Abstract
Macrophage foam cells are integral in the development of atherosclerotic lesions. Gene expression analysis of lesional macrophage foam cells is complicated by the cellular heterogeneity of atherosclerotic plaque and the presence of lesions of various degrees of severity. To overcome these limitations, we tested the ability of laser capture microdissection (LCM) and real-time quantitative reverse transcription PCR to selectively analyze RNA from lesional macrophages of apolipoprotein E (apoE)-deficient mice. Proximal aortic tissue sections were immunostained for macrophagespecific CD68/macrosialin by a rapid (≈15-min) protocol. Alternating sections from each animal were used to isolate RNA either from entire sections (analogous to isolation from whole tissue) or by LCM selection of CD68-positive cells. We measured the mRNA levels of CD68, a macrophage-specific marker, α-actin, a smooth muscle cell marker, and cyclophilin A, a control gene. Compared with whole sections, CD68 mRNA levels were greatly enriched (33.6-fold) in the laser-captured lesional macrophages. In contrast to whole sections, LCM-derived RNA had undetectable levels of α-actin. To illustrate the ability of this method to measure changes in lesional macrophage gene expression, we injected 100 μg of lipopolysaccharide i.p. into apoE-deficient mice and detected in laser-captured lesional macrophages increased mRNA expression for vascular cell adhesion molecule-1, intercellular cell adhesion molecule-1, and monocyte chemoattractant protein-1 (11.9-, 32.5-, and 31.0-fold, respectively). By selectively enriching foam cell RNA, LCM provides a powerful approach to study the in situ expression and regulation of atherosclerosis-related genes. This approach will allow the study of macrophage gene expression under various conditions of plaque formation, regression, and response to genetic and environmental perturbations.
Monocyte-derived macrophage foam cells are essential contributors to the development of atherosclerosis (1). At all stages of human disease—from early “fatty streaks” to more “advanced plaques”—macrophages are a major cellular component of atherosclerotic lesions. Circulating monocytes adhere to activated endothelium and transmigrate into the intima, where they proliferate, differentiate, and accumulate lipoproteins, leading to their characteristic “foam cell” phenotype (2, 3). Foam cells contribute to the growth and vulnerability of the plaque by secreting numerous growth factors, cytokines, and matrix-degrading metalloproteinases, and by interacting with surrounding endothelium, lymphocytes (4), and smooth muscle cells (SMC; refs. 5 and 6).
A better understanding of changes in foam-cell-associated gene expression in lesion progression and regression has become an important goal for designing potential therapies and interventions (7, 8). To this end, efforts have normally been directed to cultured macrophage cell lines (9, 10) or elicited peritoneal monocyte/macrophages (11, 12) loaded with cholesterol-rich lipoproteins. However, the genetic information derived from cultured cells may not accurately reflect the molecular events taking place in the actual lesion milieu. On the other hand, the study of macrophage foam cell gene expression in the aorta is hampered by the cellular heterogeneity of arterial tissue, which also includes lymphocytes, SMC, endothelial cells, and adventitial fibroblasts. Furthermore, analysis of macrophage gene expression in lesions of different sizes or in different regions of a given lesion are not possible with homogenized arterial samples.
To overcome these technical obstacles, we report the use of laser capture microdissection (LCM) (13, 14) navigated by cellular expression of a cell-specific marker, CD68/macrosialin scavenger receptor (15). Lesional macrophages from apolipoprotein E (apoE)-deficient mice (16, 17) were thereby procured and used as a source of RNA for quantifying specific gene transcripts by real-time reverse transcription (RT)-PCR. To validate the use of these methods to detect in vivo transcriptional regulation in a specific population of macrophage foam cells, we also measured changes in the expression of genes encoding proatherogenic factors after stimulation by the bacterial endotoxin lipopolysaccharide (LPS).
Methods
Animals and Tissue Processing.
Animals were housed in the Center for Laboratory Animal Science and procedures were approved by the Institutional Animal Use and Care Committee. ApoE−/− on the C57BL/6J background mice (The Jackson Laboratory) were fed a standard chow diet for either 20 or 25 weeks. All reagents were maintained under RNase-free sterile conditions with, in some cases, additional use of RNase Inhibitors (SUPERaseIn, Ambion, Austin, TX). Mice were killed by exsanguination by intravascular perfusion with PBS under general anesthesia (isoflurane, Baxter, Deerfield, IL). The portion of the heart containing the proximal aorta (aortic root) was excised, embedded in OCT compound (VWR Scientific), frozen on dry ice, and the tissue blocks were stored at −80°C as described (18). Frozen sections were cut at 6-μm thickness and mounted on positively charged slides (Color Frost Plus; Fisher Scientific). For the group of mice studied at 25 weeks, two mice were administered an i.p. injection of the bacterial endotoxin LPS (100 μg; Sigma catalog no. L-6636) and two mice were injected with vehicle only. Four hours after treatment, the proximal aortas were processed for each mouse separately as described above.
Immunohistochemistry and Morphometry.
