufacturer's specifications. Primary antibodies were detected by incubation with horseradish peroxidase-conjugated anti-IgG or anti-Fab antibodies and detected by enhanced chemiluminescence (Amersham Pharmacia).
Immune Precipitation.
Immunoprecipitations were performed as described (23). In short, four whole testes from wild-type male mice were lysed by Polytron disruption for 15 sec at 4°C in TBS with Complete protease inhibitors (Roche Molecular Biochemicals) and incubated for 1 h at 4°C. The crude lysate was divided and modified for final lysis in either Nonidet P-40 buffer (150 mM NaCl/1% Nonidet P-40/50 mM Tris, pH 8.0), high salt (Nonidet P-40 buffer except 500 mM NaCl), or RIPA (Nonidet P-40 buffer plus 0.5% deoxycholate and 0.1% SDS) for 1 h at 4°C. Soluble lysates were precleared with protein-A agarose (Life Technologies) for 16 h at 4°C and aliquoted for incubation with anti-Styx antibodies (above) for 1 h. Immune complexes were immobilized on protein-A agarose for 1 h at 4°C and washed four times in lysis buffer. Captured proteins were resolved by SDS/PAGE and identified as described above.
Results and Discussion
In mouse and humans, the Styx gene family is comprised of a functional intron-containing gene, Styx, and at least two processed pseudogenes, Styx-rs1 and Styx-ps1 (loci symbols are registered with mouse and human genome databases; M.J.W. and J.E.D., unpublished data). The vector used to ablate Styx expression disrupts endogenous gene structure by in-frame insertion of the lacZ sequence into the phosphatase-like active site exon (Fig. 1A). Southern blot analysis confirmed the specificity of targeting the Styx locus (Fig. 1B). Germ line transmission of the targeting construct in ES cell-derived animals was verified by PCR with genomic DNA (Fig. 1C) and reverse transcription–PCR of mRNA (data not shown). Three independently targeted ES cell lines were used to generate chimeric males and separate heterozygous offspring lines. All heterozygous animals were viable and phenotypically normal.
Figure 1.
Strategy for targeted replacement of mouse Styx. (A) Partial Styx gene structure is depicted with numbered exons (red). Relevant restriction enzyme cleavage sites are shown: B, BamHI; RV, EcoRV; H, HindIII; N, NcoI. The active site exon 7 contains either the wild-type glycine codon (Gly) or a glycine to cysteine mutation (Cys). The β-gal reporter (blue), G418 resistance marker (yellow), and bacteriophage loxP sites (black) are boxed. The arrows denote PCR primers for identification of Styx alleles. (B) Genotyping by Southern blot of BamHI-digested genomic DNA. The blots were probed for the Styx locus outside of the targeting construct as shown in A. Relative size standards are in kb. (C) Genotyping for StyxfZC (−) in ES cells and transgenic animals using the PCR primers shown in A. Relative size standards are in kb. (D and G) Northern blots of whole adult testis mRNA from StyxfZC (−) and StyxloxC mice probed with 3′-Styx cDNA. Relative size standards are in kb. (E and H) Western blot of whole testis protein probed with anti-Styx antibodies. Relative size standards are in kDa. (F) Genotyping for StyxloxC in adult mice using the PCR primers shown in A. (I) Partial exon structure, encoded amino acid sequence, and resulting phenotype of targeted Styx alleles. The Styxwt active site glycine is highlighted. Products of reverse transcription–PCR from StyxloxC/loxC mice do not contain the active site exon 7 because of alternative splicing and frameshift-induced premature termination of the Styx coding sequence.
