Abstract
Studies of SOCS-1-deficient mice have implicated Socs-1 in the suppression of JAK-STAT (Janus tyrosine kinase-signal transducers and activators of transcription) signaling and T cell development. It has been suggested that the levels of Socs-1 protein may be regulated through the proteasome pathway. Here we show that Socs-1 interacts with members of the Pim family of serine/threonine kinases in thymocytes. Coexpression of the Pim kinases with Socs-1 results in phosphorylation and stabilization of the Socs-1 protein. The protein levels of Socs-1 are significantly reduced in the Pim-1−/−, Pim-2−/− mice as compared with wild-type mice. Similar to Socs-1−/− mice, thymocytes from Pim-1−/−, Pim-2−/− mice showed prolonged Stat6 phosphorylation upon IL-4 stimulation. These data suggest that the Pim kinases may regulate cytokine-induced JAK-STAT signaling through modulation of Socs-1 protein levels.
One of the mechanisms by which many cytokines exert their effects is through activation of the JAK-STAT (Janus tyrosine kinase-signal transducers and activators of transcription) signaling pathway. In this pathway, cytokines initiate signaling by inducing the oligomerization of their receptor chains. This oligomerization in turn activates the associated JAKs. The activated JAKs then phosphorylate specific tyrosine residues within the cytoplasmic domains of their receptors. These phosphorylated tyrosine motifs can act as docking sites for signaling molecules such as the STAT family of Src homology 2 (SH2) domain-containing proteins. The JAK kinases then phosphorylate the recruited STAT proteins, which leads to their dimerization and subsequent translocation to the nucleus where they activate transcription of target genes (reviewed in ref. 1).
Several mechanisms have been identified that can control the intensity and duration of JAK-STAT activation. A new protein, Socs-1/JAB/SSI-1 (for simplicity, referred to as Socs-1 hereafter), was recently identified as a potent inhibitor of JAK activation (2–4). Sequence comparison revealed that Socs-1 belongs to a large family of proteins (reviewed in ref. 5). All SOCS family members share a conserved C-terminal SOCS box plus either an SH2 or other domain (e.g., WD40 repeats, ankyrin repeats, etc.) capable of mediating protein–protein interaction (6). The central SH2 domain of Socs-1 is required for binding to JAKs. A subdomain of approximately 24 aa immediately N-terminal to the SH2 domain is critical for maximum inhibition of JAK activity (7, 8). The levels of Socs-1 seem to be tightly controlled by several mechanisms. Transcription of SOCS-1 mRNA is rapidly induced by many cytokines. Previous work also suggests that Socs-1 protein stability is regulated. Two groups have reported stabilization of SOCS family proteins by inhibitors of the proteasome, suggesting that cells may regulate Socs-1 level through the proteasomal pathway (9, 10). Elongin BC complex, which has been implicated in ubiquitin-mediated degradation, binds to Socs-1 through the SOCS box (10, 11). Association of Elongin BC and the SOCS box has been suggested to alter the stability of the Socs-1 protein.
The Pim serine/threonine kinase family was first identified as a common proviral insertion site in T and B cell lymphomas in mice (12). Three family members have been identified: Pim-1, Pim-2, and Pim-3. Transcription of the Pim kinases is induced by T cell antigen receptor cross-linking and by cytokines such as IL-4, IL-6, and IFN-γ (13–16). Pim-1 and Pim-2 are highly expressed in cells of hematopoietic origin, whereas Pim-3 is undetectable in activated thymocytes and splenocytes (unpublished observation). Forced expression of Pim-1 reconstitutes thymic cellularity in mice lacking IL-7 or the common γ-chain of cytokine receptors (17). These data suggest that the Pim kinases may play an important role in signaling downstream of cytokine receptors. However, despite extensive investigation, the physiological substrates of the Pim kinases remain unknown. Here we report Socs-1 as a potential target of the Pim kinases. We have identified Pim-2 as a binding partner of Socs-1 and have shown that all three members of the Pim family of kinases can bind and phosphorylate Socs-1. Phosphorylation of Socs-1 by the Pim kinases prolongs the half-life of the Socs-1 protein and potentiates the inhibitory effect of Socs-1 on JAK-STAT activation.
Materials and Methods
Yeast Two-Hybrid Screen.
Full-length murine SOCS-1 was subcloned into the pAS2 vector (CLONTECH) and transformed into the yeast strain Y190. A murine macrophage cDNA library on the pGADNOT was a gift from S. Goff (Columbia University). Yeast transformation was performed by using the EZ Yeast Transformation kit from Zymo Research (Orange, CA), and yeast two-hybrid screen was conducted essentially as described by Durfee et al. (18).
Generation of Socs-1 Antibodies.
Recombinant glutathione S-transferase (GST)-Socs-1 fusion protein (1.5 mg) was expressed and purified from bacteria and injected into a rabbit for production of α-Socs-1 serum. The third bleed that had the highest titer was incubated overnight with glutathione beads (Sigma) cross-linked to GST to eliminate antibodies specific for GST. The precleared serum was then loaded at a flow rate of 10 ml/h onto an affinity column packed with glutathione beads cross-linked to GST-Socs-1 fusion protein. The column was washed at a rate of 1 ml/min with 10 times the column volume of 1× TBS (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl) and then with 10 times the column volume of 0.1× TBS. After last wash, Socs-1-specific antibodies were eluted with one column volume of 0.1 M glycine⋅HCl, pH 2.5, into tubes preloaded with an equal volume of 1 M Tris base.