Standard immunohistochemical staining protocols usually require prolonged incubation periods in aqueous media, which result in significant degradation of RNA. To overcome this limitation, a modified rapid immunostaining protocol that does not significantly affect RNA yields was developed for a macrophage-specific marker (<12% reduction of total RNA in immunostained versus nonimmunostained whole tissue sections) (19). Frozen proximal aortic sections were fixed in 70% ethanol for 15 s followed by cold acetone for 5 min. The slides were washed in PBS, and incubated with rat anti-mouse CD68/macrosialin IgG (Serotec, 1:10 dilution) for 1 min, washed in PBS, and exposed to biotinylated rabbit-anti-rat IgG mouse-adsorbed secondary antibody (Vector Laboratories, 20 μg/ml). The slides were washed in PBS, incubated with Vectastain ABC alkaline phosphatase enzyme complex, and the staining was detected with the Vector Red chromogenic substrate (Vector Laboratories). Sections were counterstained with Harris modified hematoxylin (Fisher Scientific) and the CD68-immunostain-positive areas were quantified by computer-aided morphometric analysis (IMAGE PRO PLUS 3.0 software; Media Cybernetics, Silver Spring, MD) performed on digitized microscopic images.
LCM and RNA Extraction.
Sections were subsequently dehydrated in graded ethanol solutions (95% twice, 5 min, 100% three times, 5 min) and cleared in xylene (three times, 5 min). After air-drying for 30 min, laser capture was performed under direct microscopic visualization on the CD68-positive stained areas by melting of selected regions onto a thermoplastic film mounted on optically transparent LCM caps (Arcturus Engineering, Mountain View, CA). The PixCell II LCM System (Arcturus Engineering) was set to the following parameters: 15-μm laser spot size, 40-mW power, 3.0-ms duration. The thermoplastic film containing the microdissected cells was incubated with 200 μl of 4 M guanidinium isothiocyanate and 1.6 μl of 2-mercaptoethanol for ≈10 min on ice. The transfer film was examined under the microscope to ensure complete cell lysis. Total RNA was extracted by the phenol-chloroform-based method (MicroRNA Isolation Kit, Stratagene) per the manufacturer's instructions with some minor modifications (protocol outlined in http://dir.nichd.nih.gov/lcm/Protocol.htm). RNA was precipitated with sodium acetate aided by 1 μl of glycogen carrier (GlycoBlue, Ambion; 10 μg/μl). To eliminate potential genomic DNA contamination, RNA samples were treated with DNase (4U; DNA-free, Ambion) in the presence of RNase inhibitor (RnaseOut, Invitrogen Life Technologies) at 37°C for 1 h. RNA concentration was measured by a sensitive nucleic-acid dye binding assay (Ribogreen, Molecular Probes, Eugene, OR) according to manufacturer's instructions. The integrity of the LCM-derived RNA was assessed by RT-PCR and subsequent PCR with oligo(dT) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers, respectively, yielding a 450-bp amplicon (data not shown). Because the amplified region was 642 bp upstream of the polyadenylation signal, it can be inferred that at least 52% of the total length of GAPDH transcript (1.228 kb) remained intact. Such a size of intact RNA transcripts is similar to that achieved for construction of highly representative cDNA libraries (20).
Analysis of Gene Expression by Real-Time Quantitative RT-PCR.
For measurement of the degree of macrophage selection and enrichment achieved by LCM, real-time quantitative RT-PCR (ABI Prism 7700 Sequence Detection System, Applied Biosystems, Foster City, CA) was used to determine the mRNA levels of CD68, a macrophage-specific marker, α-actin, a SMC marker, and cyclophilin A, a standard control gene (21), in total RNA samples extracted either from laser-captured lesional macrophages or from alternating whole sections (analogous to isolation from whole tissue). To generate the standard curves for CD68 and α-actin, total RNA was prepared from thioglycollate-elicited i.p. macrophages (22) and cultured primary aortic SMC (23), respectively, as described.
To test whether RNA derived by LCM of macrophage foam cells can be used to quantitatively assess the transcriptional regulation of target genes implicated in atherosclerosis, laser-captured lesional macrophage RNA from proximal aortas of control and LPS-stimulated apoE-deficient mice was used to measure the levels of the following genes: vascular cell adhesion molecule-1 (VCAM-1), intercellular adhesion molecule-1 (ICAM-1), and monocyte chemoattractant protein-1 (MCP-1).
