Abstract
One of the characteristics of the host defense of insects is the rapid synthesis of a variety of potent antibacterial and antifungal peptides. To date, seven types of inducible antimicrobial peptides (AMPs) have been characterized in Drosophila. The importance of these peptides in host defense is supported by the observation that flies deficient for the Toll or Immune deficiency (Imd) pathway, which affects AMP gene expression, are extremely susceptible to microbial infection. Here we have developed a genetic approach to address the functional relevance of a defined antifungal or antibacterial peptide in the host defense of Drosophila adults. We have expressed AMP genes via the control of the UAS/GAL4 system in imd; spätzle double mutants that do not express any known endogenous AMP gene. Our results clearly show that constitutive expression of a single peptide in some cases is sufficient to rescue imd; spätzle susceptibility to microbial infection, highlighting the important role of AMPs in Drosophila adult host defense.
Antimicrobial peptides (AMPs) are a key component of innate immunity. Their distribution throughout the animal and plant kingdom is ubiquitous, reflecting the importance of these molecules in host defense (1, 2). In insects, systemic infection induces the synthesis of combinations of AMPs that are secreted from the immune organs, mainly the fat body, an analogue of the mammalian liver, into the hemolymph, where they reach high concentrations. In Drosophila, at least seven types of AMPs (plus isoforms) have been described (3–5). Their activities have been either determined in vitro by using peptides directly purified from flies or produced in heterologous systems, or deduced by comparison with homologous peptides isolated in other insect species: (i) Drosomycin and Metchnikowin show antifungal activity (6, 7); (ii) Cecropins have both antibacterial and antifungal activities (8); (iii) Drosocin and Defensin are predominantly active against Gram-negative and -positive bacteria, respectively (9, 10); and (iv) Attacins and Diptericins are similar to peptides from other insects that show antibacterial activity (11, 12).
Analysis of the in vivo roles of each AMP on microbial infection is complicated by the numerous AMP genes present in the fly, as well as the redundant defense mechanisms within the innate immune system. The importance of AMPs, however, is supported by the sensitive phenotype of mutants that do not express AMP-encoding genes (13–15). A clear correlation is observed between the lack of expression of antibacterial peptide genes in mutants of the Immune deficiency (Imd) pathway and their susceptibility to Gram-negative bacteria (13). Conversely, mutations in the Toll pathway block Drosomycin expression and result in susceptibility to fungal infection (14). Finally, mutants deficient in both the Imd and Toll pathways failed to express any known AMP genes after infection and are extremely susceptible to both fungal and bacterial infections (14). These evidences of the importance of AMPs in fighting infection, however, are still indirect, because we cannot exclude that these mutations affect other defense reactions. The Toll pathway, for example, has also been reported to regulate hemocyte proliferation (16). To study unambiguously the in vivo role of each AMP in Drosophila host defense, we have generated imd; spätzle (spz) flies deficient for both the Imd and Toll pathways but that constitutively express different AMPs under the control of a noninducible promoter. These flies express only one AMP on infection and, consequently, we can use a simple survival experiment to monitor the contribution of this peptide in resistance to infection by various microorganisms. This powerful assay allowed us to analyze, in vivo, the spectrum of activity of each peptide and, by combining two different transgenes, any potential synergy among them. Our results clearly show that expression of a single peptide, in some cases, is sufficient to rescue the imd; spz susceptibility to microbial infection, highlighting the important role of AMPs in Drosophila adult host defense.
Materials and Methods
Plasmids.
The fusion constructs used for P-element-mediated transformation were: UAS-Drs, which contains a 691-bp BamHI-XbaI Drosomycin fragment of the gene (6) inserted between the pUAST BglII-XbaI sites; UAS-Def, which contains a 398-bp HindIII-KpnI fragment of the Defensin gene (9) inserted between the pUAST BglII (blunted)-KpnI sites; UAS-Cec A, which contains a 459-bp EcoRV-XbaI fragment of pSK-Cecropin A (17) inserted between the pUAST BglII (blunted)-XbaI sites; UAS-Att A, which contains a 894-bp BamHI-KpnI fragment of pSK-Attacin A (12) inserted between the pUAST BglII-KpnI sites; UAS-Dpt, which contains a 350-bp EcoRI-EcoRI fragment of the Diptericin gene (11) inserted in the pUAST EcoRI site; and UAS-Drc, which contains a 347-bp EcoRI-KpnI fragment of the Drosocin gene (10) inserted between the pUAST EcoRI-KpnI sites.
Fly Stocks.