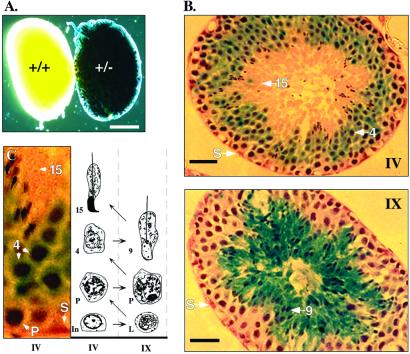
The timing and scope of Styx expression were determined by β-gal activity of the fusion protein in multistage heterozygous embryos, pups, and adult animals. Reporter activity was restricted to the testes of mutant males (Fig. 2A) commencing at ≈13–14 days after birth, in which histological sections of adult testis showed that expression was limited to the seminiferous tubule (Fig. 2B). Styx:β-gal activity was initially juxtanuclear within round spermatids and eventually extended throughout the cytoplasm of elongating spermatids (Fig. 2 B and C). Only faint expression was observed within the cytoplasm of condensing spermatids and pachytene spermatocytes (Fig. 2C), and no staining above background was seen in Leydig cells (data not shown), spermatogonia, spermatozoa, or somatic Sertoli cells (Fig. 2 B and C). Overall, the pattern of reporter expression correlated with alternative splicing and differential expression of Styx mRNA on whole-tissue Northern blots (13).
Figure 2.
The Styx reporter allele is differentially expressed during spermiogenesis of mouse spermatids. (A) β-gal staining of whole adult testis from wild-type and heterozygous males. (Scale bar, 2 mm.) (B) β-gal-stained cryosections of testicular seminiferous tubules (above). The approximate developmental staging (C) of tubules (roman numerals) and spermatids (arrows) are indicated. (Scale bar, 50 μm.) (C) Selected section of Styx± seminiferous tubule (Left) and schematic representation of the relative position of cellular cohorts in normal mouse tubule stages IV and IX (Right; refs. 20 and 24). The arrows indicate the direction of gamete differentiation from intermediate spermatogonia (In), through leptotene (L) and pachytene (P) spermatocytes, into spermatids: round (step 4), elongating (step 9), and condensed (step 15). A somatic Sertoli cell (S) is shown relative to the germ cell layers.
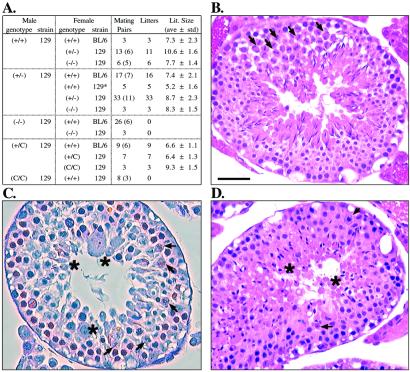
To determine the physiological consequence of the loss of Styx function, heterozygous animals were interbred to homozygosity. Genotype analysis of the offspring (Fig. 1 B and C) showed that +/+, +/−, and −/− (nullizygous) mice arose at expected Mendelian ratios despite the ablation of native Styx mRNA and protein in nullizygous animals (Fig. 1 D and E, respectively). All F2 offspring were viable and macroscopically indistinguishable from birth into adulthood. Nullizygous females exhibited no obvious impairment of growth, development, or reproductive potential (Fig. 3A). In contrast, nullizygous males failed to reproduce (Fig. 3A) despite normal mating behavior and frequency of copulatory plug formation. The testes and seminiferous tubule epithelium of nullizygous males contained normal placement and numbers of spermatogonia, primary spermatocytes, Sertoli (Fig. 3C), and Leydig cells (data not shown). However, a dramatic deficiency in the spermatid population was noted first in all seminiferous tubules with round spermatids (step 8) and peaked to nearly complete absence of elongating spermatids and successive cell types in ≈10% of tubule sections (Fig. 3C). Loss of immature spermatids resulted from premature shedding into the tubule lumen, as observed in epididymal sections (below), as well as increased cellular death along the luminal epithelium (data not shown). Beyond the cellular losses, residual postmeiotic cells displayed an irregular luminal orientation, tubule distribution, and morphological abnormality (Fig. 3C). The most striking anomaly was a failure of nuclear reorganization that characterizes spermatid differentiation (20, 24). Distinct atypical spermatid forms included multinucleated cells (step 5), acrosome-conjoined symplasts (steps 5–8), and giant cells with round and elongating nuclei (Fig. 3C). In addition, nearly all round spermatids were >50% larger than wild-type cells. Likewise, elongating spermatids (steps 9–12) were positioned irregularly in the epithelium, with aberrant anterior acrosomal structures often accompanied by posterior cytoplasmic swelling (Fig. 3C). The few condensed spermatids (step 16) often failed to complete spermiation, resulting in increased Sertoli phagocytosis.