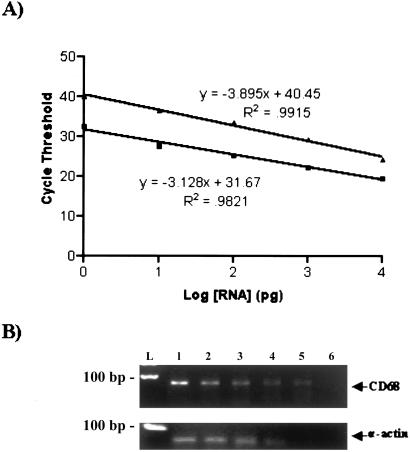
Detection of amplification is contingent on the specific hybridization of the fluorogenic probe and the flanking forward and reverse primers. The probe, labeled at the 5′ and 3′ ends with 6-carboxyfluorescein (6-FAM) reporter and 6-carboxytetramethylrhodamine (TAMRA) quencher, respectively, is hydrolyzed by the 5′ exonuclease activity of Taq DNA polymerase, causing an increase in fluorescent signal that is measured in “real time” after each cycle of PCR amplification. Fluorescence thresholds were set to 10 standard deviations above baseline fluorescence. Standard curves were constructed by plotting log10 RNA starting quantity versus cycle threshold (Fig. 2A). On the basis of appropriate serially diluted standard RNA, the amount of input standard RNA yielding the same amount of PCR product measured from an unknown sample was calculated. Measured RNA levels were normalized to cyclophilin A and expressed as a ratio of CD68/cyclophilin A. The primer pair/probe sequences for each of the specific RNA transcripts assayed are listed in Table 1.
Figure 2.
Measurement of RNA transcripts by real-time quantitative RT-PCR. (A) Representative standard curves from real-time quantitative RT-PCR of CD68 (▴) and α-actin (■). The plot of cycle threshold versus log10 of input starting mass of RNA shows a linear relationship for five orders of magnitude of starting RNA (the square of the correlation coefficients for CD68 and α-actin are 0.99 and 0.98, respectively). The mean is shown for each RNA mass from duplicate standards (the SD bars are not depicted because they are smaller than the figure symbol). (B) Electrophoretic analysis of the quantitative RT-PCR products. The products for CD68 (Upper) and α-actin (Lower) verify the specific amplification of appropriately sized amplicons (67 and 66 bp, respectively). The semiquantitative relationship (i.e., graded intensity of ethidium bromide stained bands) for the serial diluted standards (10 ng to 1 pg; lanes 1–5) corresponds to that measured in A. A water blank in place of the RNA was used as a negative control (lane 6).
Table 1.
Sequences of real-time quantitative RT-PCR primers and probes of measured gene transcripts
| Gene/primer | Sequence (5′ → 3′) | Position | Amplicon length, bp |
|---|---|---|---|
| Cyclophilin A (NM_008907) | |||
| Fwd | GGCCGATGACGAGCCC | 74–89 | 64 |
| Probe | TGGGCCGCGTCTCCTTCGA | 91–109 | |
| Rev | TGTCTTTGGAACTTTGTCTGCAA | 115–148 | |
| CD68/macrosialin (X68273) | |||
| Fwd | TTGGGAACTACACACGTGGGC | 472–490 | 67 |
| Probe | CGGCTCCCAGCCTTGTGTTCAGC | 494–516 | |
| Rev | CGGATTTGAATTTGGGCTTG | 519–538 | |
| α-Actin (NM_007392) | |||
| Fwd | AACGCCTTCCGCTGCCC | 814–829 | 66 |
| Probe | AGACTCTCTTCCAGCCATCTTTCATTGGGA | 832–861 | |
| Rev | CGATGCCCGCTGACTCC | 863–879 | |
| VCAM-1 (NM_011693) | |||
| Fwd | CCCCAAGGATCCAGAGATTCA | 402–422 | 63 |
| Probe | TTCAGTGGCCCCCTGGAGGTTG | 424–445 | |
| Rev | ACTTGACCGTGACCGGCTT | 448–466 | |
| ICAM-1 (X52264) | |||
| Fwd | ATCTCAGGCCGCAAGGG | 600–616 | 66 |
| Probe | TGGCATTGTTCTCTAATGTCTCCGAGGC | 617–645 | |
| Rev | CGAAAGTCCGGAGGCTCC | 648–665 | |
| MCP-1 (M19681) | |||
| Fwd | TTCCTCCACCACCATGCAG | 535–553 | 64 |
| Probe | CCCTGTCATGCTTCTGGGCCTGC | 556–578 | |
| Rev | CCAGCCGGCAACTGTGA | 582–598 | |
The probes are labeled at the 5′ and 3′ positions with 6-carboxyfluorescein reporter and a 6-carboxytetramethylrhodamine quencher, respectively. The positions of the primers and probes are annotated according to the sequences derived from GenBank (accession numbers given in parenthesis). Fwd, forward; Rev, reverse.
RT-PCR and subsequent PCR were both carried out in a single sealed optical tube using gene-specific primers and fluorogenic probes. Two microliters of sample (100 pg) and appropriate standard RNA (10 ng to 1 pg) were added separately into 23 μl of reaction mix containing 1× first strand buffer (50 mM Tris⋅HCl, pH 8.3/75 mM KCl/3 mM MgCl2), 5 mM DTT, 0.3 mM dNTP mix, 20 units of SuperScript II reverse transcriptase enzyme, 2.5 units of Taq DNA polymerase, 40 units of RnaseOut RNase inhibitor, 0.625 μg of acetylated BSA, and 1× 5-carboxy-X-rhodamine internal reference dye (Invitrogen Life Technologies). The thermal cycling conditions consisted of an RT-PCR stage (95°C, 10 s; 45°C, 50 min; 95°C, 2 min) immediately followed by 40 cycles of PCR amplification (denaturation: 95°C, 15 s; annealing/extension: 60°C, 1 min). The reaction products were run on a 2% Nusieve 3:1 (BioWhittaker) agarose gel to verify the appropriate size of the amplicons (≈63–67 bp).