Fly cultures and crosses were grown on standard fly medium at 25°C. Oregon R flies were used as wild-type standard. spzrm7 is a strong mutation in the spätzle gene that encodes the putative Toll ligand (18). Flies homozygous for spzrm7 failed to express the Drosomycin gene after septic injury and are highly susceptible to fungal infection (14). Flies homozygous for imd failed to express antibacterial peptide genes after septic injury and are highly susceptible to Gram-negative bacterial infection (13). The da-GAL4 driver expresses GAL4 ubiquitously and strongly (19). w1118 flies were used as recipients for transformation, and transgenic lines were established as described (20). We obtained at least two independent lines for each UAS-Pep construct, with transposon located on the autosomes. By standard crosses and recombination, we generated UAS-Pep fly lines of the following genotype: w; imd, UAS-Pepx/imd, UAS-Pepx; spzrm7, UAS-Pepy/TM6C. These flies were crossed with w; imd/imd; da-Gal4, spzrm7/TM6C flies to obtain the flies [w; imd, UAS-Pepx/imd; spzrm7, UAS-Pep(x or y)/da-GAL4, spzrm7] that we used in our survival experiments.
Microorganisms.
Bacteria were precultured in LB medium, with the exceptions of Staphyloccocus aureus and Agrobacterium tumefaciens, which were cultured in brain heart infusion (Difco) and yeast extract/peptone media, respectively. The following are the bacterial strains used in this study: Pseudomonas aeruginosa (CFBP2466), Escherichia coli (MG1655), A. tumefaciens (EHA 105); Erwinia carotovora (21), S. aureus (gifts from H. Monteil, University of Strasbourg, Strasbourg, France); Bacillus subtilis (168), Microccocus luteus (gifts from J. Millet and A. Klier, Pasteur Institute, Paris). Fungi were grown on malt–agar medium with the exceptions of Neurospora crassa and Fusarium oxysporum, which were cultured on 1/2 potato dextrose agar and on 6CA medium, respectively. Spores and hyphae were harvested as described in ref. 22. The strains used in this study were: F. oxysporum (MUCL 909), N. crassa (CBS 327–54), Beauvaria bassiana (80.2), and Aspergillus fumigatus (gift from A. Brakhage, Technische Universität, Darmstadt, Germany).
Northern Blot Analysis.
Total RNA extraction and Northern blot experiments were performed as described in ref. 23.
Mass Spectrophotometer.
A PerSeptive Biosystems (Framingham, MA) Voyager Eite matrix-assisted spectrometer was used with laser desorption time-of-flight with delayed extraction and high-sensitivity linear detector (24). Ten UAS-Drs imd; spzrm7 adults were bled with a hand-pulled capillary to collect hemolymph directly into 0.1% trifluoroacetic acid/acetonitrile (1:1 vol/vol). Zip-Tip (Millipore) was used to purify the hemolymph sample according to the manufacturer's protocol. Final resuspension was in 25% acetonitrile (total 10 μl), and 2.5 μl was spotted on a glass plate for analysis.
Survival Experiments.
Microbial challenge was performed by pricking adult flies in the thorax with a sharpened tungsten needle (<0.2 mm) dipped into a concentrated bacterial or fungal pellet. Natural infections with B. bassiana were performed by shaking anesthetized flies for 30 sec in a Petri dish containing a sporulating fungal culture (23). Survival experiments were carried out in the same conditions for each line tested. Three groups of 20 young adults from each line were challenged, then incubated at 29°C and transferred to fresh vials every 3 days. Flies that died within the first hour of challenge were not considered in the analysis. The number of bacteria injected was estimated by crushing five flies immediately after bacterial challenge (or clean septic injury for control) in LB medium under sterile conditions and plating in appropriate dilutions on LB plates. For more information, see ref. 22.
Results
Generation of Fly Lines That Express One Single AMP.
