Abstract
In vivo phage display identifies peptides that selectively home to the vasculature of individual organs, tissues, and tumors. Here we report the identification of a cyclic nonapeptide, CPGPEGAGC, which homes to normal breast tissue with a 100-fold selectivity over nontargeted phage. The homing of the phage is inhibited by its cognate synthetic peptide. Phage localization in tissue sections showed that the breast-homing phage binds to the blood vessels in the breast, but not in other tissues. The phage also bound to the vasculature of hyperplastic and malignant lesions in transgenic breast cancer mice. Expression cloning with a phage-displayed cDNA library yielded a phage that specifically bound to the breast-homing peptide. The cDNA insert was homologous to a fragment of aminopeptidase P. The homing peptide bound aminopeptidase P from malignant breast tissue in affinity chromatography. Antibodies against aminopeptidase P inhibited the in vitro binding of the phage-displayed cDNA to the peptide and the in vivo homing of phage carrying the peptide. These results indicate that aminopeptidase P is the receptor for the breast-homing peptide. This peptide may be useful in designing drugs for the prevention and treatment of breast cancer.
Keywords: endothelial cells‖vascular targeting‖breast cancer
The vascular beds of individual tissues differ in structure and metabolic function, and in expression of organ-specific adhesion molecules (1). Such specialization is particularly striking in lymphoid tissues where the high endothelia are composed of cells that express unique adhesion molecules for lymphocyte homing (2). Furthermore, metastasis into preferred organs by certain tumors may depend on interactions between tumor cells and organ-specific molecules in the vasculature of the target organ (3).
Our laboratory has shown that peptide libraries displayed on phage can be screened in vivo for phage that home to a specific target, and we and others have shown that peptides capable of homing to the vasculature of various individual tissues can be isolated in this manner (4–6). In vivo selections have also successfully identified peptides that home to tumor vasculature and the vasculature of other pathological lesions (7, 8). Some of the tumor-homing peptides recognize known markers of sprouting endothelial cells and angiogenic tumor vasculature, such as the αvβ3, αvβ5, and α5β1 integrins (7, 9), matrix metalloproteases (10–12), and NG2 proteoglycan (13, 14). A group of tumor-homing peptides containing the sequence motif NGR were used to identify aminopeptidase N as a new marker of angiogenic vasculature (15). Only one homing peptide receptor in the vasculature of normal tissues has been identified so far; membrane dipeptidase is selectively expressed in lung vasculature and serves as a receptor for a set of lung-homing peptides (16).
Homing peptides show promise in the delivery of drugs and other therapies into designated target sites. Thus, peptides identified by phage display have been used to target doxorubicin, proapoptotic peptides, and tumor necrosis factor-α into tumors, with a resulting enhancement of drug activity and reduction of side effects (7, 17–19). To begin to develop a strategy for the selective delivery of drugs to the breast vasculature, we have explored the specialization of the vasculature in breast tissue.
Here we screened a phage-displayed peptide library in vivo and identified a cyclic nonapeptide that selectively homes to breast vasculature in mice and identified a membrane-bound proline-specific aminopeptidase P (APaseP) as the receptor for this peptide. We also show that the nonapeptide recognizes the vasculature of hyperplastic mammary tissue and primary breast tumors in transgenic breast cancer mice. It may be possible to use this peptide to selectively target therapies to breast tissue and to precancerous breast lesions.
Experimental Procedures
Material and Animals.
Anti-APaseP antibody was a gift of J. Lasch, University of Halle, Germany (20). ICR CD-1 mice were purchased from Harlan Sprague–Dawley. MMTV PyMT mice (21) were provided by W. Muller (McMaster University, Hamilton, ON, Canada) and R. Oshima (The Burnham Institute). The MMTV PyMT mice develop breast cancers under the influence of a polynoma middle T antigen that is driven by the mouse tumor virus promoter.
Phage Libraries.
A peptide library with the general structure of CX7C, where C is cysteine and X is any amino acid, was constructed in T7 phage. We annealed complementary oligonucleotides, encoding a random peptide insert as NNK codons (N is A,T,C, or G; to eliminate some of the stop codons, K is G or T). The resulting double-stranded DNA had 5′ EcoRI and 3′ HindIII overhangs which were phosphorylated with T4 polynucleotide kinase (Novagen) and ligated into 1 μg of T7 Select 415–1b vector arms (Novagen). The ligate was directly added to 50 μl of packaging extract, and incubated for 2 h. The total number of recombinants obtained was ≈108 as measured by formation of plaque-forming units (pfu). The recombinants were amplified in 500 ml of liquid culture. Purification of phage particles and sequencing of single-stranded phage DNA was performed as described (22). The human breast carcinoma cDNA library was from Novagen.
