Abstract
The innate immune system includes antimicrobial peptides that protect multicellular organisms from a diverse spectrum of microorganisms. β-Defensins comprise one important family of mammalian antimicrobial peptides. The annotation of the human genome fails to reveal the expected diversity, and a recent query of the draft sequence with the blast search engine found only one new β-defensin gene (DEFB3). To define better the β-defensin gene family, we adopted a genomics approach that uses hmmer, a computational search tool based on hidden Markov models, in combination with blast. This strategy identified 28 new human and 43 new mouse β-defensin genes in five syntenic chromosomal regions. Within each syntenic cluster, the gene sequences and organization were similar, suggesting each cluster pair arose from a common ancestor and was retained because of conserved functions. Preliminary analysis indicates that at least 26 of the predicted genes are transcribed. These results demonstrate the value of a genomewide search strategy to identify genes with conserved structural motifs. Discovery of these genes represents a new starting point for exploring the role of β-defensins in innate immunity.
The β-defensins are defined by a six-cysteine motif [usual spacing, C-X6-C-X4-C-X9-C-X6-C-C (1, 2)] and a large number of basic amino acid residues. Their coding sequences consist of two exons. The first exon includes the 5′ untranslated region and encodes the leader domain of the preproprotein; the second exon encodes the mature peptide with the six-cysteine domain. As this project began, five human β-defensin genes (3–9) and six mouse β-defensin genes (10–14) were reported or submitted to GenBank (accession no. AF318068). However, we suspected the existence of additional β-defensin genes because of the high frequency of gene duplication within β-defensin clusters (15). Thus, we hypothesized that analyzing the genomic sequence (16–18) with multiple computational tools (19, 20) would identify novel defensin genes.
Materials and Methods
blast-Based Searches.
Genomic search strategies for human gene discovery were applied to the GenBank nonredundant, high throughput genomic sequences, and expressed sequence tag (EST) databases by using the blastp and tblastn programs (19), using National Center for Biotechnology Information (NCBI) web site tools (www.ncbi.nlm.nih.gov/blast/). Similar approaches were used to query the Celera mouse genome assembly (www.celera.com). The initial queries for the search used the amino acid sequences for the known human defensins (DEFB1, DEFB2, DEFB3, and DEFB4) (3–7) and two HE2/EP2 sequences (8, 9) and the known mouse β-defensins (Defb1, Defb2, Defb3, Defb4, Defb5, and Defb6) (10–14) and GenBank (accession no. AF318068). NCBI and Celera default parameters were used in the searches, and any potential hits were curated manually.
For each novel β-defensin gene identified by using the hmmsearch program (described below), additional iterative blast searches were performed against the human and mouse databases to identify additional related sequences and search for ESTs to confirm that the sequences were transcribed.
Construction of Hidden Markov Models (HMMs) for the Six-Cysteine β-Defensin Motif.
The complementary strategy used to identify β-defensin genes used a quantitative sequence analysis with the HMM (20). For this purpose, we defined core human and mouse β-defensin amino acid sequences containing the six-cysteine motif and sorted them according to their scores in HMMs trained on defensin motifs. Initially, 12 second-exon, 6-cysteine motifs derived from human and mouse β-defensin sequences were defined by manual inspection of full-length β-defensin domain sequences. These motifs were aligned by using the clustalw program (21) and trimmed of extra amino acids extending on both sides of a 38-aa core. We used these 12 aligned sequences as input for the HMMER 2.1.1 suite of software at Washington University (St. Louis) (http://hmmer.wustl.edu; ref. 20) to build the first of our HMM β-defensin models. We constructed the first model by using the program hmmbuild and then used hmmcalibrate to calibrate E-value scores. Ultimately, three models were built: all 33 human second-exon sequences, all 36 mouse second exons, and a combined model of human and mouse second exons (69 sequences). These models were used to search all of the contigs of the Univ. of California (Santa Cruz) Golden Path assembly (http://genome.cse.ucsc.edu/goldenPath/01apr2001) and the July 30, 2001, NCBI assembly (ftp://ncbi.nlm.nih.gov/genomes/H_sapiens) by using the hmmsearch program. The assemblies, once downloaded to a local file server, were translated into all six reading frames for searching, as hmmer does not currently have the native capability to search nucleotide databases. We followed the same procedure for all contigs from the mouse genomic sequence that contained β-defensin genes as revealed by blast analysis. A Linux cluster consisting of 32 dual-Pentium-III (550 MHz) processors, each with 2 giga-bytes of RAM, was used to perform the searching in parallel, taking roughly 90 min. We used the default cutoff values for filtering the results of these queries and performed postprocessing of the results (using custom Perl programs) to remove motifs that did not contain the six-cysteine target motif. Before this filtering, hundreds of false domains were reported. Also, we observed multiple hmmer hits of the same gene, indicating that the present state of the assembled draft sequence contains multiple copies of the same regions.