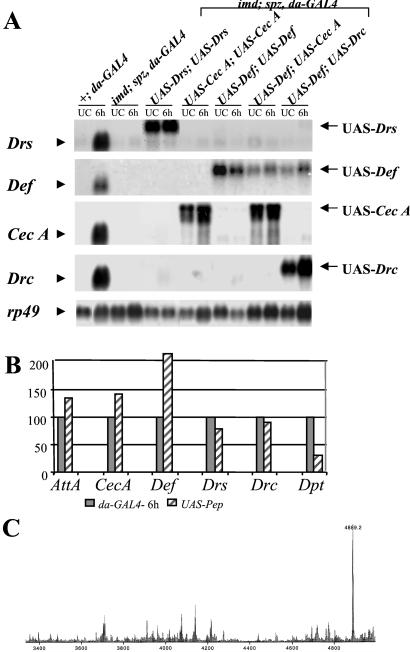
We used the UAS/GAL4 system (25) to construct flies that constitutively express a single AMP gene. We generated fly lines that carry a P transgene containing a fusion between the cDNA of each of the six AMP genes (Diptericin, Drosocin, Cecropin A, Attacin A, Drosomycin, and Defensin) under the control of the binding sites for the GAL4 transactivator (referred to as UAS-Pep). To drive the expression of the UAS-Pep construct, we used daughterless (da)-GAL4 that produces GAL4 protein ubiquitously to high levels. We then recombined each of the UAS-Pep and da-GAL4 with either the imd or spz mutation. By standard genetic crosses, we generated imd; spz double mutant flies carrying two copies of a UAS-Pep and one copy of da-GAL4 (referred to as UAS-Pepx, imd;UAS-Pepx, spz). imd; spz double mutant flies, as previously reported, do not express any of the known AMP genes (Fig. 1A). In contrast, UAS-Pepx, imd; UAS-Pepx, spz flies constitutively produce UAS-Pep-derived AMP [Fig. 1A for UAS-Drosomycin (Drs), UAS-Cecropin A (Cec A), and UAS-Defensin (Def); data are not shown for others]. Importantly, Fig. 1 A and B show that the levels of AMP gene expression in flies carrying two copies of UAS-Pep and one copy of da-GAL4 were similar to those observed in 6-h bacterial-challenged wild-type flies. The two exceptions are the UAS-Diptericin (Dpt) and UAS-Def constructs, which are expressed to approximately 30 and 200% of the respective endogenous gene after infection. The endogenous Defensin has been reported to express at lower levels after bacterial injection than the other AMP genes (9). Thus, the 2-fold higher levels of expression of UAS-Def suggest that the UAS-Pep lines (except UAS-Dpt) are expressed to similar levels.
Figure 1.
Generation of imd; spz flies that express one or two AMP genes. (A) Northern blot analysis showing that imd; spz flies carrying two copies of UAS-Pep and the da-GAL4 driver produce only the intended AMP genes. Total RNA was collected from wild-type, imd/imd; spz, da-GAL4/spz, da-GAL4 or imd, UAS-Pepx/imd; spz. UAS-Pepx or UAS-Pepy/spz, da-GAL4 adult flies either unchallenged (UC) or 6 h after injection of E. coli and M. luteus (6 h). In contrast to wild-type, imd; spz flies express no AMP genes after bacterial challenge. imd; spz flies carrying both the UAS-Pep (UAS-Drs, UAS-Def, UAS-Drc, or UAS-Cec A; data not shown for the others) and da-GAL4 express only the UAS-Pep-derived transcript (arrows). The additional sequences of the UAS-Pep fusion construct result in a transcript with lower mobility than that generated by the endogenous genes (arrowheads). The blot was successively probed with Drosomycin (Drs), Defensin (Def), Cecropin A1 (CecA), Drosocin (Drc), and ribosomal protein 49 (rp49) as loading control. (B) The quantification of AMP gene expression shows that flies carrying two UAS-Pep insertions and da-GAL4 express the UAS-Pep fusion transcript (solid bar) at a level comparable to the level observed in 6-h bacterial-challenged wild-type flies (striped bar). The two exceptions are Diptericin and Defensin, which are expressed at 30 and 220% of the wild-type level, respectively. The signals from a Northern blot performed as in A were quantified by a Bio-Imager system, and the levels of AMP gene expression were normalized by the corresponding values of the rp49 signal. This experiment was repeated and yielded similar results. (C) Differential matrix-assisted laser desorption ionization time-of-flight analysis of hemolymph from imd; spz adult flies carrying both UAS-Drs and da-GAL4 collected in the absence of infection. A unique peak at the exact molecular mass of the putative Drosomycin is observed in flies overexpressing the UAS-Drs.
To test for potential synergy between two peptides, we also constructed a series of imd; spz lines that contained two different UAS-Pep constructs and a da-GAL4 driver (referred to as UAS-Pepx, imd;UAS-Pepy, spz). Fig. 1A shows an illustration of imd; spz flies that express either Defensin and Cecropin A or Defensin and Drosocin. All of the UAS-Pep flies that we generated exhibited a wild-type viability, even though da-GAL4 is reported to have strong and ubiquitous expression throughout all developmental stages of Drosophila (19). This observation indicates that chronic expression of AMP genes is not harmful to the flies.
To ascertain that AMPs can indeed be made by the UAS/GAL4 expression system, we have monitored by matrix-assisted laser desorption ionization time of flight in the presence of Drosomycin from imd; spz adult flies carrying two copies of UAS-Drs and da-GAL4. We show that flies expressing Drosomycin via UAS/GAL4 display a unique peak at 4,889.2 mass units that corresponds exactly to the calculated mass of Drosomycin. The absence of this peak in wild-type control flies (data not shown) indicates that the expected peptide was produced by the UAS/GAL4 system. These data also demonstrate that posttranscriptional mechanisms that might control Drosomycin maturation do not depend on either the Imd or Toll pathway.
Resistance Against Gram-Positive Bacterial Infection.