In Vivo Phage Selection.
In vivo phage selection was performed as described (22), with a few modifications. Mice were anesthetized with avertin and then injected intravenously with 109 pfu of the CX7C library. Seven minutes after the injection, the mice were perfused through the heart with 10 ml of PBS. Mammary tissue was excised and weighed, and a cell suspension was made by using Medimachine (Dako). The resulting single cells were centrifuged at 1,500 rpm and washed five times with 5 ml PBS. The cell-adherent phage were rescued by infecting BL21 bacteria (Novagen), and the number of recovered phage was quantified by plaque assay.
cDNA Cloning.
A cDNA library displayed on T7 phage (23–25) was used to clone proteins binding to the CPGPEGAGC peptide. The peptide was synthesized in a Symphony synthesizer (Rainin Instruments) at our peptide facility, and cyclized and purified by HPLC. The peptide showed the correct mass by matrix-assisted laser desorption ionization–time of flight mass spectroscopy and was >95% pure. The peptide was immobilized on a 96-well Reacti-Bind polystyrene strip plate (Pierce). The wells were then treated with 3 × 200 μl SuperBlock blocking buffer (Pierce). The human breast carcinoma cDNA library displayed on T7 phage was amplified in a liquid BLT 5615 bacterial culture. Phage suspension (100 μl, 106 pfu/μl) in PBS was incubated in the wells for 1 h; the wells were washed five times with 200 μl PBS and once with buffer containing 1% SDS (Novagen) to elute phage bound with low and intermediate affinity. Phage bound to the immobilized peptide were then recovered by incubating a BLT 5615 culture in the wells for 10 min at room temperature.
Immunohistochemistry.
Phage proteins were detected in tissue sections as described (5). Anesthetized mice were injected with 1 × 109 pfu of phage and perfused as described above. The organs were removed and fixed in 4% paraformaldehyde, and phage was detected with a rabbit anti-T7 antiserum (P. Laakkonen, J. A. Hoffman, K. Pokka, and E.R., unpublished work) and a peroxidase-coupled secondary anti-rabbit IgG antibody (Sigma).
Immunoblotting of APaseP.
Mouse tissues were weighed, minced with a scalpel and homogenized with Medimachine. Cells were centrifuged at 1,200 × g and the pellet was resuspended in lysis buffer (200 mM N-octylglucoside/3 mM PMSF in PBS) at 4°C. The lysates were passed 10 times through a 24-gauge needle. Lysates were mixed with 2× Laemmli buffer (NOVEX, San Diego), boiled for 5 min, and applied to SDS/PAGE by using precast 4–20% Tris-glycine gradient gels (NOVEX). The proteins were electroblotted onto poly(vinylidene difluoride) membranes. The membranes were blocked with TBST (Tris-buffered saline/0.3% Tween-20) containing 20% FBS, incubated with a 1/1000 dilution of anti-APaseP antibody, washed three times with TBST, and incubated with 1/5000 horseradish peroxidase-conjugated goat anti-rabbit antibody (Bio-Rad). Blots were developed with Western Blotting Luminol Reagent (Santa Cruz Biotechnology).
Tumor Extracts.
MMTV PyMT tumors were minced with a scalpel and homogenized with Medimachine. The resulting cell suspension was centrifuged at 3,000 rpm for 5 min (4°C). The cell pellet was treated with lysis buffer (2.5 ml/g tissue) for 2 h (4°C). The lysates were centrifuged for 30 min at 12,000 × g to remove debris and applied to a CPGPEGAGC-Sepharose column.
Affinity Chromatography.
Peptide affinity chromatography was performed essentially as described (26). All steps were performed at 4°C. The CPGPEGAGC peptide was coupled to N-hydroxysuccinimide-activated Sepharose (Pierce) according to the manufacturer's instructions. An extract from an MMTV PyMT tumor was applied to 1 ml of the affinity matrix equilibrated in column buffer (50 mM N-octylglucoside/100 μM PMSF in PBS) for 90 min. The column was washed with 20 column volumes of the buffer, and eluted with 2 ml of 2 mg/ml of the CPGPEGAGC peptide in column buffer over 1 h, and then with 8 M urea. The eluates were dialyzed against column buffer and concentrated 5-fold by using a Centricon 10,000 molecular weight cut-off column (Millipore). The column fractions were separated by SDS/PAGE, and APaseP was detected by immunoblotting.