Figure 3.
Styx-deficient males are infertile and exhibit a disruption of normal spermatid differentiation. (A) Homozygous Styx−/− and StyxC/C male mice are not fertile. Mouse strains are either C57BL/6 (BL/6) or 129/SvIMJ × BL/6 (129). An asterisk denotes the 129/SvJ strain. Individual males (parentheses) were mated to age-matched fertile females for 2 weeks before separation and scoring for fertility. (B–D) Cross sections of stage XII seminiferous tubules from wild-type (B), Styx−/− (C), and StyxC/C (D) mice. Spermatocytes containing meiotic bodies are indicated (arrowheads). Representative spermatids exhibiting aberrant distribution, shape, and size are shown (asterisks). (Scale bar, 50 μm.)
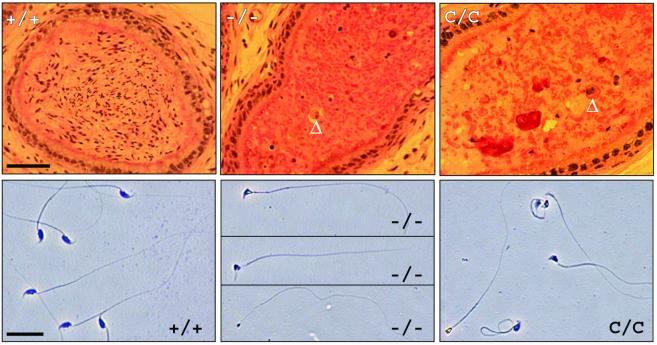
As a result of the germ cell losses in nullizygous testes, individual histological sections of cauda epididymidis were devoid of mature luminal spermatozoa, with the few cells present appearing as immature, round, or elongating spermatids (Fig. 4 Upper). Recovery of total epididymal content confirmed a >1,000-fold decrease in spermatozoa-like cells, with all mature sperm forms exhibiting either aberrant rounded heads, blunted acrosomes with misattached flagellum, gaps in detached acrosomes, or pinhead morphologies (Fig. 4 Lower). Furthermore, all residual sperm either were immobile or possessed a nonprogressive, uncoordinated motility, although <50% of these cells contained tail abnormalities.
Figure 4.
Loss of Styx results in abnormal sperm production. (Upper) Sections of adult mouse caudal epididymis. The arrowheads indicate multinucleated, round, spermatid-like cells. (Scale bar, 50 μm.) (Lower) Representative epididymal sperm morphologies. (Scale bar, 5 μm.)
To determine whether the infertility of Styx−/− males resulted from reporter protein expression, a second knockout allele was created by excision of the Styx:β-gal reporter cassette (Fig. 1 A and F). The excised allele expressed normal levels of Styx mRNA (Fig. 1G); however, protein levels were attenuated (Fig. 1H) by alternative splicing and premature truncation at the active site exon (Fig. 1I). In the absence of the Styx exon7:lacZ sequence, a preference for using the splice acceptor site of exon 8 in vivo likely resulted from the residual loxP site only ≈250 bp upstream of the endogenous exon 7 acceptor site (Fig. 1A). Thus, because of the absence of Styx protein, mice homozygous for the excised allele, StyxC/C, were indistinguishable phenotypically from Styx−/− mice (Figs. 3 and 4). The resulting phenotypes for all targeted alleles were consistent among littermates, between independent ES cell-derived lines, and within disparate genetic backgrounds (C57BL/6 and 129/SvIMJ). Despite lower epididymal and testis weights that correlated with the reduction in germ cells (testis mass at 60 days old in mg ± SD: −/−, 71.8 ± 8.4; +/−, 111.8 ± 0.4; +/+, 112.2 ± 10.9), other secondary reproductive organs and androgen targets such as seminal vesicles were normal in size and morphology. Moreover, total body mass did not differ between littermates (mass at 60 days old in g ± SD: −/−, 29.8 ± 4.5; +/−, 30.9 ± 3.5; +/+, 32.8 ± 3.3). Collectively, these results demonstrate that a loss of Styx protein results in an intrinsic testicular defect of mouse spermatogenesis and as such reveals Styx as a candidate fertility gene in man.