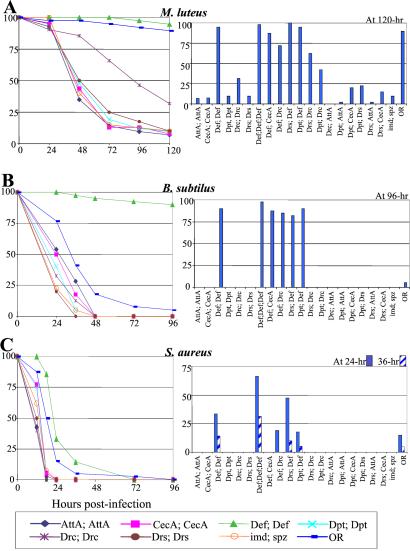
In a first set of experiments, we compared the survival rate of wild-type, imd; spz and UAS-Pepx imd; UAS-Pep(x or y), spz flies after injection by three Gram-positive bacterial species: M. luteus, S. aureus, and B. subtilis. Fig. 2 A–C Left show the survival curve of flies expressing a single peptide over 5 days. A–C Right display the percentage of survival at one critical time point for all of the UAS-Pep combinations that we tested.
Figure 2.
Immune-compromised flies overexpressing the Defensin gene resist as wild type to Gram-positive bacterial infection. The survival rates (expressed in percentage) of wild-type, imd; spz and imd; spz UAS-Pep flies after infection by three Gram-positive bacteria are shown. Sixty flies of each line aged 2–5 days were pricked by a needle dipped into a pellet (OD600 = 50) of (A) M. luteus (≈400 bacteria/fly), (B) B. subtilis (≈400 bacteria/fly), or (C) S. aureus (≈300 bacteria/fly). (Left) Survival curve of the flies that express only one single peptide (UAS-PepX, UAS-PepX) over a 5-day time course. (Right) Survival rate at one or two critical time point(s) for all of the UAS-Pep combinations that we tested (UAS-PepX,UAS-PepY). The survival rate was also monitored on flies carrying three copies of UAS-Def in combination with da-GAL4 (referred to as Def; Def; Def). The precise genotype of flies indicated below each bar as Pepx; Pepy was w; imd, UAS-Pepx/imd; spzrm7, UAS-Pepy/spzrm7, da-GAL4.
Fig. 2A shows that infection by M. luteus does not affect the viability of wild-type flies but reproducibly kills 90% of the imd; spz double mutant within 5 days. imd; spz flies overexpressing Defensin show a wild-type survival after infection by M. luteus, whereas most other peptide-expressing flies survived at less than 25% after 3 days. This result corroborates previous in vitro data showing that Defensin has strong activity against Gram-positive bacterial species (9) (26). In addition, the expression of Drosocin confers a weak protection against M. luteus (32% survival compared with 5–10% survival) by other lines after 5 days, which is also in agreement with previous in vitro studies (10) (Fig. 2A). Flies expressing Drosocin in combination with either Drosomycin or Diptericin display an increased resistance against this Gram-positive bacterium, suggesting a potential cooperative effect between these peptides (Fig. 2A Right).
We repeated this analysis by using B. subtilis and S. aureus. Fig. 2 B and C show that these two bacteria are highly pathogenic for wild-type adults. However, the observation that imd; spz double mutants succumb faster than wild-type flies indicates that the inducible immune response provides some defense against these pathogens. The overexpression of Defensin in imd; spz mutants provides resistance against these two Gram-positive bacteria to levels higher than those of wild-type flies (Fig. 2 B and C). Overexpression of a single copy of UAS-Def provides a complete resistance to B. subtilis, whereas wild-type flies succumb in less than 48 h (Fig. 2B Left), suggesting that the accumulation of Defensin produced constitutively via the UAS/GAL4 system before infection is critical for eliminating B. subtilis cells injected into the host. Fig. 2C shows that Defensin overexpression significantly delays the death of flies infected by S. aureus. We attempted to increase Drosophila resistance to this pathogenic bacterium by generating imd; spz adults carrying three copies of UAS-Def. Fig. 2C shows a clear correlation between the survival rate after infection by S. aureus and the number of UAS-Def copies, indicating a dose-dependent activity of Defensin. Despite the high level of Defensin expression, however, all of the flies succumbed to this infection. Finally, the protective effect induced by Defensin overexpression is not enhanced in the presence of other AMPs (Fig. 2 A–C Left).
Resistance Against Gram-Negative Bacterial Infection.