Biochemical Experiments.
Murine SOCS-1 was subcloned into a mammalian expression vector pcDNA3.1 HISA (Invitrogen) with an in-frame Xpress tag. Murine Pim-2 was subcloned into another mammalian expression vector pCGN (gift from S. Goff) with an in-frame hemagglutinin (HA) tag. Anti-Xpress antibodies were from Invitrogen. Rabbit polyclonal anti-HA antibodies and normal rabbit serum were from Santa Cruz Biotechnology. Coimmunoprecipitation experiments were performed as described (19). LLnL (N-acetyl-leu-leu-norleucinal) and cycloheximide were purchased from Sigma. λ-Phosphatase was from New England Biolabs.
To detect endogenous Socs-1 protein, thymi were removed from 10 wild-type BALB/c mice (4 weeks old). Total thymocytes were cultured in the presence of 50 ng/ml phorbol myristate acetate (PMA, Sigma) and 500 ng/ml ionomycin (Sigma) for 4 h. Cells were lysed in lysis buffer containing 1% Nonidet P-40, 50 mM Tris, pH 8.0, 2 mM EDTA, 5 μg/ml each of protease inhibitors leupeptin, aprotinin, pepstatin, 1 mM PMSF plus phosphatase inhibitors, 1 mM sodium orthovanadate, and 50 mM NaF. Cell lysates were subjected to immunoprecipitation by using normal rabbit serum or an affinity-purified rabbit anti-Socs1 antibodies. The immunoprecipitates were loaded to an SDS-12% polyacrylamide gel and immunoblotted with a goat anti-Socs1 antibody C20 (Santa Cruz Biotechnology).
GST Pull-Down Experiments.
Pim-2 was subcloned into the pGEX vector (Amersham Pharmacia). GST fusion proteins were expressed in the bacteria DH5α. The conditions for GST expression GST pull-down experiment were as described (20). Socs-1 ΔN, Socs-1 ΔC, and Socs-1 ΔSH2 mutants were generated by PCR followed by subcloning into the pcDNA3.1 vector.
In Vitro Kinase Assay.
Various SOCS-1 mutants were subcloned into the pGEX vector (Amersham Pharmacia). Expression of GST fusion proteins was as described (20), except for the GST-Socs-1 fusion proteins, which were induced at 30°C. GST fusion proteins of Pim-2 and Socs-1 were incubated together with [γ-32P]ATP essentially as described (21). The reactions were washed three times and fractionated on SDS/PAGE. The gel was subjected to drying and autoradiography. A small aliquot of each reaction was analyzed by Western blot with an anti-GST antibody.
Transient Luciferase Assay.
Luciferase assay was performed as described (22) with some modifications: 293T cells were transfected by calcium phosphate precipitation method. Twenty-four hours after transfection, cells of each transfection were split into half. One half was treated with human IL-4, the other half served as control. Cells were harvested 18 h after treatment. Half of the cells were processed to determine luciferase activity, whereas the other half were subject to Western blot to compare the expression level of Pim-2 protein. NIH 3T3 cells were transfected by using the Lipofectamine reagent from Life Technologies (Rockville, MD) according to the manufacturer's manual.
Results
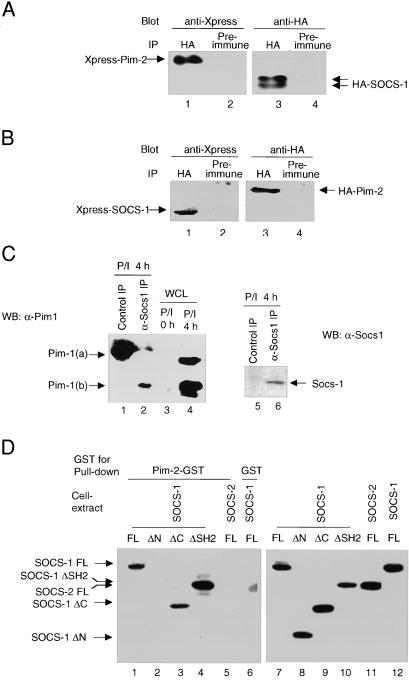
To identify proteins that interact with Socs-1, a yeast two-hybrid screen was conducted with full-length murine SOCS-1 as bait. A murine cDNA library from macrophages was screened as described by Durfee et al. (18). One of the genes identified encodes the serine/threonine kinase Pim-2. To confirm the interaction between Socs-1 and Pim-2 in mammalian cells, plasmids encoding epitope-tagged Pim-2 and SOCS-1 were transfected into 293T cells. Cell lysates were subjected to coimmunoprecipitation analysis. Pim-2 was detected in Socs-1 immunoprecipitates by immunoblot analysis (Fig. 1A). Conversely, Socs-1 was detected in the Pim-2 immunoprecipitates (Fig. 1B). In addition to Pim-2, Socs-1 interacted with Pim-1 and Pim-3 in coimmunoprecipitation experiments (data not shown). To determine whether Socs-1 can bind to Pim kinases under physiologic conditions, thymocytes were isolated from wild-type mice and stimulated with PMA and ionomycin for 4 h before being subjected to lysis and immunoprecipitation (12). As expected, two isoforms of Pim-1 [Pim-1(a) and Pim-1(b)] were induced by PMA and ionomycin treatment (Fig. 1C, lanes 3 and 4). The smaller form Pim-1(b) was associated with endogenous Socs-1 more strongly than the larger form Pim-1(a) (Fig. 1C, lane 2), because coimmunoprecipitation of Pim-1(a) was only detectable when the same blot was exposed much longer (data not shown). Thus, Socs-1 protein interacts with the Pim family of kinases in vivo.