Assembly of Human and Mouse β-Defensin Genomic Clusters.
To generate a continuous DNA sequence for some analyses, the sequences from the human and mouse defensin containing BAC clones and genomic contigs were aligned by using the sequencher program (Gene Codes, Ann Arbor, MI).
Analysis of Predicted β-Defensin Peptide Sequences: Alignment and Phylogeny.
The multiple sequence alignment and dendrogram construction were performed by using the program pileup from the Wisconsin Package software (Accelrys, San Diego). The amino acid sequences were predicted from the known, related, and predicted β-defensin genes in human and/or mouse and included two residues before and after the six-cysteine domain. The comparison matrix was set at Blosum62 with a gap creation penalty of 8 and a gap extension penalty of 2.
Results
To discover new β-defensin genes, we constructed an HMM (20) with the mature peptide sequences of known β-defensin genes. Then we used this model [hmmer software (20)] to screen ∼4 Mb of genomic DNA sequence around the β-defensin locus on human chromosome 8p23-p22. This search found 11 genes, including the five known β-defensin genes (DEFB1–4 and HE2/EP2) plus six novel genes (DEFB5–9 and DEFBp1) (Fig. 1). When these novel sequences were used in a blast search of the draft human genomic sequence, we discovered another β-defensin gene, DEFB10. Surprisingly, DEFB10 maps to chromosome 6p12, whereas all previously identified human defensin genes localized to chromosome 8p23-p22 (22, 23). We reseeded the HMM with the predicted peptide sequences from the new genes and used it to analyze the genomic DNA sequence derived from the BAC clone that contains DEFB10 at the 6p12 locus (GenBank accession no. NT_007402). This analysis revealed four more β-defensin genes (DEFB11–14) (Fig. 1). Thus, the DEFB10–14 genes represent a second β-defensin gene cluster. Subsequent iterations of the blast/hmmer process identified 15 additional β-defensins (DEFB15–29) (Fig. 1). These genes are located on two sequence contigs, one on chromosome 20q11.1 and one on chromosome 20p13, and represent two more β-defensin gene clusters.
Figure 1.
Multiple sequence alignment of human β-defensin proteins. The amino acid sequences were predicted from genomic sequence (Table 1). The chromosomal location of each gene is indicated, except where mapping was ambiguous (A). The sequences were aligned as described in Materials and Methods followed by minor manual manipulations to maximize sequence alignment and clustering of genes by chromosome. We classified β-defensin genes as known (K), if evidence exists that they are transcribed and that their protein product demonstrates antimicrobial activity; related (R), if evidence exists that they are transcribed but their protein product has not been tested for antimicrobial activity; predicted (P), if no evidence exists that they are transcribed; and pseudogenes (S), if the DNA sequence is highly similar to a β-defensin gene, but the predicted amino acid sequence lacks an ORF across the six-cysteine motif. The consensus sequence shows cysteine residues (yellow highlight), positively charged residues (blue +) and other residues (red) if they are represented in greater than 50% of all predicted β-defensin proteins.