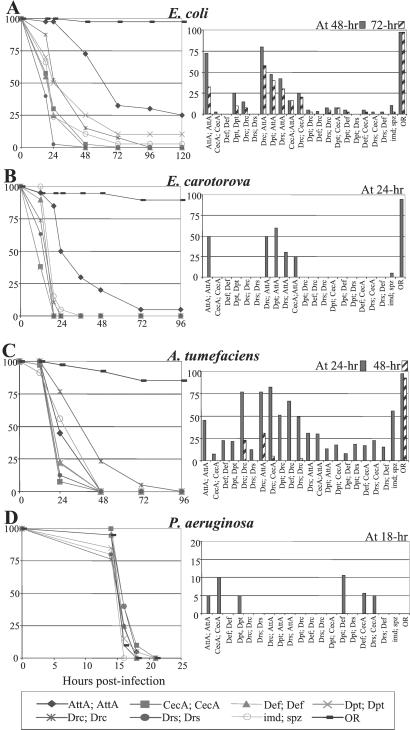
We repeated this analysis by using four Gram-negative bacterial species: E. coli, E. carotovora, A. tumefaciens, and P. aeruginosa. Infection by E. coli or E. carotovora, two bacterial species of the Enterobacteriaceae family, does not affect the viability of wild-type adults but does kill imd; spz double mutants within 72 and 48 h after infection, respectively (Fig. 3 A and B). Attacin A expression significantly increases the resistance of imd; spz double mutant flies to these bacteria. At 24 h after infection, 50–75% of the imd; spz flies expressing Attacin A survive, whereas the imd; spz flies are nearly all dead. Drosocin and Diptericin expression conferred a weak protection against E. coli. Fig. 3 A and B Left, however, show that coexpression of Attacin A with either Diptericin or Drosocin provides a better resistance to E. coli and E. carotovora than the single peptide or other combinations. This observation suggests that Diptericin and Drosocin may cooperate with Attacin A to combat some Gram-negative bacteria.
Figure 3.
Immune-compromised flies expressing Attacin A show increased resistance after infection by Gram-negative bacterial species. The survival rates of wild-type, imd; spz and imd; spz UAS-Pep-expressing flies after infection by four Gram-negative bacteria are shown. Sixty flies were treated as described in the legend to Fig. 2 with a pellet of (A) E. coli (pellet OD600 = 20, ≈400 bacteria/fly), (B) E. caratovora (pellet OD600 = 20, ≈ 560 bacteria/fly), (C) A. tumefaciens (pellet OD600 = 20, ≈300 bacteria/fly), and (D) P. aeruginosa (pellet OD600 = 0.2, ≈70 bacteria/fly). Because P. aeruginosa is a fast-acting pathogen, the survival curve is represented by a shorter time interval.
In contrast, Drosocin expression, but not Attacin A, increases slightly but significantly the resistance against A. tumefaciens (25% survived compared with 0% shown by other AMPs at 48 h after infection), demonstrating the differences between the activities of these two peptides (Fig. 3C). Our data indicate that the protective effect against A. tumefaciens by Drosocin expression requires two UAS-Drc copies. In addition, Drosocin coexpression with Attacin A (but not with Cecropin A or Drosomycin) provides enhanced protection against this bacterium.
P. aeruginosa, a human opportunistic pathogen, is highly pathogenic to Drosophila (27). Fig. 3D shows that injection of a few P. aeruginosa cells kills wild-type flies in less than 16 h. Surprisingly, imd; spz flies succumb with the same kinetics, suggesting that either this bacterium can exert its pathogenic effect before the induction of an immune response or the Drosophila immune response is inefficient against this species. Our analysis shows that the constitutive expression of AMPs does not confer any significant protection against P. aeruginosa, suggesting that this bacterium is more resistant to AMPs than the other three Gram-negative bacteria that we tested. Cecropin A, Attacin A, and Diptericin provide a weak protection to the infected flies (Fig. 3D Left).
Resistance to Fungal Infection.
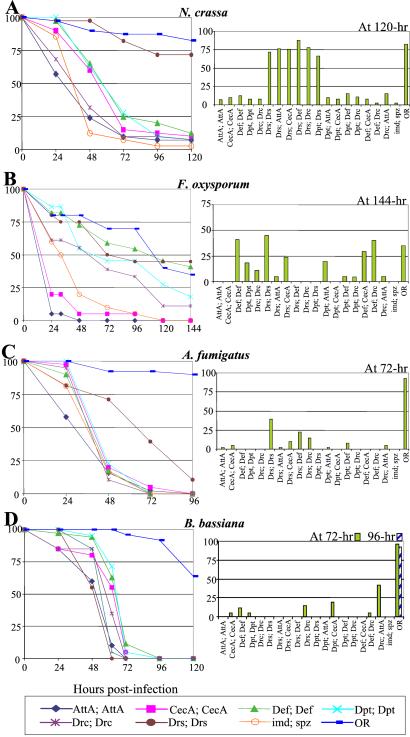
We next repeated our analysis by using three different fungal species: N. crassa, F. oxysporum, and A. fumigatus. Fig. 4 A–C show that Drosomycin was the most potent antifungal peptide (6). Drosomycin expression in imd; spz mutants restores a wild-type level of survival against both N. crassa and F. oxysporum and an increased resistance to A. fumigatus. The enhanced resistance to F. oxysporum and, to a lesser extent, A. fumigatus was observed in the lines carrying two UAS-Drs but not in the lines carrying one copy, suggesting that the concentration of this antifungal peptide must be above a certain threshold to be effective. In contrast to imd; spz mutants, N. crassa did not sporulate on imd; spz flies expressing Drosomycin that died after infection (Fig. 5), indicating that the high concentration of Drosomycin in the dead flies may also limit the sporulation and spread of this fungus. Furthermore, flies carrying one copy of UAS-Drs show resistance to F. oxysporum when coexpressed with Cecropin A (Fig. 4B Left), a peptide that has been shown to have antifungal activity (8). Flies expressing Defensin and, to a lesser extent, Diptericin and Drosocin all show increased resistance to F. oxysporum.