Figure 1.
Association of Pim-2 with Socs-1 in vivo and in vitro. (A) Immunoprecipitates of Socs-1 contain Pim-2. The 293T cells were cotransfected with plasmids carrying Xpress-tagged Pim-2 and HA-tagged SOCS-1. Lysates of transfectants were immunoprecipitated with a polyclonal anti-HA Ab (lanes 1 and 3) or normal rabbit serum as a negative control (lanes 2 and 4). Both immunoprecipitates were split, loaded onto SDS gel, and immunoblotted with a monoclonal anti-Xpress (lanes 1 and 2) or anti-HA (lanes 3 and 4) Abs, respectively. (B) Socs-1 is present in Pim-2 immunoprecipitates. Same as A, except that plasmids carrying Xpress-tagged SOCS-1 and HA-tagged Pim-2 were used for transfection. (C) Endogenous association of Socs-1 and Pim-1. Total thymocytes were isolated from wild-type BALB/c mice, stimulated with PMA and ionomycin for 4 h, and lysed. Lysates were immunoprecipitated by using either preimmune serum (lanes 1 and 5) or an affinity-purified rabbit α-Socs-1 antibody (lanes 2 and 6). The immunoprecipitates were analyzed by immunoblotting first with a goat α-Socs-1 antibody (C20) (Right) and then with a monoclonal α-Pim-1 antibody (Left). As a control, 50 μg of lysates from unstimulated and stimulated thymocytes was loaded in lanes 3 and 4. (D) The N terminus of Socs-1 is required for binding to Pim-2. Constructs of Xpress-tagged SOCS-1, full-length (FL) (lanes 1 and 6), ΔN (deleting amino acids 1–79) (lane 2), ΔC (deleting amino acids 167–212) (lane 3), or ΔSH2 (deleting amino acids 80–166) (lane 4) were transiently expressed in 293T cells. Xpress-tagged SOCS-2 was used as a control (lane 5). Whole-cell lysates of the transfected 293T cells were incubated with bacterial-expressed Pim-2-GST fusion protein (lanes 1–5) or GST alone (lane 6) bound to glutathione agarose beads (see Materials and Methods). Proteins bound to the beads were analyzed by immunoblot with anti-Xpress Ab. To ensure equal input, small aliquots of whole-cell lysates were subjected to Western blot analysis with anti-Xpress Ab (lanes 7–12).
To map the domain of Socs-1 that mediates its interaction with Pim-2, lysates of 293T cells transfected with various truncation mutants of Socs-1 were incubated with Pim-2-GST fusion protein immobilized on agarose beads. Socs-1 lacking either the SH2 domain (ΔSH2) or the C-terminal SOCS box (ΔC) could bind to Pim-2 and full-length Socs-1. In contrast, truncation of the N-terminal 79 aa of Socs-1 (ΔN) abolished its association with Pim-2 (Fig. 1D). As a control, Socs-2, another member of the SOCS family, did not bind to Pim-2. Taken together, these results indicate that Pim-2 specifically binds to Socs-1, and that this interaction is mediated by the N terminus of Socs-1.
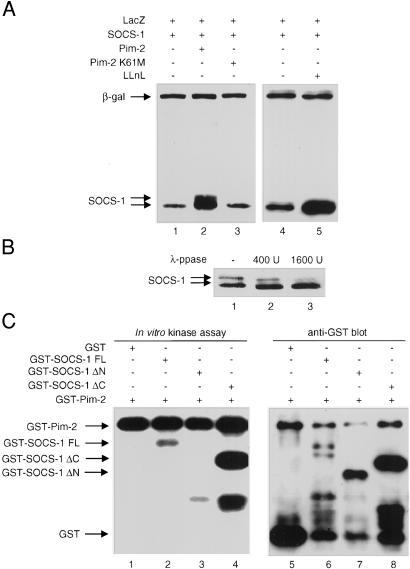
When Socs-1 and Pim-2 were coexpressed in 293T cells, a slower-migrating Socs-1 isoform was observed (Fig. 2A). Coexpression of Socs-1 with kinase-inactive Pim-2 did not result in the slower-migrating Socs-1 band, even though the expression levels of wild-type and mutant Pim-2 were similar (data not shown). Pim-1 or Pim-3 also caused a mobility shift of Socs-1 when they were coexpressed in 293T cells (unpublished results). In contrast, no mobility shift was observed when the other SOCS protein was coexpressed with the Pim kinases (data not shown), implying that Socs-1 may be a specific substrate for Pim kinases. To determine whether the slower-migrating band is a phosphorylated form of Socs-1, total cell lysates of 293T cells expressing Socs-1 and Pim-2 were incubated with λ-phosphatase. Upon phosphatase treatment, the intensity of the slower-migrating Socs-1 band decreased, whereas the levels of the faster-migrating band increased (Fig. 2B). Thus, Pim kinases are capable of phosphorylating Socs-1 in vivo, and this modification can be reversed by phosphatase treatment.