In considering the underlying molecular defect of knockout males, our previous work had shown that Styx possessed all the molecular determinants for binding serine-, threonine-, or tyrosine-phosphorylated substrates (13). Therefore, we postulated that Styx could function through proteins that have expression patterns or knockout phenotypes that were similar to Styx nullizygous males. Candidate effectors included: known kinases, Cdc2, Cdk2 and Cdk3 (25), and Ck2α and Ck2α′ (26); phosphatase PP1cγ (27); transcription factor Crem-τ (28, 29); phosphoproteins Rho-GDI (30) and Hsp-27 (31); and 14-3-3 isoforms (32). Despite the reduced number of differentiating spermatids in Styx−/− males as mentioned above, the absence of Styx protein did not affect the level of expression of the candidate effector proteins dramatically in whole nullizygous testis extracts (Fig. 5A), including the relative levels of phosphorylated Crem-τ isoforms (28, 29). Likewise, anti-Styx immune complexes failed to coprecipitate these proteins from wild-type testis (Fig. 5B), suggesting a unique mechanism for Styx action.
Figure 5.
Styx interacts with Crhsp-24 in vivo. (A) Expression of selected proteins in whole adult mouse testis. Testes proteins were isolated and resolved by SDS/PAGE. Selected proteins were detected by commercial antibodies. Relative positions of molecular mass standards are in kDa. (B Upper) Crhsp-24 is selectively retained in anti-Styx immune complexes from wild-type testes. Increasing amounts (1, none; 2, 15 μg; 3, 40 μg) of anti-Styx antibodies were used to immunoprecipitate proteins for each lysis condition (Nonidet P-40, high salt, and RIPA). After removing the immune complexes, the unbound lysate material was combined (Ext. Lysate), and all proteins were resolved by SDS/PAGE and Western blots probed with antibodies as described above. (Lower) Styx is retained in Crhsp-24 immune complexes from wild-type testes. Immune precipitation was performed as described above by using affinity-purified Crhsp-24 antisera.
Styx immune complexes did precipitate a phosphorylated, calcium-responsive heat-stable protein with a molecular mass of 24 kDa (33), Crhsp-24 (Fig. 5B). Crhsp-24 and its brain-specific paralog, PIPPin, possess cold-shock and double-stranded RNA-binding domains (34), which for PIPPin are capable of binding replacement histone H3.3 and H1° mRNAs to repress their translation in vitro (34, 35). In light of overlapping expression of Styx and Crhsp-24 in elongating spermatids (Fig. 2B and J. A. Williams, personal communication, respectively), a complex between Styx and Crhsp-24 may provide insight into a mechanism regulating chromatin protein expression during normal spermiogenesis (3, 4). Previous work has shown that dephosphorylation of the testicular RNA-binding proteins TLS/FUS (5), TB-RBP (6), and MSY2 (7) diminishes their affinity for mRNA binding and thereby relieves translational repression of bound transcripts. Although the effect of Styx on Crhsp-24 is under study, immune complexes of Crhsp-24 also precipitate Styx (Fig. 5B Lower), and regulation of specific histone subtype expression would be consistent with the prominent nuclear abnormalities of round and elongating spermatids in Styx-deficient males.