Figure 4.
Immune-compromised flies expressing Drosomycin show increased resistance to fungal infection. The survival rates of wild-type, imd; spz and imd; spz UAS-Pep flies after infection by four fungal species are shown. Sixty flies were either pricked by a needle dipped into a concentrated pellet of spores of (A) N. crassa (5.7 × 1011 spores/ml), (B) F. oxysporum (1.8 × 1011 spores/ml), or (C) A. fumigatus (4 × 1011 spores/ml), or (D) naturally infected by B. bassiana.
Figure 5.
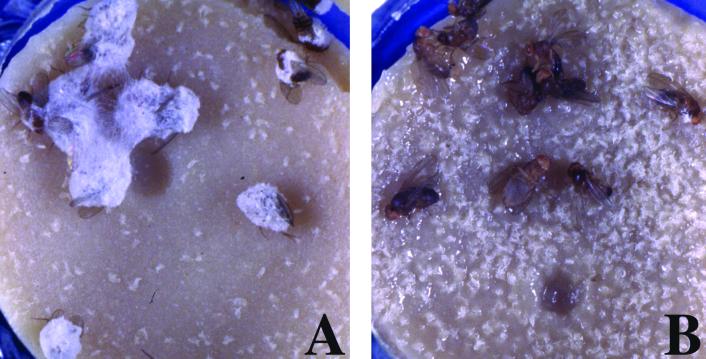
N. crassa does not sporulate on dead flies that expressed the Drosomycin gene. Injection of N. crassa spores induces fungal sporulation on dead imd; spz flies (A) but not on imd; spz flies that constitutively express Drosomycin (B). Visible germination was photographed 48 h after death of flies.
B. bassiana is a fungal entomopathogen that infects through the insect cuticle. Previous experiments have indicated the existence of a Toll-dependent host-defense reaction against this fungus, because Toll-deficient mutants survived B. bassiana infection at lower levels than wild-type flies (23). We have monitored the survival of the different UAS-Pep lines after deposition of spores on fly cuticle, in contrast to the preceding experiments in which microbes are injected by a needle. Fig. 4D shows that natural infection by this fungus kills all of the imd; spz double mutants in less than 72 h, whereas wild-type flies succumb at a slower rate (40% survived at 6 days after infection). Surprisingly, none of the UAS-Pep-expressing lines resisted B. bassiana infection better than imd; spz flies (Fig. 4D), suggesting that the Toll-mediated defense response to this fungus involves other AMP combinations or a yet uncharacterized defense reaction.
Discussion
Most of our knowledge of Drosophila AMP activity has come from studies performed in vitro. Although these studies have been successful in identifying the specificity and mode of action of some AMPs, they do not fully measure the in vivo role of each AMP in fighting infections. Here, we developed a genetic approach to address the functional relevance of a defined AMP in the host defense of Drosophila adults. We take advantage of the imd; spz double mutants that do not express any known endogenous AMP genes to generate immuno-compromised flies that express only one or two AMPs, even after infection. We show that constitutive expression of a single AMP gene such as Drosomycin or Defensin can confer a wild-type resistance to these otherwise immunodeficient flies against certain microorganisms. These results provide strong support for a critical role of AMPs in surviving infections in Drosophila, taking into account that the wild-type antimicrobial response involves the concomitant synthesis of about 15 inducible AMP genes.
In our assay, the AMP genes are expressed via the UAS/GAL4 system at a level similar to that observed in wild-type induction of the endogenous AMP genes (except Defensin and Diptericin). However, there are still some differences between our assay and the wild-type physiological condition. In the UAS-Pep flies, AMP genes are expressed ubiquitously and constitutively, contrasting to the wild-type flies in which peptides are made mainly by the fat body in an acute phase profile. The accumulation of AMP, therefore, through constitutive gene expression before infection may be critical to confer an effective protection. It is noteworthy that Defensin and Drosomycin, the two most potent AMPs shown by our analysis, contain three and four internal disulfide bonds, respectively, that may confer an increased resistance to proteases, resulting in more stable peptides.