Figure 2.
Phosphorylation of Socs-1 by Pim-2. (A) Coexpression of Socs-1 and Pim-2 results in mobility shift of Socs-1. Plasmids expressing SOCS-1 tagged with Xpress were transfected alone (lanes 1, 4, and 5) or together with Pim-2 (lane 2) or kinase-inactive Pim-2 (K61M) (lane 3) into 293T cells. An equal amount of a plasmid with the LacZ gene was included in each transfection as an internal control. Whole-cell lysates were then analyzed by immunoblot with anti-Xpress Ab. In lanes 4 and 5, DMSO control (lane 4) or 10 μM LLnL (lane 5) was added to the cells 24 h after transfection, and cells were harvested after another 24 h incubation. The expression levels of wild-type and mutant Pim-2 were comparable (data not shown). (B) The mobility shift of Socs-1 can be reversed by phosphatase. Lysate of 293T cells transfected with SOCS-1 and Pim-2 was incubated with an increasing amount of λ-phosphatase at 30°C for 90 min and analyzed by immunoblot. (C) N-terminal truncation of Socs-1 abolishes phosphorylation of Socs-1 by Pim-2. Pim-2 and various constructs of Socs-1 were expressed in bacteria as GST fusion proteins. The GST-Pim-2 fusion protein was incubated with full-length (lane 2), N-terminal truncated (lane 3), or C-terminal truncated (lane 4) Socs-1 for in vitro kinase assay (see Materials and Methods). GST alone (lane 1) was used as a negative control. A small aliquot of each sample from the kinase assay was analyzed by Western blot with anti-GST Ab to ensure that the amounts of proteins loaded in each lane were comparable (lanes 5–8)
Because the slower-migrating Socs-1 species is likely the result of phosphorylation by serine/threonine kinases, we sought to determine whether Socs-1 is a direct substrate of the Pim-2 kinase. An in vitro kinase assay was performed by using GST-Pim-2 and GST-Socs-1 fusion proteins. Although Pim-2 did not phosphorylate the GST protein, it phosphorylated the full-length Socs-1-GST fusion protein in vitro. The C-terminal SOCS box is dispensable for phosphorylation as a Socs-1 mutant lacking the SOCS box (ΔC) was still phosphorylated (Fig. 2C). Deletion of the N-terminal 79 aa of Socs-1 (ΔN) abolished its phosphorylation by Pim-2 completely (Fig. 2C, lane 3). Although the N terminus of Socs-1 is required for its interaction with Pim-2, the GST moiety, through GST dimerization, was presumably able to mediate an interaction between Pim-2 and N-terminal truncated Socs-1. Therefore, abrogation of phosphorylation is likely not caused by a lack of interaction between Socs-1 and Pim-2, rather, it is most likely a result of the loss of phosphorylation sites at the N terminus of Socs-1. Consistent with the in vitro kinase assay, Pim-2 failed to cause a mobility shift of the Socs-1 ΔN mutant when they were coexpressed in 293T cells (unpublished observation). These results suggest that Socs-1 is a direct substrate for Pim-2 kinase and that, at least in vitro, the major phosphorylation sites are within the N-terminal 79 aa of Socs-1.
Coexpression of Pim-2 and Socs-1 in 293T cells drastically increased the steady-state levels of Socs-1, an effect that resembles the stabilization of Socs-1 protein by the proteasomal inhibitor LLnL (Fig. 2A, lanes 1, 2, 4, and 5). The kinase activity of Pim-2 was required for this stabilization effect, because a kinase-inactive mutant of Pim-2 failed to increase the protein levels of Socs-1 (Fig. 2A). As a control for transfection efficiency, a plasmid carrying the LacZ gene was included in each transfection. The levels of β-galactosidase encoded by the LacZ gene did not change, but the protein levels of Socs-1 increased in the presence of Pim-2 or LLnL.