Because the cellular affectors of RNA-binding proteins are largely unknown, a protein complex of Styx and Crhsp-24 provides new insight into components of this important biological process. In a larger context, this work fundamentally demonstrates an essential physiological function for noncatalytic or “dead” PTPs (14). It also underscores the importance of determining the roles of noncatalytic, structural analogs of protein tyrosine kinases, caspases, carbonic anhydrases, and other PTPs (14, 15) emerging from genome-sequencing projects (36, 37).
Acknowledgments
We thank Drs. J. A. Williams, K. L. Guan, and H. Guardiola-Diaz for helpful discussion, the University of Michigan (UM) Transgenic Animal Model Core for blastocyst injection and assistance with ES cell culture, Dr. A. K. Christiansen, and R. K. Brabec for expertise on testis development and histology, Dr. S. Camper for use of Cre-transgenic mice, and the veterinarian and animal husbandry staff of the UM Unit for Lab Animal Medicine. We were supported in part by grants from the National Institutes of Health and the Walther Cancer Institute (to J.E.D.), Systems and Integrated Biology Training Grant 5T32GM08322-07, UM Horace H. Rackham Distinguished Research Partnership Award, and UM Department of Physiology Horace W. Davenport Fellowship (to M.J.W.). Funding for generating transgenic mice was provided in part by the UM Cancer Center, Arthritis Center, and Molecular Biology core of the Diabetes Center.
Abbreviations
- PTP
protein tyrosine phosphatase
- Styx
phosphoserine, -threonine, or -tyrosine interaction protein
- Crhsp-24
calcium-responsive heat-stable protein with a molecular mass of 24 kDa
- ES
embryonic stem
- β-gal
β-galactosidase
References
- 1.Zhong J, Peters A H F M, Lee K, Braun R E. Nat Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- 2.Kuroda M, Sok J, Webb L, Baechtold H, Urano F, Yin Y, Chung P, de Rooij D G, Akhmedov A, Ashley T, Ron D. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee K, Haugen H S, Clegg C H, Braun R E. Proc Natl Acad Sci USA. 1995;92:12451–12455. doi: 10.1073/pnas.92.26.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecht N B. BioEssays. 1998;20:555–561. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 5.Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Lozzo R, Cooper D, Calabretta B. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales C R, Wu X Q, Hecht N B. Dev Biol. 1998;201:113–123. doi: 10.1006/dbio.1998.8967. [DOI] [PubMed] [Google Scholar]
- 7.Steger K. Anat Embryol. 1999;1999:471–487. doi: 10.1007/s004290050245. [DOI] [PubMed] [Google Scholar]
- 8.Barinaga M. Science. 1999;283:1247. doi: 10.1126/science.283.5406.1247. [DOI] [PubMed] [Google Scholar]
- 9.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 10.Yaffe M B, Cantley L C. Nature (London) 1999;402:30–31. doi: 10.1038/46925. [DOI] [PubMed] [Google Scholar]
- 11.van der Greer P, Pawson T. Trends Biochem Sci. 1995;20:277–280. doi: 10.1016/s0968-0004(00)89043-x. [DOI] [PubMed] [Google Scholar]
- 12.Kuriyan J, Cowburn D. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 13.Wishart M J, Denu J M, Williams J A, Dixon J E. J Biol Chem. 1995;270:26782–26785. doi: 10.1074/jbc.270.45.26782. [DOI] [PubMed] [Google Scholar]
- 14.Wishart M J, Dixon J E. Trends Biochem Sci. 1998;23:301–306. doi: 10.1016/s0968-0004(98)01241-9. [DOI] [PubMed] [Google Scholar]