Results
Phage Expressing the Cyclic Nonapeptide CPGPEGAGC Homes to Breast Tissue.
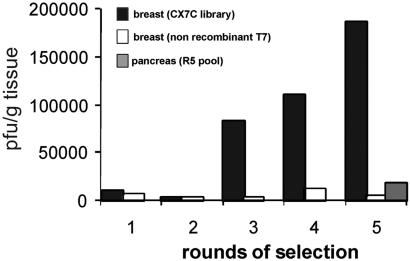
To identify phage that selectively home to mammary vasculature, we injected a phage-displayed peptide library intravenously into mice and subsequently rescued bound phage from breast tissue. Fig. 1 shows the enrichment profile obtained for five rounds of selection. The number of phage recovered from breast tissue increased about 100-fold in rounds 3–5 of the selection. The number of phage recovered from the pancreas, which was used as a control tissue, remained unaffected. Nonrecombinant T7 phage could not be enriched by in vivo selection for breast homing.
Figure 1.
Isolation of a breast-homing phage by in vivo screening of a phage library. A CX7C library (109 pfu) was injected into the tail vein of mice, and 7 min later the mice were perfused through the heart and phage was rescued from breast tissue. The rescued phage was amplified and reinjected in four consecutive rounds. The number of pfu recovered from breast tissue is shown (black bars). As a control, nonrecombinant T7 phage was injected (white bars). In round 5 (R5), the titer of phage recovered from the pancreas was also determined (gray bar).
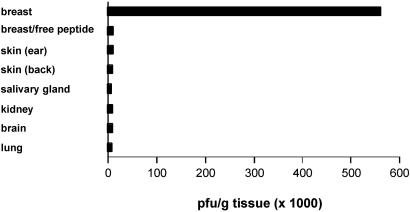
Sequence analysis showed that phage displaying the nonapeptide CPGPEGAGC were enriched, accounting for 14% of phage present in the pool. When tested individually, the CPGPEGAGC-displaying phage homed to breast tissue about 100 times more effectively than nonrecombinant T7 phage (Fig. 2). This phage did not home to other tissues, including the pancreas, brain, kidney, lung, or skin from parts of the body other than the breast fat pad. The breast homing of the CPGPEGAGC phage was also specific by the criterion of ligand inhibition, because co-injecting synthetic free peptide with the phage inhibited phage recovery from breast tissue (Fig. 2).
Figure 2.
Homing specificity of CPGPEGAGC phage. CPGPEGAGC phage (109 pfu) was injected into mice and recovered as in Fig. 1 from the indicated organs. The number of pfu recovered is shown. Selective phage homing to the breast is seen. Coinjection of free CPGPEGAGC peptide (1 mg) blocked the breast homing of the phage.
Tissue Localization of CPGPEGAGC Phage.
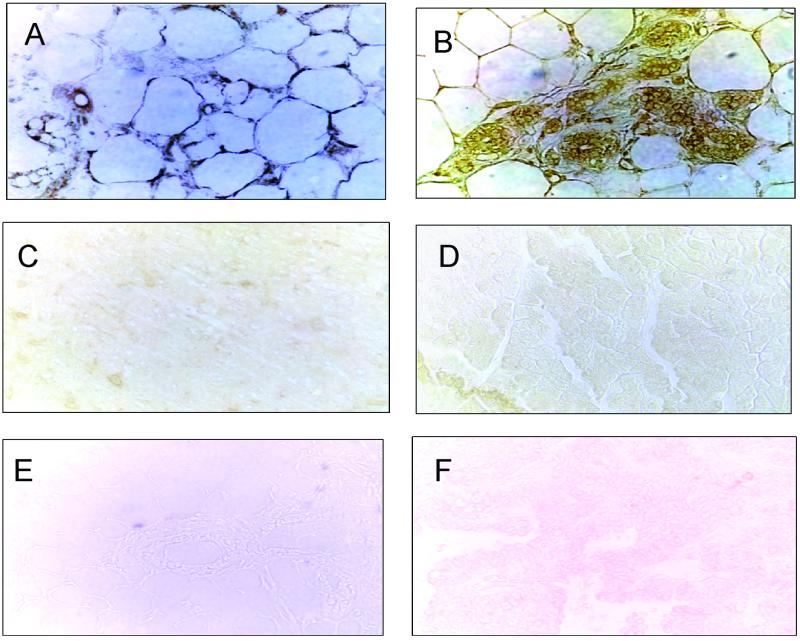
Immunoperoxidase staining revealed phage in the vasculature of normal breast (Fig. 3A) and mammary carcinoma (Fig. 3B) 15 min after i.v. injection of the CPGPEGAGC phage. Phage staining also occurred in the parenchymal tissue. No CPGPEGAGC phage was detected in the brain (Fig. 3C) or the pancreas (Fig. 3D). Nonrecombinant T7 phage injected similarly could not be detected in normal or cancerous breast tissue (Fig. 3 E and F).