Finally, to analyze the entire human genome with hmmer, we used all 31 human β-defensin genes for the HMM. This analysis revealed 40 more sequences that encode a 6-cysteine domain. However, the position of the cysteines and the cationic charge density were consistent with β-defensin genes in only two cases, DEFB30 and DEFB31 (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). In addition, these two genes are located on the same BAC clone that ambiguously maps to multiple regions of the genome, including chromosome 8p23-p22 (Fig. 1). Because the sequence of this clone is not contiguous with the 8p23-p22 contigs containing the known β-defensin locus, it may represent a fifth cluster in the human genome. Significantly, this hmmer search detected only 13 of the 31 previously identified β-defensin genes. These results suggest that, like blast searches, genomewide searches with hmmer alone are not sufficient for identifying all β-defensin genes. Further blast and hmmer analyses did not detect additional sequences in the human genome. In total, this iterative, computational method discovered 28 novel human β-defensin genes (Fig. 1 and Table 1).
To search for novel β-defensin genes in the mouse genome, we used this hybrid process to screen assembled mouse sequence contigs in the Celera database (18). This analysis discovered 43 new genes (Table 1) in five clusters, on chromosomes 1, 8, and 14, and two on 2. Significantly, these regions are syntenic to the regions in the human genome that contain the β-defensin clusters (www.ncbi.nlm.nih.gov/Homology), indicating that the syntenic clusters have a common origin. Supporting this hypothesis, we found the highest sequence similarity between gene products from syntenic clusters (Fig. 2 and Table 1). Moreover, the order and orientation for many genes in the syntenic clusters are conserved (Fig. 3). Given the syntenic relationship between the human and mouse clusters and the conservation of genes within them, we conclude that each pair originated from a common ancestral gene cluster (13, 22). In addition to the six-cysteine domain and density of cationic residues that define the β-defensin peptides, comparisons of the predicted amino acid sequences for the new human (Fig. 1) and mouse (Table 1) β-defensin genes reveal two new features, a conserved glycine between cysteines 1 and 2 and a previously unrecognized glutamic acid between cysteines 3 and 4 (Fig. 1).
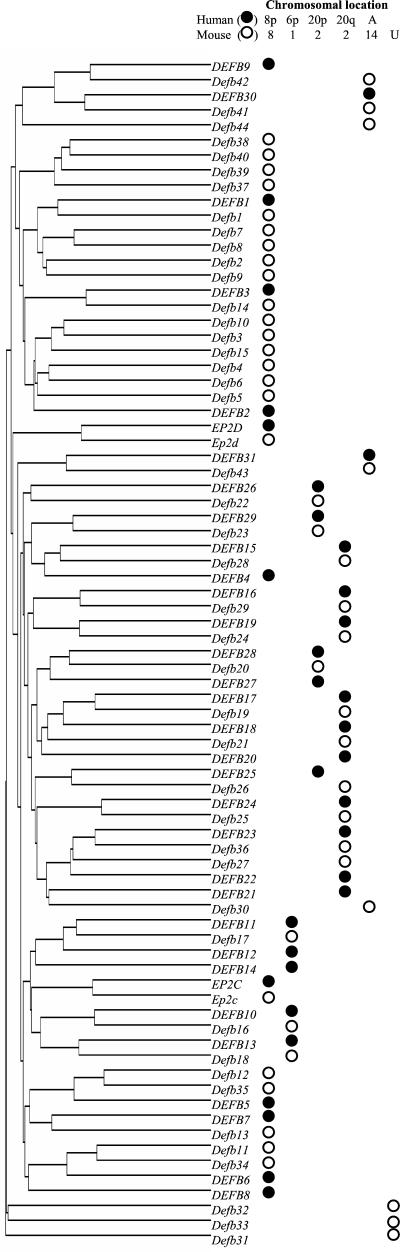
Figure 2.
Dendrogram of predicted β-defensin proteins. The length of each branch is inversely related to their similarity. The tree was constructed with the partial amino acid sequences predicted from the indicated human (●) and mouse (○) known, related, and predicted genes (Table 1). The genomic location for each gene is indicated. In some cases, the location of the genes was ambiguous (A) or unknown (U).