Statistical Analysis.
The data are expressed as a ratio of the quantity of specific transcripts to the quantity of the control gene cyclophilin A in mean arbitrary units. In some cases, as indicated in Results, data were analyzed by an unpaired, two-tailed Student's t test with statistical significance attributed to P < 0.05.
Results
Laser Capture of CD68-Immunopositive Macrophage Foam Cells from Atherosclerotic Lesions.
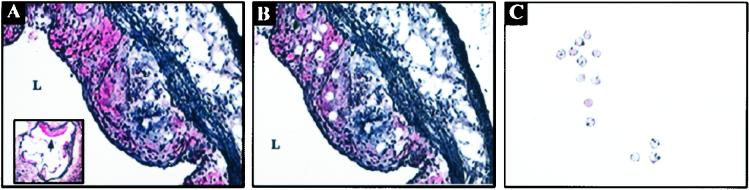
To aid in the identification of foam cells for laser capture, aortic sections were immunostained for the CD68/macrosialin antigen (Fig. 1). LCM was performed on 30 proximal aortic sections from each apoE-deficient mouse in regions immunostained positive for CD68/macrosialin and with morphologically identifiable cells having the characteristic “foamy” appearance. Approximately 300 laser pulses were performed on each section with a 15-μm laser diameter (equivalent to ≈9,000 laser pulses per proximal aorta per animal). RNA extraction of the laser-captured material yielded 3.52 ± 0.18 ng of total RNA per animal (mean ± SD).
Figure 1.
LCM of macrophage foam cells from atherosclerotic lesions of apoE-deficient mice. Selection of cells for laser capture was guided by immunohistochemical detection of macrophage specific marker CD68/macrosialin (red staining). (A) A proximal aortic lesion is shown before laser capture (200×; Inset: 30×). The CD68-positively stained cells are targeted for laser capture with a 15-μm laser diameter. Arrow in Inset denotes a representative lesion from which CD68-positive cells were isolated by LCM. (B) After the thermoplastic film is removed, the punched holes left after laser capture are seen in the remaining heterogeneous tissue. (C) The homogeneity of the captured material is confirmed under microscopic visualization before processing for RNA extraction. L, lumen.
LCM Enriches RNA from Lesional Macrophage Foam Cells As Assessed By Real-Time Quantitative RT-PCR.
For the different gene transcripts measured, the quantitative RT-PCR assay was highly reproducible (median intra-assay coefficients of variation, based on >20 samples run in duplicate, ranged between 2.4% and 7.7%) and highly sensitive (10 pg of starting total RNA resulted in detectable product formation). In Fig. 2A, representative standard curve plots for CD68 (upper line) and α-actin (lower line) are shown. For all of the genes measured, a linear relationship between the log of the initial mass input RNA (pg) and the threshold cycle held true over a range of five orders of magnitude variation in the starting RNA mass (for all experiments, the correlation coefficients were between 0.980 and 0.999). RT-PCR products from the serially diluted standards and samples were analyzed by gel electrophoresis to confirm the presence of the specific amplicon. In Fig. 2B, representative RT-PCR amplicons of CD68 and α-actin are shown, which demonstrate the specificity of the primers and the increasing yield of product with greater amounts of starting sample.
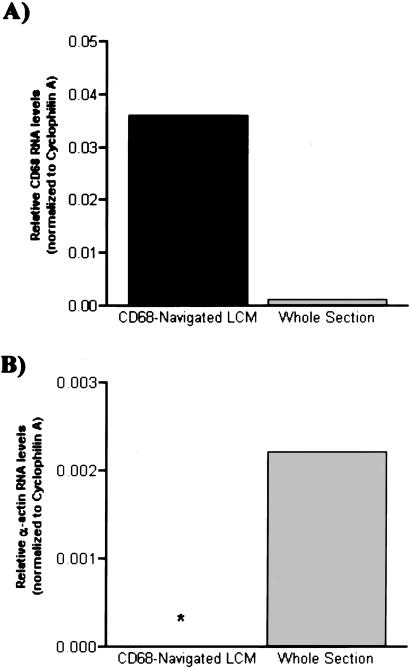
To show that LCM enriches macrophage-specific transcripts, RNA was extracted either from whole sections (analogous to homogenized tissue) or from LCM-derived CD68 immunopositive macrophage foam cells. Real-time quantitative RT-PCR for CD68 was performed on equivalent amounts of RNA (100 pg), and the results were normalized to the control gene, cyclophilin A. There was no significant difference in cyclophilin A mRNA levels between the two sample types, demonstrating equal RNA loading and comparable RNA quality (relative levels in LCM-derived and whole-section RNA were 11,518 ± 2,061 and 12,231 ± 1,821 units per 100 pg of total RNA, respectively, P = not significant). In contrast, as shown in Fig. 3A, compared with RNA derived from whole aortic sections, the ratio of CD68 to cyclophilin mRNA levels in the LCM-derived material was significantly greater [mean (range): 0.036 (0.035–0.037) vs. 0.0011 (0.00096–0.0014)]. In other words, the LCM-derived RNA was significantly enriched in the mRNA for the macrophage-specific marker, CD68, compared with whole section RNA (33.6-fold increase with normalization to cyclophilin A).