Our study provides an alternative method for monitoring and comparing the antimicrobial activity of the various Drosophila AMPs. We show that Defensin is the most potent peptide against Gram-positive bacteria (Fig. 2), whereas Attacin A and Drosomycin are active against Gram-negative bacteria and fungi, respectively (Figs. 3 and 4). We observe that one copy of UAS-Def is sufficient to protect flies to wild-type level against M. luteus, B. subtilis, and S. aureus. The efficiency of Defensin may explain why the endogenous Defensin gene is transcribed to lower levels than the other AMP genes after infection (9). We also observe that one copy of UAS-Drs was sufficient to protect against N. crassa, whereas two copies are required to induce a complete and partial protection against F. oxysporum and A. fumigatus, respectively. These results are consistent with the Minimum Inhibitory Concentration assay of Drosomycin required in vitro to kill these three fungi: 0.3–0.6 μM for N. crassa, 1.2–2.5 μM for F. oxysporum, and 20–40 μM for A. fumigatus (28). In addition, we show, to our knowledge for the first time, that Diptericin in Drosophila contributes to resistance against some Gram-negative bacteria, although its activity is probably underestimated because of the low levels of Diptericin expression generated by our constructs. Surprisingly, we did not detect a clear protective effect of Cecropin A in our assay, whereas this peptide shows strong in vitro activity (8). We cannot exclude the possibility that in our lines, Cecropin A was not effectively produced or well processed to the active form. Alternatively, a higher level of Cecropin A expression may be required to generate a protective effect, considering that the Drosophila genome contains three other inducible Cecropin genes.
Our results also underline the differential activities of Drosophila AMPs, such is the case of Attacin A and Drosocin in resistance to some Gram-negative bacterial species (Fig. 3). Thus the existence of numerous AMPs may help widen the protection against a large number of microorganisms. In the case of Gram-negative bacterial infection, none of the peptides were able to restore a wild-type resistance in imd; spz double mutants. Our results and the observation that the Drosophila genome encodes a high number of AMP genes with activity directed against Gram-negative bacteria suggest that the elimination of this class of bacteria may require the global toxicity generated by multiple, rather than one or two, AMPs.
Our study does not reveal a striking synergistic activity among any pair of AMPs tested. In some cases, we observed a rather cooperative effect between two AMPs such as Attacin A when coexpressed with either Diptericin or Drosocin in resistance to some Gram-negative bacteria. These observations suggest that the multiple Drosophila AMPs may function in an additive way, rather than synergistically.
Host–pathogen interactions are antagonistic relationships in which the success of each organism depends on its ability to overcome the other. The production of AMPs is a common strategy to eliminate the invading microbes and, consequently, pathogens have evolved strategies to prevail over these defenses (1). Our assay provides a powerful tool to compare the resistance of various bacteria to different AMPs, because in our experiments, microbes were injected in an environment previously enriched in peptides. The time race between pathogen and the host defense is clearly illustrated by our observation that a preexisting level of Defensin is sufficient to ensure a complete resistance against B. subtilis, a Gram-positive bacterium highly pathogenic for flies (Fig. 2B). This observation indicates that this bacterial species is sensitive to Drosophila AMP but nevertheless can overtake the Drosophila immune response by its rapid growth. The observation that “immunizing” flies with nonpathogenic bacteria fully protects Drosophila from a subsequent infection by B. subtilis (data not shown) is consistent with this hypothesis. Our results also show that the kinetics of infection by P. aeruginosa or B. bassiana, two highly entomopathogenic microbes, were not delayed in flies expressing AMP genes, suggesting that these microbes have developed some mechanisms to escape the AMP activity. The observation that Drosomycin expression does not confer any protection against B. bassiana is unexpected, because Toll-mediated defense against this pathogen has been reported (23). This observation suggests that other antifungal peptides (e.g., Metchnikowin) or a yet uncharacterized defense reaction may be required to resist this fungus. Finally, the human pathogen, S. aureus, is also highly pathogenic to Drosophila and shows a better resistance to a high level of Defensin compared with other Gram-positive bacteria. These results underline the correlation between pathogenicity and increased resistance to AMPs.
Acknowledgments
We thank Dr. C. Parker for his interest and support in this study and the Protein/Peptide MicroAnalytical Laboratory facility at California Institute of Technology for help with mass spectrometry. We thank F. Boccard, M. Dushay, E. De Gregorio, and P. Bulet for helpful discussions and the members of our laboratories, especially R. Khush, for valuable comments on the manuscript. The technical assistance of Annie Meunier and Brigitte Maroni is gratefully acknowledged. P.T. was supported by the Fondation pour la Recherche Médicale (FRM). This project was funded by the Action Thématique et Incitative sur Programme et Equipe Centre National de la Recherche Scientifique, FRM, and Program Microbiologie (PRMMIP98).