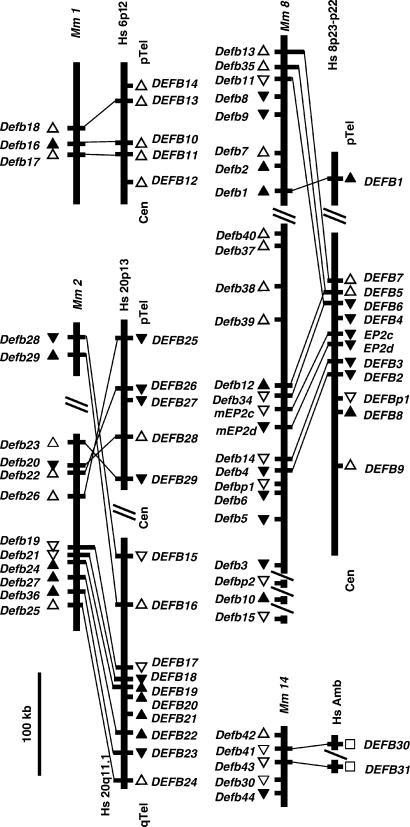
Figure 3.
Genomic structure of five conserved β-defensin gene clusters. Vertical bars represent assembled genomic DNA sequence contigs (see Table 1 for GenBank and Celera accession nos.) from the indicated human (Hs) and mouse (Mm) chromosome. Slanted lines represent gaps in the genomic sequence of known (single) or unknown (double) length. The telomere (Tel) and centromere (Cen) orientation of human DNA sequence contigs was deduced from the position of genetic markers. The orientation of the mouse DNA sequence contigs was deduced from the alignment of human and mouse gene homologs. The direction of transcription is indicated for known and related genes (▴) and predicted genes and pseudogenes (▵) or is unknown (▫). Thin lines connect human and mouse genes with highest sequence similarity (Fig. 2).
To test whether these predicted genes are transcribed, we performed blast searches against the six-frame translation of the EST database (dbEST). At least one EST was found for 12 human and 14 mouse predicted genes (Table 1), representing all clusters. The absence of some β-defensin genes in dbEST is not surprising. For example, the β-defensin gene DEFB3 is not found in dbEST; transcripts from this gene are rare until inflammatory stimuli induce expression (5, 6). We note that presence of β-defensin transcripts alone may not accurately predict protein expression and additional studies are required to verify the production of functional β-defensin peptides by these new genes.
We aligned the human and mouse EST sequences with the genomic sequence to verify that the EST sequences encode full-length β-defensin proteins. Eight human EST sequences split into the familiar two-exon structure of a β-defensin gene. The first exon encoded the leader domain and the second exon encoded the six-cysteine domain. The other four human EST sequences failed to extend into an exon 1 sequence (DEFB25) or the first exon did not encode a leader domain (DEFB8, DEFB22, DEFB31). The latter three genes may represent transcribed pseudogenes or EP2-like genes. The EP2 gene (also called HE2) has two promoters and produces multiple transcripts. Each alternative EP2 transcript encodes a different protein isoform, including predicted proteins with a six-cysteine β-defensin domain (8, 24, 25). Further expression studies are needed to distinguish between these two possibilities. We observed similar results with the mouse EST sequences—12 ESTs split into the expected two-exon structure and two ESTs lacked a leader domain in the first exon. Finally, we identified ESTs with partial identity to the human DEFB9 and mouse Defb23. However, the intron/exon sequences indicated that the transcripts were produced from the opposite strand. Consequently, we do not know whether a β-defensin gene is produced at these two loci. These preliminary expression studies together with the conservation of the five sequence clusters suggest that many of the 28 human and 39 mouse predicted β-defensin genes encode functional products.