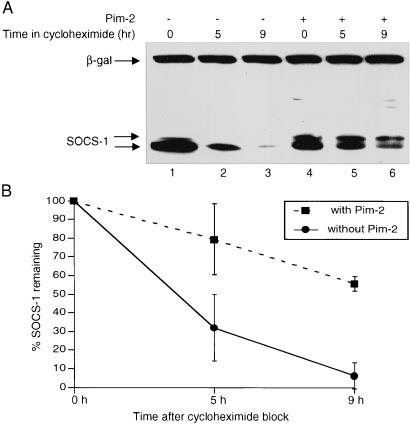
The increase in Socs-1 protein levels in the presence of the Pim kinases can be attributed to augmented production or decreased degradation of the Socs-1 protein. To distinguish between these two possibilities, the levels of Socs-1 were monitored after addition of cycloheximide to block protein synthesis. SOCS-1 was transiently transfected into 293T cells in the absence or presence of Pim-2. Thirty-six hours after transfection, cycloheximide was added to the culture to block further protein synthesis, and the decay of the Socs-1 protein was measured by immunoblotting. In the absence of Pim-2, less than 10% of the Socs-1 protein remained after 9 h in cycloheximide. In contrast, when Pim-2 was present, more than 60% of Socs-1 remained after the same period (Fig. 3 A and B). As a control for transfection efficiency and protein loading, the levels of β-galactosidase encoded by a cotransfected plasmid remained steady. The slower-migrating band of Socs-1 persisted longer than the band with faster mobility (Fig. 3A, lanes 4–6), indicating that phosphorylation renders Socs-1 more stable. Consistent with the cycloheximide experiment, the half-life of Socs-1 protein was prolonged by coexpression of the Pim kinases in a pulse-chase experiment (data not shown). Moreover, the stabilization of Socs-1 by Pim kinases seems to be specific, because the decay of Socs-3 was not significantly affected by coexpression of the Pim kinases under the same conditions (data not shown). Together, these data suggest that phosphorylation by the Pim kinases down-regulates degradation of Socs-1 protein, thus augmenting the levels of Socs-1 protein in the cells.
Figure 3.
Phosphorylation by Pim kinases protects Socs-1 from degradation. (A) Phosphorylated form of Socs-1 decays more slowly than unphosphorylated form. The 293T cells were transfected with Xpress-tagged SOCS-1 alone (lanes 1–3) or together with Pim-2 (lanes 4–6). A plasmid carrying the LacZ gene was used as a control. Cycloheximide (100 μg/ml) was added to the media 36 h after transfection to block new protein synthesis. Cells were harvested at 0-, 4.5-, and 9-h time points, and total lysates were analyzed by immunoblot with anti-Xpress antibody. (B) The blot in A was scanned and quantitated by using the software nih image 1.6.2. The results from three independent experiments are plotted such that the protein level at 0-h time point is 100%.
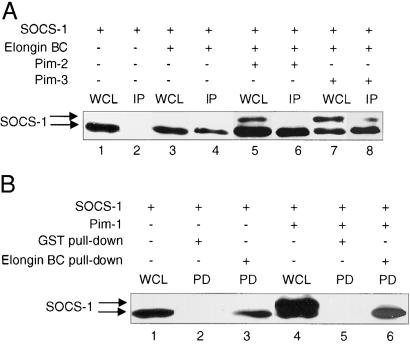
The stability of the Socs-1 protein has been reported to be altered by its association with Elongin BC (10, 11). We thus sought to determine whether the Pim kinases have any effect on the association of Socs-1 and Elongin BC. Socs-1 was coexpressed in 293T cells with Elongin BC in the presence or absence of Pim-2 or Pim-3. The formation of the Socs-1 doublet was not altered by coexpression of Elongin BC (Fig. 4A, lanes 1, 3, 5, and 7). When the cell lysates were subjected to immunoprecipitation using α-Elongin C antibodies, the fast-migrating band of Socs-1 preferentially coimmunoprecipitated with Elongin BC (Fig. 4A, lanes 6 and 8). To confirm this result, a GST pull-down experiment was also conducted. When bacterial-expressed GST-Elongin C fusion protein was incubated with lysates of 293T cells transfected with SOCS-1 alone or together with the Pim kinases, GST-Elongin C associated specifically with the fast-migrating band of Socs-1 (Fig. 4B, lanes 4 and 6). Because the slow-migrating band likely represents a phosphorylated form of Socs-1, these results suggest that phosphorylation of Socs-1 by the Pim kinases decreases the interaction between Socs-1 and Elongin BC.
Figure 4.
Phosphorylation of Socs-1 decreases its binding to Elongin BC. (A) Phosphorylated Socs-1 does not bind Elongin BC as well as unphosphorylated Socs-1 in coimmunoprecipitation experiments. The 293T cells were transfected with plasmids carrying Xpress-tagged SOCS-1 alone (lanes 1 and 2) or together with plasmids expressing HA-tagged Elongin B and Elongin C (lanes 3–8), in the absence (lanes 3 and 4) or presence of Pim-2 (lanes 5 and 6) or Pim-3 (lanes 7 and 8). Total cell lysates were immunoprecipitated with anti-HA antibody to pull down Elongin BC. The immunoprecipitates (IP) were then analyzed by Western blot by using anti-Xpress antibody to detect proteins that bind to Elongin BC (lanes 2, 4, 6, and 8). For comparison, whole-cell lysates (WCL) (lanes 1, 3, 5, and 7) were analyzed on the same blot. (B) GST-Elongin C fusion protein associates preferentially with the faster-migrating band of Socs-1. The 293T cells were transfected with plasmids carrying Xpress-tagged SOCS-1 either in the absence (lanes 1–3) or presence (lanes 4–6) of Pim-1. Total cell lysates from the transfectants were incubated with bacterial expressed GST (lanes 2 and 5) or GST-Elongin C (lanes 3 and 6) immobilized on glutathione beads. The beads were washed four times before being subjected to SDS/PAGE analysis (lanes 3 and 4). Lanes 1 and 4 were small aliquots of whole-cell lysates (WCL) before incubation with glutathione beads.