Figure 3.
LCM selectively enriches lesional macrophage RNA, as assessed by measurements of mRNA transcript levels of cell-specific markers. (A) Mean levels of macrophage-specific marker CD68 (relative to cyclophilin A) were assessed in LCM-derived lesional foam cell RNA (100 pg) and compared with that extracted from alternate whole sections. The levels of CD68 were markedly increased in the LCM sample (range: 0.035–0.037) compared with the whole-section RNA sample (range: 0.00096–0.0014). The 33.6-fold increase in CD68 levels in the LCM-procured sample attests to the enrichment of macrophage cell-derived RNA. (B) Selectivity of LCM as assessed by SMC α-actin mRNA content. The α-actin levels [mean (range): 0.0022 (0.0019–0.0025); normalized to cyclophilin A] in LCM-derived foam cells are compared with those in whole-section RNA. Although the levels of the control reference gene, cyclophilin A, were comparable between the two sample groups, there was no detectable amplification for α-actin in the LCM foam cell-derived RNA (the asterisk indicates where the data bar would have been).
The Specificity of Macrophage LCM as Assessed by Real-Time Quantitative RT-PCR.
To determine the potential cellular contamination of the laser-captured cells by adjacent medial SMC either from nonspecific tissue adherence to the thermoplastic film or from imprecise laser beam positioning, SMC-specific α-actin was measured by real-time quantitative RT-PCR. As shown in Fig. 3B, in contrast to the level of α-actin in whole section extracted RNA [mean (range): 0.0022 (0.0019–0.0025), in the LCM-derived RNA, α-actin was at background levels (24) after 40 cycles of amplification. These results show that lesional macrophage foam cells can be precisely and selectively isolated from atherosclerotic vessels by CD68-guided LCM.
Induction of VCAM-1, ICAM-1, and MCP-1 RNA in Laser-Captured Macrophage Foam Cells Isolated from Atherosclerotic Lesions of LPS-Stimulated ApoE-Deficient Mice.
Both the LPS-stimulated and control apoE-deficient mice had comparable total cholesterol (455.1 ± 142.2 vs. 523.0 ± 204.4 mg/dl, respectively; P = not significant; average of two samples of each type) and high density lipoprotein levels (23.1 ± 4.1 vs. 20.1 ± 1.8 mg/dl, respectively, P = not significant). Levels of VCAM-1, ICAM-1, and MCP-1 were consistently greater in laser-captured lesional macrophage foam cells from LPS-stimulated mice than from control mice (Table 2). Compared with the mRNA levels measured from lesional macrophages from control mice, in LPS-treated mice VCAM-1 (11.9-fold), ICAM-1 (32.5-fold), and MCP-1 (31.0-fold) were all increased [mean (range): VCAM-1, 22.1 (11.9–32.3) vs. 1.85 (1.6–2.1); ICAM-1, 9.8 (4.1–15.4) vs. 0.3 (0.2–0.4); MCP-1: 3.9 (2.4–5.5) vs. 0.1 (0.126 - 0.128)].
Table 2.
Induction of proatherogenic mRNAs by LPS
| mRNA species | Normalized level
|
Ratio of means (LPS/Control) | |
|---|---|---|---|
| Control | LPS | ||
| VCAM-1 | 2.1 (1.6–2.1) | 22.1 (11.9–32.3) | 11.9 |
| ICAM-1 | 0.3 (0.2–0.4) | 9.8 (4.1–15.4) | 32.5 |
| MCP-1 | 0.1 (0.126–0.128) | 3.9 (2.4–5.5) | 31.0 |
Real-time quantitative RT-PCR measurements (normalized to the control gene cyclophilin A) of RNA derived from LCM-isolated lesional macrophages from control and LPS-treated apoE-deficient animals are given as a mean (range) calculated from two animals per treatment condition.
Discussion
We demonstrate that LCM can be used to selectively isolate lesional macrophage foam cell RNA. We show that this RNA is enriched ≈30-fold for a macrophage-specific marker, CD68, with undetectable contamination by SMC RNA. We also show in laser-captured lesional macrophages that VCAM, ICAM, and MCP-1 were up-regulated after in vivo treatment with LPS.