- 15.Hunter T. Nat Genet. 1998;18:303–305. doi: 10.1038/ng0498-303. [DOI] [PubMed] [Google Scholar]
- 16.Chui D, Oh-Eda M, Liao Y F, Panneerselvam K, Lal A, Marek K W, Freeze H H, Moremen K W, Fukuda M N, Marth J D. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 17.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kendall S K, Samuelson L C, Saunders T L, Wood R I, Camper S A. Genes Dev. 1995;9:2007–2019. doi: 10.1101/gad.9.16.2007. [DOI] [PubMed] [Google Scholar]
- 19.Hill D P, Wurst W. In: Guide to Techniques in Mouse Development. Wassarman P, DePamphilis M, editors. Vol. 225. San Diego: Academic; 1993. pp. 664–681. [Google Scholar]
- 20.Russel L D, Ettlin R A, Hikim A P, Clegg E D. Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River; 1990. [Google Scholar]
- 21.Hess R A, Moore B J. In: Male Reproductive Toxicology. Chapin R, Heindel J, editors. 3A. San Diego: Academic; 1993. pp. 52–85. [Google Scholar]
- 22.Filler R. In: Male Reproductive Toxicology. Chapin R, Heindel J, editors. 3A. San Diego: Academic; 1993. pp. 334–343. [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 24.Oakberg E F. Am J Anat. 1956;99:391–414. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- 25.Rhee K, Wolgemuth D J. Dev Dyn. 1995;204:406–420. doi: 10.1002/aja.1002040407. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Toselli P A, Russell L D, Seldin D C. Nat Genet. 1999;23:118–121. doi: 10.1038/12729. [DOI] [PubMed] [Google Scholar]
- 27.Varmuza S, Jurisicova A, Okano K, Hudson J, Boekelheide K, Shipp E B. Dev Biol. 1999;205:98–110. doi: 10.1006/dbio.1998.9100. [DOI] [PubMed] [Google Scholar]
- 28.Blandy J A, Kaestner K H, Weinbauer G F, Nieschlag E, Schutz G. Nature (London) 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 29.Nantel F, Monaco L, Foulkes N S, Masquilier D, LeMeur M, Henriksen K, Dierich A, Parvinen M, Sassone-Crosi P. Nature (London) 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 30.Togawa A, Miyoshi J, Ishizaki H, Tanaka M, Takakura A, Nishioka H, Yoshida H, Doi T, Mizoguchi A, Matsuura N, et al. Oncogene. 1999;18:5373–5380. doi: 10.1038/sj.onc.1202921. [DOI] [PubMed] [Google Scholar]
- 31.Biggiogera M, Tanquay R M, Marin R, Wu Y, Martin T E, Fakan S. Exp Cell Res. 1996;229:77–85. doi: 10.1006/excr.1996.0345. [DOI] [PubMed] [Google Scholar]
- 32.Perego L, Berruti G. Mol Reprod Dev. 1997;47:370–379. doi: 10.1002/(SICI)1098-2795(199708)47:4<370::AID-MRD3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 33.Groblewski G E, Yoshida M, Bragado M J, Ernst S A, Leykam J, Williams J A. J Biol Chem. 1998;273:22738–22744. doi: 10.1074/jbc.273.35.22738. [DOI] [PubMed] [Google Scholar]
- 34.Nastasi T, Scaturro M, Bellafiore M, Raimondi L, Beccari S, Cestelli A, Di Liegro I. J Biol Chem. 1999;274:24087–24093. doi: 10.1074/jbc.274.34.24087. [DOI] [PubMed] [Google Scholar]
- 35.Nastasi T, Muzi P, Beccari S, Bellafiore M, Dolo V, Bologna M, Cestelli A, Di Liegro I. NeuroReport. 2000;11:2233–2236. doi: 10.1097/00001756-200007140-00034. [DOI] [PubMed] [Google Scholar]
- 36.Ijkel W F J, van Strien E A, Heldens J G M, Broer R, Zuidema D, Goldbach R W, Vlak J M. J Gen Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- 37.Uwanogho D A, Hardcastle Z, Balogh P, Mirza G, Thornburg K L, Ragoussis J, Sharpe P T. Genomics. 1999;62:406–416. doi: 10.1006/geno.1999.5950. [DOI] [PubMed] [Google Scholar]