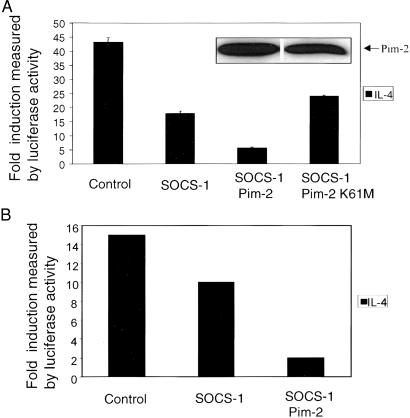
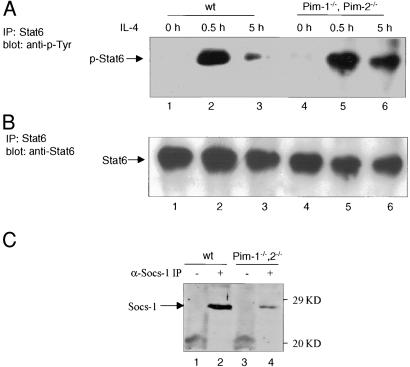
To determine the functional significance of the interaction between the Pim kinases and Socs-1, the effect of Pim-2 on IL-4-mediated Stat6 activation was evaluated by transient luciferase assays. We have reported that Socs-1 can inhibit IL-4-induced Stat6 activation in 293T cells (23). Cotransfection of Pim-2 with SOCS-1 further inhibited Stat6-mediated reporter expression, whereas kinase inactive Pim-2 had no effect (Fig. 5A). The same effect by Pim-2 was observed in NIH 3T3 cells (Fig. 5B). To elucidate the biochemical mechanisms by which Pim kinases affect cytokine signaling, primary thymocytes from Pim-1−/−, Pim-2−/− mice, or wild-type littermates were treated with IL-4, and the status of Stat6 tyrosine phosphorylation was assessed. Five hours after IL-4 stimulation, tyrosine phosphorylation of Stat6 was almost completely abrogated in wild-type cells. In contrast, significant amounts of Stat6 remained phosphorylated in the Pim-1−/−, Pim-2−/− cells after the same period (Fig. 6 A and B). Thymocytes from SOCS-1−/− mice also exhibit prolonged activation of Stat6 after IL-4 treatment (24). This defect in down-regulating JAK-STAT signaling may be caused by the lack of adequate amounts of Socs-1 when both Pim-1 and Pim-2 are absent. We thus examined the levels of Socs-1 protein in these mice. Thymocytes from either wild-type or Pim-1−/−, Pim-2−/− mice were treated with PMA and ionomycin for 4 h. Cells were lysed and subjected to α-Socs-1 immunoprecipitation and immunoblot analysis. The levels of Socs-1 protein were significantly higher in wild type than in the mutant mice (Fig. 6C). As a control, α-Lck immunoprecipitation was performed with the same lysates, and no significant differences of Lck levels were observed (data not shown). Taken together, these findings suggest that the Pim kinases help maintain the levels of Socs-1 protein and thus potentiate Socs-1 inhibition of JAK-STAT activation.
Figure 5.
Pim-2 enhances Socs-1 inhibition of IL-4-induced Stat6 activation. (A) Pim-2 potentiates Socs-1 inhibition of JAK-STAT activation in 293T cells. The 293T cells were transfected with 2 μg of p(Iɛ-IL4RE)4-Luc reporter, 1 μg of pSV40-LacZ, and 0.6 μg of human Stat6 expression vector by calcium phosphate precipitation. Plasmid DNA (0.005 μg) carrying SOCS-1 and 2 μg of plasmid DNA carrying either wild-type Pim-2 or kinase-inactive Pim-2 were used. The total amount of transfected DNA was kept constant by addition of vector DNA as described (see Materials and Methods). Shown on the y axis is the ratio of luciferase activity between treated and untreated cells. Values reflect means of three independent experiments. (Inset) The expression levels of wild-type Pim-2 and kinase-inactive Pim2, respectively. (B) Pim-2 potentiates Socs-1 inhibition of JAK-STAT activation in NIH 3T3 cells. NIH 3T3 cells were transfected with 2 μg of p(Iɛ-IL4RE)4-Luc reporter, 1 μg of pSV40-LacZ as described in Materials and Methods. Used were 0.02 μg of plasmid DNA carrying SOCS-1 and 10 μg of Pim-2 plasmid DNA. Luciferase assay was performed essentially as described in A.
Figure 6.