Abbreviation
- AMP
antimicrobial peptide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hancock R E, Scott M G. Proc Natl Acad Sci USA. 2000;97:8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehrer R I, Ganz T. Curr Opin Immunol. 1999;11:23–27. doi: 10.1016/s0952-7915(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann J, Reichhart J. Trends Cell Biol. 1997;7:309–316. doi: 10.1016/S0962-8924(97)01087-8. [DOI] [PubMed] [Google Scholar]
- 4.Khush R S, Lemaitre B. Trends Genet. 2000;16:442–449. doi: 10.1016/s0168-9525(00)02095-3. [DOI] [PubMed] [Google Scholar]
- 5.Hedengren M, Borge K, Hultmark D. Biochem Biophys Res Commun. 2000;279:574–581. doi: 10.1006/bbrc.2000.3988. [DOI] [PubMed] [Google Scholar]
- 6.Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broeckaert W, Hetru C, Hoffmann J. J Biol Chem. 1994;269:33159–33163. [PubMed] [Google Scholar]
- 7.Levashina E, Ohresser S, Bulet P, Reichhart J, Hetru C, Hoffmann J. Eur J Biochem. 1995;233:694–700. doi: 10.1111/j.1432-1033.1995.694_2.x. [DOI] [PubMed] [Google Scholar]
- 8.Ekengren S, Hultmark D. Insect Biochem Mol Biol. 1999;29:965–972. doi: 10.1016/s0965-1748(99)00071-5. [DOI] [PubMed] [Google Scholar]
- 9.Dimarcq J, Hoffmann D, Meister M, Bulet P, Lanot R, Reichhart J, Hoffmann J. Eur J Biochem. 1994;221:201–209. doi: 10.1111/j.1432-1033.1994.tb18730.x. [DOI] [PubMed] [Google Scholar]
- 10.Charlet M, Lagueux M, Reichhart J, Hoffmann D, Braun A, Meister M. Eur J Biochem. 1996;241:699–706. doi: 10.1111/j.1432-1033.1996.00699.x. [DOI] [PubMed] [Google Scholar]
- 11.Wicker C, Reichhart J M, Hoffmann D, Hultmark D, Samakovlis C, Hoffmann J A. J Biol Chem. 1990;265:22493–22498. [PubMed] [Google Scholar]
- 12.Asling B, Dushay M, Hultmark D. Insect Biochem Mol Biol. 1995;25:511–518. doi: 10.1016/0965-1748(94)00091-c. [DOI] [PubMed] [Google Scholar]
- 13.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J, Hoffmann J. Proc Natl Acad Sci USA. 1995;92:9365–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemaitre B, Nicolas E, Michaut L, Reichhart J, Hoffmann J. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 15.Hedengren M, Asling B, Dushay M S, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 16.Qiu P, Pan P C, Govind S. Development (Cambridge, UK) 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- 17.Samakovlis C, Kimbrell D, Kylsten P, Engstrom A, Hultmark D. EMBO J. 1990;9:2969–2976. doi: 10.1002/j.1460-2075.1990.tb07489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morisato D, Anderson K V. Cell. 1994;76:677–688. doi: 10.1016/0092-8674(94)90507-x. [DOI] [PubMed] [Google Scholar]
- 19.Giebel B, Stuttem I, Hinz U, Campos-Ortega J A. Mech Dev. 1997;63:75–87. doi: 10.1016/s0925-4773(97)00029-4. [DOI] [PubMed] [Google Scholar]
- 20.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 21.Basset A, Khush R, Braun A, Gardan L, Boccard F, Hoffmann J, Lemaitre B. Proc Natl Acad Sci USA. 2000;97:3376–3381. doi: 10.1073/pnas.070357597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzou P, Meister M, Lemaitre B. In: Methods in Microbiology. Sansonetti P, Zychlinsky A, editors. Vol. 31. London: Academic; 2002. pp. 507–529. [Google Scholar]
- 23.Lemaitre B, Reichhart J, Hoffmann J. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann J A, Bulet P. Proc Natl Acad Sci USA. 1998;95:11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Vizioli J, Richman A M, Uttenweiler-Joseph S, Blass C, Bulet P. Insect Biochem Mol Biol. 2001;31:241–248. doi: 10.1016/s0965-1748(00)00143-0. [DOI] [PubMed] [Google Scholar]
- 27.Boman H G, Nilsson I, Rasmuson B. Nature (London) 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- 28.Lamberty M, Ades S, Uttenweiler-Joseph S, Brookhart G, Bushey D, Hoffmann J A, Bulet P. J Biol Chem. 1999;274:9320–9326. doi: 10.1074/jbc.274.14.9320. [DOI] [PubMed] [Google Scholar]