Macrophage foam cell accumulation in the subendothelial space is a critical event in the development of atherosclerotic lesions. Several studies have provided evidence that macrophages play roles both in early atherogenesis as well as in late lesional events (1, 25). Foam cells secrete cytokines, growth factors, pro-oxidants, and matrix-degrading metallo-proteases that are thought to contribute to plaque instability and acute thrombotic events (2, 26). The ability to accurately quantify gene expression during changes in the vessel wall is necessary to better define the temporal, spatial, and dynamic regulation of such genes and their role in atherosclerosis. However, differentially expressed transcripts may not be detected in homogenized tissue because of a dilution effect from RNA from other cell types. Similarly, the conditions of cultured cells cannot duplicate the environment of the cells in the actual tissue from which they were derived, because the cells are not in contact with the in vivo factors that regulate gene expression (e.g., soluble factors, extracellular matrix molecules, and cell–cell communication). Toward this goal, we show that sufficient amounts of RNA could be extracted from laser-captured foam cells (3.5 ng from 30 6-μm thick proximal aortic sections, equivalent to CD68-immunostain-positive volume of 1.35 × 106 μm3). Furthermore, high sensitivity was achieved by real-time RT-PCR in which transcripts could be reliably amplified from 100 pg of input LCM RNA, making gene expression analysis of a panel of potential candidates quite feasible.
We also demonstrated that these methods can be applied to the study of regulated gene expression. By real-time quantitative RT-PCR, the transcriptional induction of VCAM-1, ICAM-1, or MCP-1 in lesional macrophage RNA procured from LPS-stimulated apoE-deficient mice was easily demonstrated (Fig. 3). These three genes were chosen for study because their products are implicated in atherogenesis, as reflected by being up-regulated in human coronary plaque (27–29) and in lesions of animal models of atherosclerosis (18, 30, 31). More direct support for causative roles has come from recent mouse studies. For example, the proatherogenic effect of VCAM-1 has been demonstrated by the decreased lesion size in low density lipoprotein receptor-deficient mice containing a targeted deletion of the VCAM-1 gene (32). Similarly, ICAM-1 deficiency reduced atherosclerosis in both diet-induced (33) and apoE-deficient animals (34). The importance of MCP-1 has been demonstrated by a decrease in atherosclerosis in mice that lack either MCP-1 (35) or its receptor, CCR2 (25). In peritoneal macrophages, LPS has been previously shown to induce MCP-1 mRNA [up to 40-fold, measured 4 h after treatment, in good agreement with our results (36)] and ICAM-1 protein (37). Although VCAM-1 is expressed by macrophages in mouse atherosclerotic lesions (38, 39), the induction of VCAM-1 by LPS has been examined only in endothelial cells (40). Our data, therefore, establish that VCAM-1 is also responsive in macrophage foam cells.
Recent findings by Lehr et al. (41) have shown that repeated LPS injections in rabbits on a hypercholesterolemic diet caused increased atherosclerosis. The authors speculated that this reflected a link between the innate immune response and accelerated atherosclerosis. Our data suggest that the increase in atherosclerosis observed by Lehr et al. might actually be caused by the up-regulation of inflammatory and cell adhesion molecules expressed by macrophage foam cells in atherosclerotic lesions. LPS binds to cell surface receptors (CD14/Toll-like receptors) and exerts its downstream effects, in part, by nuclear factor (NF)-κB activation (42, 43). The presence of NF-κB consensus DNA-binding sites in the 5′ flanking regions of VCAM-1 (44), ICAM-1 (45), and MCP-1 (46) suggests a shared means of regulation that is consistent with the concurrent up-regulation of all three genes by LPS.
As valuable as it is to quantify the expression of candidate genes in specific cell types by LCM/real-time quantitative RT-PCR, a remaining advance is to adapt these methods to a more global analysis, such as DNA microarrays. Recent studies related to foam cell formation and atherosclerosis have indeed taken a microarray approach, but have used either homogenized tissue (47) or cultured lipid-loaded macrophage cell lines (10). Although it is clear that LCM can be used to procure cell-specific RNA, it is limited by the amount of RNA that can be realistically obtained from captured populations of cells, making it likely that the yield will be insufficient for the commonly used gene array platforms. To overcome this obstacle, linear amplification methods have been devised to increase the yield of RNA without distorting the relative differences in transcripts (48, 49). Recently, several studies reported the use of laser-captured RNA that was linearly amplified before microarray analysis (50, 51), and it should be possible to translate these successes in other systems to the study of gene expression in lesional macrophage foam cells.
In conclusion, the present study demonstrates that by selectively obtaining cells from atherosclerotic lesions by LCM, dramatic enrichment in macrophage foam cell-specific mRNA is achieved. These methods make possible the quantitative analysis of basal and induced gene expression in specific cells in atherosclerotic lesions and add a powerful dimension to the study of factors that regulate plaque progression and regression.