IL-4 induced Stat6 activation is prolonged in thymocytes from Pim-1−/−, Pim-2−/− mice. (A) Thymocytes from Pim-1−/−, Pim-2−/− mice and wild-type littermates were stimulated with α-CD3 (PharMingen) (1 μg/ml) plus IL-4 (10 ng/ml) and harvested at the indicated time points. Cells were then washed and lysed in 1× Nonidet P-40 lysis buffer containing inhibitors of proteases and phosphatases as described (28). Immunoprecipitates were obtained by using anti-Stat6 antibody M20 (Santa Cruz Biotechnology) and blotted with anti-phosphor-tyrosine antibody 4G10 (Upstate Biotechnology). (B) The same blots as in A were stripped and reblotted with anti-Stat6 antibody M20. (C) The levels of endogenous Socs-1 were reduced in the Pim-1−/−, Pim-2−/− mice. Thymocytes were isolated from wild-type (lanes 1 and 2) or Pim-1−/−, Pim-2−/− (lanes 3 and 4) mice, and cultured in the presence of PMA and ionomycin for 4 h. Cells were harvested and lysed, and the protein concentration was determined. Equal amounts of lysates were subjected to immunoprecipitation analysis by using preimmune serum (lanes 1 and 3) or an α-Socs-1 antibody (lanes 2 and 4) as described in Fig. 1C.
Discussion
Socs-1 was first identified as an autofeedback inhibitor of JAKs. The current model is that cytokine stimulation activates JAK-STAT signaling, which in turn triggers the transcription of SOCS-1. The resultant Socs-1 protein translocates to the cytokine receptors and suppresses the kinase activity of JAK. As a potent inhibitor of JAKs, the levels of Socs-1 must be tightly regulated. Here we show that Socs-1 is a labile protein and that its degradation is regulated by Pim-mediated serine/threonine phosphorylation.
In thymocytes, the steady-state levels of Socs-1 are increased by LLnL (unpublished observation), suggesting that the levels of Socs-1 protein may be regulated through the proteasome pathway. Although the exact mechanism by which the Socs-1 protein is degraded is not well understood, studies on the interaction between Socs-1 and Elongin BC have suggested a role for Elongin BC in the degradation of Socs-1 (10, 11). However, the precise role of Elongin BC remains controversial. Although Kamura et al. (11) have provided evidence that Elongin BC complex helps stabilize Socs-1, Zhang et al. (10) have suggested that Elongin BC complex targets Socs-1 to the proteasomal degradation pathway. Our data demonstrate that phosphorylation of Socs-1 by the Pim kinases decreases the binding between Socs-1 and Elongin BC and down-regulates Socs-1 degradation. Moreover, in preliminary experiments, a Socs-1 mutant (L175P, C179F) that fails to bind to Elongin BC is more stable than wild-type Socs-1 (unpublished observation), suggesting that the Elongin BC complex negatively affects the stability of Socs-1 protein.
Socs-1 mRNA is expressed at the highest level in the thymus, and SOCS-1-deficient mice manifest defects in cytokine signaling and T cell development (24–26). Both Pim-1 and Pim-2 have been implicated in thymocyte development (17, 27). Forced expression of Pim-1 rescues the defects in thymocyte development caused by deficiency in RAG, IL-7, or the common γ-chain of cytokine receptors (17). Moreover, although Pim kinases are up-regulated by cross-linking of T cell receptors, Socs-1 transcription does not seem to be induced by T cell antigen receptor signaling (26). However, when thymocytes were activated with PMA and ionomycin or anti-CD3, which mimic cross-linking of T cell antigen receptor, the protein levels of both Socs-1 and Pim-1 were augmented (unpublished observation). Northern blot analysis indicated that the levels of Socs-1 mRNA were unchanged, suggesting that the levels of Socs-1 may be posttranscriptionally regulated. The hypothesis that Pim kinases stabilize Socs-1 posttranscriptionally was further strengthened by the observation that the protein levels of Socs-1 are much lower in the Pim-1−/−, Pim-2−/− mice than in wild-type mice.
Although we consistently observe a mobility shift of the Socs-1 protein when it is coexpressed with the Pim kinases, endogenous Socs-1 is not detected as a doublet even when Pim kinases are abundant. It is possible that the single band of endogenous Socs-1 is phosphorylated and mobility-shifted, because it migrates slower than would be predicted from the calculated molecular weight. The unphosphorylated form of endogenous Socs-1 is not detectable because it may be rapidly degraded under physiologic conditions. Alternatively, endogenous Socs-1 may be phosphorylated by Pim kinases to an extent that does not result in such a dramatic mobility shift as observed in the overexpression system.
The Pim kinases and Socs-1 all are induced by a variety of cytokines, and the interaction between Pim and Socs-1 may represent a mechanism by which cytokines cross-regulate one another. For example, expressions of Pim-1, Pim-2, and Socs-1 all are induced by IFN-γ (4, 16), and the interplay between Pim kinases and Socs-1 may be important for IFN-γ-induced inhibition of IL-4 signaling (28–31). In summary, the interaction between Socs-1 and the Pim kinases may be important in regulating various signaling pathways in cells of both hematopoietic and nonhematopoietic origin.
Acknowledgments
We thank C. Schindler, M. Harris, and A. Banerjee for comments on the manuscript; B. Lu and S. Goff for sharing of reagents, mice, and protocols; T. A. Wilson and N. A. Nicola for plasmids encoding Elongin B and C; H. Niu, G. Wang, and J. Kitajewski for reagents and helpful discussion; and members of the Rothman laboratory for help and support. P.R. is a Scholar of the Leukemia Society of America and a Cancer Research Institute Investigator. This work was also supported by National Institutes of Health Grant RO1 AI33541.