Figure 3.
Localization of CPGPEGAGC phage by immunohistochemistry. CPGPEGAGC phage or nonrecombinant control phage (both at 109 pfu) were injected into the tail veins of mice. After 15 min of circulation, mice were perfused through the heart. The indicated organs were dissected, fixed, and cryo-sections were stained for T7 phage proteins by using a rabbit anti-T7 antibody and a peroxidase-coupled anti-rabbit IgG. CPGPEGAGC phage was detected in the blood vessels and surrounding tissue of the mammary fat pad (A) and early MMTV PyMT breast carcinomas (B). No phage staining occurred in the brain (C) and the pancreas (D). The nonrecombinant phage was not detected in normal breast (E) or in breast carcinomas (F).
APaseP-Related Clone Binds to the CPGPEGAGC Peptide.
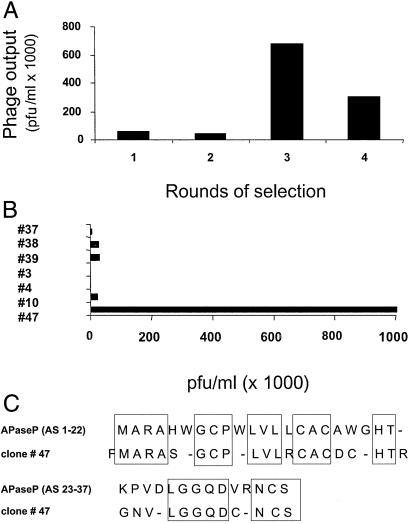
To identify a receptor for the CPGPEGAGC peptide in the breast vasculature, we screened a human breast cancer cDNA library on immobilized CPGPEGAGC peptide. Phage recovery increased about 50-fold in five rounds of selection on the peptide (Fig. 4A). Among the individual phage clones from the selected pool, one clone avidly bound to the peptide-coated surface (Fig. 4B), but not to a surface treated with the blocking buffer only (not shown). Sequence analysis showed that this clone encodes a peptide that is homologous to the signal peptide plus the N-terminal 14 amino acids of the mature human APaseP (Fig. 4C).
Figure 4.
Isolation of cDNA clones encoding CPGPEGAGC-binding proteins. (A) The CPGPEGAGC peptide was covalently linked to microtiter wells and a phage-displayed cDNA library was screened for CPGPEGAGC-binding clones by performing four consecutive rounds of selection. The number of pfu recovered from the wells is shown. (B) Clones from the screening shown in A were individually tested for binding to wells coated with the CPGPEGAGC peptide. Phage clone 47 avidly bound to the CPGPEGAGC-coated wells. (C) Alignment of the sequences of clone 47 and human membrane-bound APaseP (XNPEP2; GenBank U90724). The dashes denote gaps in the alignment of the sequences. The signal sequence of ApaseP is shown on the first line, and the N-terminal sequence of the mature protein is shown on the second line.
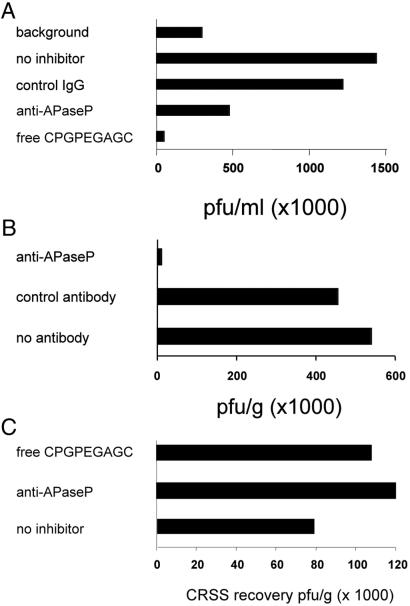
The binding of the APaseP clone phage to immobilized CPGPEGAGC peptide could be blocked by coincubation of the phage with the free peptide or with an anti-APaseP antiserum (Fig. 5A). A control antibody had no effect. Furthermore, coinjection into mice of the CPGPEGAGC phage with the anti-APaseP antibody reduced the number of phage rescued from breast tissue by about 90%. A control antibody did not affect the breast homing of the CPGPEGAGC phage (Fig. 5B). The antibody did not block the breast homing of another phage (insert sequence, CRSS) identified in the screening for breast homing. The free CPGPEGAGC peptide also had no effect on the recovery of this phage from breast tissue (Fig. 5C).