Acknowledgments
We thank Mr. Muzaffar Akram and Dr. Carlos Cordon-Cordo of Memorial Sloan–Kettering Cancer Center for initial technical assistance with LCM. We thank Mr. Raymond Soccio of The Rockefeller University for early help with real-time quantitative RT-PCR. This study was supported by National Institutes of Health Grants HL-61814 (to E.A.F.) and HL-33714 (to J.L.B.), and by a Sigma Xi Grant-In-Aid (to E.T.). E.T. was supported by an National Institutes of Health Training Grant in Molecular and Cellular Cardiology (HL-07824).
Abbreviations
- LCM
laser capture microdissection
- SMC
smooth muscle cells
- apoE
apolipoprotein E
- LPS
lipopolysaccharide
- VCAM-1
vascular cell adhesion molecule-1
- ICAM-1
intercellular cell adhesion molecule-1
- MCP-1
monocyte chemoattractant protein-1
- RT
reverse transcription
References
- 1.Smith J D, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Proc Natl Acad Sci USA. 1995;92:8264–8268. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lusis A J. Nature (London) 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross R. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 4.Hansson G K, Holm J, Jonasson L. Am J Pathol. 1989;135:169–175. [PMC free article] [PubMed] [Google Scholar]
- 5.Stein O, Stein Y. Curr Opin Lipidol. 1995;6:269–274. doi: 10.1097/00041433-199510000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Bruemmer D, Riggers U, Holzmeister J, Grill M, Lippek F, Settmacher U, Regitz-Zagrosek V, Fleck E, Graf K. Am J Cardiol. 2001;87:21–27. doi: 10.1016/s0002-9149(00)01266-2. [DOI] [PubMed] [Google Scholar]
- 7.Chong P H, Bachenheimer B S. Drugs. 2000;60:55–93. doi: 10.2165/00003495-200060010-00005. [DOI] [PubMed] [Google Scholar]
- 8.Brewer H B., Jr J Clin Invest. 2000;105:703–705. doi: 10.1172/JCI9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi Y, Miyata M, Zheng P, Imazato T, Horwitz A, Smith J D. Biochim Biophys Acta. 2000;1492:385–394. doi: 10.1016/s0167-4781(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 10.Shiffman D, Mikita T, Tai J T, Wade D P, Porter J G, Seilhamer J J, Somogyi R, Liang S, Lawn R M. J Biol Chem. 2000;275:37324–37332. doi: 10.1074/jbc.M004732200. [DOI] [PubMed] [Google Scholar]
- 11.Yao P M, Tabas I. J Biol Chem. 2000;275:23807–23813. doi: 10.1074/jbc.M002087200. [DOI] [PubMed] [Google Scholar]
- 12.Venkateswaran A, Repa J J, Lobaccaro J M, Bronson A, Mangelsdorf D J, Edwards P A. J Biol Chem. 2000;275:14700–14707. doi: 10.1074/jbc.275.19.14700. [DOI] [PubMed] [Google Scholar]
- 13.Emmert-Buck M R, Bonner R F, Smith P D, Chuaqui R F, Zhuang Z, Goldstein S R, Weiss R A, Liotta L A. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 14.Bonner R F, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta L A. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 15.Ramprasad M P, Fischer W, Witztum J L, Sambrano G R, Quehenberger O, Steinberg D. Proc Natl Acad Sci USA. 1995;92:9580–9584. doi: 10.1073/pnas.92.21.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plump A S, Smith J D, Hayek T, Aalto-Setala K, Walsh A, Verstuyft J G, Rubin E M, Breslow J L. Cell. 1992;71:343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S H, Reddick R L, Piedrahita J A, Maeda N. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 18.Nakashima Y, Plump A S, Raines E W, Breslow J L, Ross R. Arterioscler Thromb. 1994;14:133–140. doi: 10.1161/01.atv.14.1.133. [DOI] [PubMed] [Google Scholar]
- 19.Fend F, Emmert-Buck M R, Chuaqui R, Cole K, Lee J, Liotta L A, Raffeld M. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson L A, Brown M R, Carlisle A J, Kohn E C, Liotta L A, Emmert-Buck M R, Krizman D B. Cancer Res. 1998;58:5326–5328. [PubMed] [Google Scholar]
- 21.Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. J Biotechnol. 1999;75:291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 22.Gargalovic P, Dory L. J Biol Chem. 2001;276:26164–26170. doi: 10.1074/jbc.M011291200. [DOI] [PubMed] [Google Scholar]
- 23.Pauly R R, Bilato C, Cheng L, Monticone R, Crow M T. Methods Cell Biol. 1997;52:133–154. doi: 10.1016/s0091-679x(08)60377-5. [DOI] [PubMed] [Google Scholar]
- 24.Bustin S A. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 25.Boring L, Gosling J, Cleary M, Charo I F. Nature (London) 1998;394:894–897. doi: 10.1038/29788. [DOI] [PubMed] [Google Scholar]