Abbreviations
- JAK
Janus tyrosine kinase
- STAT
signal transducers and activators of transcription
- SH2
Src homology 2
- GST
glutathione S-transferase
- HA
hemagglutinin
- PMA
phorbol myristate acetate
- LLnl
N-acetyl-leu-leu-norleucinal
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Leonard W J, O'Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 2.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 3.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 4.Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 5.Chen X P, Losman J A, Rothman P. Immunity. 2000;13:287–290. doi: 10.1016/s1074-7613(00)00028-5. [DOI] [PubMed] [Google Scholar]
- 6.Hilton D J, Richardson R T, Alexander W S, Viney E M, Willson T A, Sprigg N S, Starr R, Nicholson S E, Metcalf D, Nicola N A. Proc Natl Acad Sci USA. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson S E, Willson T A, Farley A, Starr R, Zhang J G, Baca M, Alexander W S, Metcalf D, Hilton D J, Nicola N A. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle J N, Yoshimura A. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narazaki M, Fujimoto M, Matsumoto T, Morita Y, Saito H, Kajita T, Yoshizaki K, Naka T, Kishimoto T. Proc Natl Acad Sci USA. 1998;95:13130–13134. doi: 10.1073/pnas.95.22.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J G, Farley A, Nicholson S E, Willson T A, Zugaro L M, Simpson R J, Moritz R L, Cary D, Richardson R, Hausmann G, et al. Proc Natl Acad Sci USA. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamura T, Sato S, Haque D, Liu L, Kaelin W G, Jr, Conaway R C, Conaway J W. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuypers H T, Selten G, Quint W, Zijlstra M, Maandag E R, Boelens W, van Wezenbeek P, Melief C, Berns A. Cell. 1984;37:141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 13.Lilly M, Le T, Holland P, Hendrickson S L. Oncogene. 1992;7:727–732. [PubMed] [Google Scholar]
- 14.Dautry F, Weil D, Yu J, Dautry-Varsat A. J Biol Chem. 1988;263:17615–17620. [PubMed] [Google Scholar]
- 15.Domen J, van der Lugt N M, Laird P W, Saris C J, Clarke A R, Hooper M L, Berns A. Blood. 1993;82:1445–1452. [PubMed] [Google Scholar]
- 16.Yip-Schneider M T, Horie M, Broxmeyer H E. Blood. 1995;85:3494–3502. [PubMed] [Google Scholar]
- 17.Jacobs H, Krimpenfort P, Haks M, Allen J, Blom B, Demolliere C, Kruisbeek A, Spits H, Berns A. J Exp Med. 1999;190:1059–1068. doi: 10.1084/jem.190.8.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 19.Danial N N, Pernis A, Rothman P B. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 20.Danial N N, Losman J A, Lu T, Yip N, Krishnan K, Krolewski J, Goff S P, Wang J Y, Rothman P B. Mol Cell Biol. 1998;18:6795–6804. doi: 10.1128/mcb.18.11.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Lugt N M, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, Berns A. EMBO J. 1995;14:2536–2544. doi: 10.1002/j.1460-2075.1995.tb07251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu B, Reichel M, Fisher D A, Smith J F, Rothman P. J Immunol. 1997;159:1255–1264. [PubMed] [Google Scholar]
- 23.Losman J A, Chen X P, Hilton D, Rothman P. J Immunol. 1999;162:3770–3774. [PMC free article] [PubMed] [Google Scholar]
- 24.Naka T, Matsumoto T, Narazaki M, Fujimoto M, Morita Y, Ohsawa Y, Saito H, Nagasawa T, Uchiyama Y, Kishimoto T. Proc Natl Acad Sci USA. 1998;95:15577–15582. doi: 10.1073/pnas.95.26.15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander W S, Starr R, Fenner J E, Scott C L, Handman E, Sprigg N S, Corbin J E, Cornish A L, Darwiche R, Owczarek C M, et al. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 26.Marine J C, Topham D J, McKay C, Wang D, Parganas E, Stravopodis D, Yoshimura A, Ihle J N. Cell. 1999;98:609–616. doi: 10.1016/s0092-8674(00)80048-3. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt T, Karsunky H, Rodel B, Zevnik B, Elsasser H P, Moroy T. EMBO J. 1998;17:5349–5359. doi: 10.1093/emboj/17.18.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.te Velde A A, Rousset F, Peronne C, De Vries J E, Figdor C G. J Immunol. 1990;144:3052–3059. [PubMed] [Google Scholar]
- 29.Lee C E, Yoon S R, Pyun K H. Mol Immunol. 1993;30:301–307. doi: 10.1016/0161-5890(93)90058-j. [DOI] [PubMed] [Google Scholar]
- 30.Venkataraman C, Leung S, Salvekar A, Mano H, Schindler U. J Immunol. 1999;162:4053–4061. [PubMed] [Google Scholar]
- 31.Dickensheets H L, Venkataraman C, Schindler U, Donnelly R P. Proc Natl Acad Sci USA. 1999;96:10800–10805. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]