Discussion
These findings are a proof of principle for a pioneering genomewide search strategy that identifies genes with conserved structural motifs. The study highlights the complementary nature of the blast and hmmer analysis tools and demonstrates their potential synergy for mining genomic databases and identifying new members of gene families. None of the novel sequences identified were included in previous annotations of the human and mouse genomes, indicating that they were missed. Based on this example, the current estimates of 30,000–40,000 genes in the human and mouse genome (16, 17) may be a significant underestimate. We speculate that novel genes in other families are not counted with conventional strategies but could be detected by using this approach. Excellent candidates include gene families whose products are secreted, such as α-defensins, chemokines, growth factors, and neuropeptides. Like the β-defensin gene products, such peptides use disulfide bridges that are stable in the high oxidation environment outside the cell. This bonding strategy allows the primary structure of a peptide to be short and diverse, because only a few amino acids are required to maintain tertiary and quaternary structure. Consequently, gene families that encode such peptides may be refractory to conventional search strategies.
Some of the predicted protein products of the new β-defensin genes exhibit unique features. Many have subtle variations in the typical spacing between the cysteine residues (see Table 1). However in DEFB7, there are 21 amino acid residues between the first and second cysteines rather than the more typical 6, a finding of unknown functional significance. A unique feature of the sequences within the chromosome 20 cluster and their mouse homologs is the longer C-terminal tails following the six-cysteine domain (Fig. 1). Further studies are needed to learn whether these C-terminal sequences are posttranslationally modified in the mature peptides. This structure is reminiscent of some chemokine gene products that share both functional and structural features with the β-defensins (26, 27). Both have a structural core of three anti-parallel β-sheets secured by disulfide linkages (28). The β-defensins and the chemokine CCL20 (MIP-3α) engage the CCR6 receptor on selected immune effector cells (27), and some chemokines exhibit antimicrobial activity (29). Chemokines are also encoded by small genes that exist in the genome as gene clusters, with functionally related sequences present within a cluster, suggesting that the gene family expanded by duplication events (30). Thus, it is possible that features of functional significance are conserved within this larger family of β-defensin sequences and the chemokines.
Expansion of the β-defensin gene family has significant implications for future studies of innate host defense. The genomic conservation between human and mouse clusters suggests functional conservation and supports use of the mouse as a model organism to study this gene family. However, the size of the gene family provides the potential for redundant functions and may limit studies of mice that lack the function of a single gene or cluster. The search strategy outlined here focused on finding β-defensin second exons—the second exon encodes the mature peptide. A similar approach could be used to discover all first exon-coding sequences and the associated regulatory elements that confer cell specificity and responsiveness to inflammatory stimuli and pathogens. Addition of a large number of proteins to this family should also provide new opportunities to investigate the relationship between antimicrobial peptide structure, function, and specificity. Thus, this study helps lay the groundwork to understand better the complex interaction between innate host defense and the diversity of microorganisms in our environment.
Supplementary Material
Acknowledgments
We thank Margaret Malik, Andrea Penisten, and Autumn Bradley for technical assistance. We thank John Stokes, Beverly Davidson, and Tomas Ganz for critically reviewing the manuscript. These data were generated in part through the use of the Celera Discovery System and Celera Genomics-associated databases. We acknowledge the support of National Institutes of Health Grant NHLBI HL-61234 (to M.J.W., P.B.M., and B.C.S.), National Institutes of Health/National Institute of Child Health and Human Development Grant P30-HD27748 (to B.C.S.), and the Howard Hughes Medical Institute. T.L.C. is supported by National Institutes of Health/National Cancer Institute Grant CA-85188 and by a Cystic Fibrosis Genome Analysis grant. M.J.W. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- HMM