- 26.Lendon C L, Davies M J, Born G V, Richardson P D. Atherosclerosis. 1991;87:87–90. doi: 10.1016/0021-9150(91)90235-u. [DOI] [PubMed] [Google Scholar]
- 27.Davies M J, Gordon J L, Gearing A J, Pigott R, Woolf N, Katz D, Kyriakopoulos A. J Pathol. 1993;171:223–229. doi: 10.1002/path.1711710311. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien K D, Allen M D, McDonald T O, Chait A, Harlan J M, Fishbein D, McCarty J, Ferguson M, Hudkins K, Benjamin C D, et al. J Clin Invest. 1993;92:945–951. doi: 10.1172/JCI116670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yla-Herttuala S, Lipton B A, Rosenfeld M E, Sarkioja T, Yoshimura T, Leonard E J, Witztum J L, Steinberg D. Proc Natl Acad Sci USA. 1991;88:5252–5256. doi: 10.1073/pnas.88.12.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos C L, Huo Y, Jung U, Ghosh S, Manka D R, Sarembock I J, Ley K. Circ Res. 1999;84:1237–1244. doi: 10.1161/01.res.84.11.1237. [DOI] [PubMed] [Google Scholar]
- 31.Rayner K, Van Eersel S, Groot P H, Reape T J. J Vasc Res. 2000;37:93–102. doi: 10.1159/000025720. [DOI] [PubMed] [Google Scholar]
- 32.Cybulsky M I, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos J C, Connelly P W, Milstone D S. J Clin Invest. 2001;107:1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nageh M F, Sandberg E T, Marotti K R, Lin A H, Melchior E P, Bullard D C, Beaudet A L. Arterioscler Thromb Vasc Biol. 1997;17:1517–1520. doi: 10.1161/01.atv.17.8.1517. [DOI] [PubMed] [Google Scholar]
- 34.Bourdillon M C, Poston R N, Covacho C, Chignier E, Bricca G, McGregor J L. Arterioscler Thromb Vasc Biol. 2000;20:2630–2635. doi: 10.1161/01.atv.20.12.2630. [DOI] [PubMed] [Google Scholar]
- 35.Gu L, Okada Y, Clinton S K, Gerard C, Sukhova G K, Libby P, Rollins B J. Mol Cell. 1998;2:275–281. doi: 10.1016/s1097-2765(00)80139-2. [DOI] [PubMed] [Google Scholar]
- 36.Kopydlowski K M, Salkowski C A, Cody M J, van Rooijen N, Major J, Hamilton T A, Vogel S N. J Immunol. 1999;163:1537–1544. [PubMed] [Google Scholar]
- 37.Bernatchez S F, Atkinson M R, Parks P J. Biomaterials. 1997;18:1371–1378. doi: 10.1016/s0142-9612(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 38.Iiyama K, Hajra L, Iiyama M, Li H, DiChiara M, Medoff B D, Cybulsky M I. Circ Res. 1999;85:199–207. doi: 10.1161/01.res.85.2.199. [DOI] [PubMed] [Google Scholar]
- 39.Reis E D, Li J, Fayad Z A, Rong J X, Hansoty D, Aguinaldo J G, Fallon J T, Fisher E A. J Vasc Surg. 2001;34:541–547. doi: 10.1067/mva.2001.115963. [DOI] [PubMed] [Google Scholar]
- 40.Haraldsen G, Kvale D, Lien B, Farstad I N, Brandtzaeg P. J Immunol. 1996;156:2558–2565. [PubMed] [Google Scholar]
- 41.Lehr H A, Sagban T A, Ihling C, Zahringer U, Hungerer K D, Blumrich M, Reifenberg K, Bhakdi S. Circulation. 2001;104:914–920. doi: 10.1161/hc3401.093153. [DOI] [PubMed] [Google Scholar]
- 42.Guha M, Mackman N. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 43.Jones B W, Means T K, Heldwein K A, Keen M A, Hill P J, Belisle J T, Fenton M J. J Leukoc Biol. 2001;69:1036–1044. [PubMed] [Google Scholar]
- 44.Collins T, Cybulsky M I. J Clin Invest. 2001;107:255–264. doi: 10.1172/JCI10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voraberger G, Schafer R, Stratowa C. J Immunol. 1991;147:2777–2786. [PubMed] [Google Scholar]
- 46.Ueda A, Ishigatsubo Y, Okubo T, Yoshimura T. J Biol Chem. 1997;272:31092–31099. doi: 10.1074/jbc.272.49.31092. [DOI] [PubMed] [Google Scholar]
- 47.Wuttge D M, Eriksson P, Sirsjo A, Hansson G K, Stemme S. Am J Pathol. 2001;159:417–423. doi: 10.1016/S0002-9440(10)61712-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Gelder R N, von Zastrow M E, Yool A, Dement W C, Barchas J D, Eberwine J H. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang E, Miller L D, Ohnmacht G A, Liu E T, Marincola F M. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 50.Luo L, Salunga R C, Guo H, Bittner A, Joy K C, Galindo J E, Xiao H, Rogers K E, Wan J S, Jackson M R, Erlander M G. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 51.Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley J F, Koontongkaew S, Liotta L A, Emmert-Buck M, Gutkind J S. Oncogene. 2000;19:3220–3224. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]