hidden Markov model
- EST
expressed sequence tag
References
- 1.Huttner K M, Bevins C L. Pediatr Res. 1999;45:785–794. doi: 10.1203/00006450-199906000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Tang Y Q, Selsted M E. J Biol Chem. 1993;268:6649–6653. [PubMed] [Google Scholar]
- 3.Bensch K W, Raida M, Magert H J, Schulz-Knappe P, Forssmann W G. FEBS Lett. 1995;368:331–335. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 4.Harder J, Bartels J, Christophers E, Schroder J-M. Nature (London) 1997;387:861–862. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 5.Harder J, Bartels J, Christophers E, Schroder J M. J Biol Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 6.Jia H P, Schutte B C, Schudy A, Linzmeier R, Guthmiller J M, Johnson G K, Tack B F, Mitros J P, Rosenthal A, Ganz T, McCray P B., Jr Gene. 2001;263:211–218. doi: 10.1016/s0378-1119(00)00569-2. [DOI] [PubMed] [Google Scholar]
- 7.Garcia J R, Krause A, Schulz S, Rodriguez-Jimenez F J, Kluver E, Adermann K, Forssmann U, Frimpong-Boateng A, Bals R, Forssmann W G. FASEB J. 2001;5:1819–1821. [PubMed] [Google Scholar]
- 8.Kirchhoff C, Osterhoff C, Habben I, Ivell R, Kirchhoff C. Int J Androl. 1990;13:155–167. doi: 10.1111/j.1365-2605.1990.tb00972.x. [DOI] [PubMed] [Google Scholar]
- 9.Frohlich O, Po C, Murphy T, Young L G. J Androl. 2000;21:421–430. [PubMed] [Google Scholar]
- 10.Huttner K M, Kozak C A, Bevins C L. FEBS Lett. 1997;413:45–49. doi: 10.1016/s0014-5793(97)00875-2. [DOI] [PubMed] [Google Scholar]
- 11.Morrison G M, Davidson D J, Dorin J R. FEBS Lett. 1999;442:112–116. doi: 10.1016/s0014-5793(98)01630-5. [DOI] [PubMed] [Google Scholar]
- 12.Bals R, Wang X, Meegalla R L, Wattler S, Weiner D, Nehls M C, Wilson J M. Infect Immun. 1999;67:3542–3547. doi: 10.1128/iai.67.7.3542-3547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia H P, Wowk S A, Schutte B C, Lee S K, Vivado A, Tack B F, Bevins C L, McCray P B., Jr J Biol Chem. 2000;275:33314–33320. doi: 10.1074/jbc.M006603200. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi Y, Fukuhara S, Nagase T, Tomita T, Hitomi S, Kimura S, Kurihara H, Ouchi Y. J Biol Chem. 2001;14:31510–31514. doi: 10.1074/jbc.M104149200. [DOI] [PubMed] [Google Scholar]
- 15.Hughes A L, Yeager M. J Mol Evol. 1997;44:675–682. doi: 10.1007/pl00006191. [DOI] [PubMed] [Google Scholar]
- 16.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 17.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 18.Marshall E. Science. 2001;292:822–823. doi: 10.1126/science.292.5518.822. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Eddy S R. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, Zhao C, Heng H H Q, Ganz T. Genomics. 1997;43:316–320. doi: 10.1006/geno.1997.4801. [DOI] [PubMed] [Google Scholar]
- 23.Bevins C L, Jones D E, Dutra A, Schaffzin J, Muenke M. Genomics. 1996;31:95–106. doi: 10.1006/geno.1996.0014. [DOI] [PubMed] [Google Scholar]
- 24.Hamil K G, Sivashanmugam P, Richardson R T, Grossman G, Ruben S M, Mohler J L, Petrusz P, O'Rand M G, French F S, Hall S H. Endocrinology. 2000;141:1245–1253. doi: 10.1210/endo.141.3.7389. [DOI] [PubMed] [Google Scholar]
- 25.Frohlich O, Po C, Young L G. Biol Reprod. 2001;64:1072–1079. doi: 10.1095/biolreprod64.4.1072. [DOI] [PubMed] [Google Scholar]
- 26.Zlotnik A, Yoshie O. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 27.Yang D, Chertov O, Bykovskaia S N, Chen Q, Buffo M J, Shogan J, Anderson M, Schroder J M, Wang J M, Howard O M, Oppenheim J J. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Canadillas J M, Zaballos A, Gutierrez J, Varona R, Roncal F, Albar J P, Marquez G, Bruix M. J Biol Chem. 2001;276:28372–28379. doi: 10.1074/jbc.M103121200. [DOI] [PubMed] [Google Scholar]
- 29.Cole A M, Ganz T, Liese A M, Burdick M D, Liu L, Strieter R M. J Immunol. 2001;167:623–627. doi: 10.4049/jimmunol.167.2.623. [DOI] [PubMed] [Google Scholar]
- 30.Rollins B J. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.