Figure 5.
Anti-APaseP antibody and free CPGPEGAGC peptide block CPGPEGAGC phage binding and homing. (A) The binding of phage displaying the APaseP cDNA fragment (108 pfu) to microtiter wells coated with CPGPEGAGC peptide was tested in the presence of the free CPGPEGAGC peptide (2 mg/ml), purified IgG from an anti-APaseP antiserum, or from normal rabbit serum, each at 10 μg/ml, or buffer. As a control, microtiter wells were coated with an unrelated peptide. (B) CPGPEGAGC-displaying phage was injected into the tail veins of mice together with 2.5 mg of the anti-APaseP IgG or control IgG. Anti-APaseP blocked the breast homing, whereas the control antibody had no effect. (C) Recovery of a phage carrying another breast-homing peptide, CRSS, could not be blocked by the anti-APaseP antiserum (2.5 mg) or by free CPGPEGAGC peptide (1 mg).
APaseP Specifically Binds to a CPGPEGAGC Peptide Affinity Column.
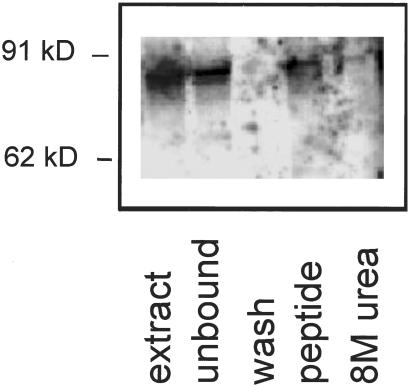
To determine whether the breast-homing peptide binds to intact APaseP, we fractionated MMTV PyMT tumor lysates on a CPGPEGAGC peptide affinity matrix. A 90-kDa band that reacted with the anti-APaseP antiserum was partially removed from the extract by the matrix and eluted from the affinity matrix by the CPGPEGAGC peptide (Fig. 6). This result confirms the specificity of the breast-homing peptide for APaseP.
Figure 6.
APaseP specifically binds to a CPGPEGAGC peptide affinity column. An extract of MMTV PyMT tumor tissue was fractionated on a CPGPEGAGC peptide column. The column was washed and eluted with the CPGPEGAGC peptide. Aliquots from the tumor extract before (extract) and after (unbound) application to the column, the wash fraction, CPGPEGAGC-peptide eluate, and 8 M urea eluate were analyzed by immunoblotting with anti-APaseP.
Discussion
We report that phage displaying the cyclic nonapeptide CPGPEGAGC selectively homes to breast vasculature in mice. This phage also homes to premalignant breast tissue and to primary breast tumors. We also provide evidence that the receptor for the homing peptide is the membrane-bound peptidase, APaseP.
These results support the notion that the vasculature in individual organs expresses specific marker molecules. Peptides that home to the vasculature of many other normal tissues have been isolated in our laboratory (4, 5) and by others (6). The numerous tissues that have been successfully targeted with in vivo phage display suggests that tissues differ in their vascular markers. Furthermore, tumor-homing phage that bind to angiogenesis-specific molecular markers (7) and phage homing to atherosclerotic plaques (8) have been isolated by in vivo phage library screening.
Regardless of this earlier evidence for vascular heterogeneity, the specificity of the breast-homing peptide we describe here is remarkable. Most of the breast tissue consists of fat, and one might expect that the blood vessels of fat tissue in various locations would be identical. Yet our phage homes only to the blood vessels in the breast, not to other fat-rich tissues. This finding agrees with the apparent specialization of the mammary fat pad as evidenced by the accelerated growth of experimental breast cancers when implanted in the mammary fat pad versus s.c. implantation, and the secretion of specific growth factors by the breast tissue (27).
Only a few organ- and tumor-specific receptors for homing peptides have been reported, but it is striking that several of them are peptidases or proteases. A lung-homing peptide recognizes membrane dipeptidase, which is selectively expressed in lung blood vessels (16), and aminopeptidase N (15) and matrix metalloproteases (12) serve as receptors for two peptides that home to angiogenic vasculature. This report adds APaseP to the list of tissue-specific homing peptide receptors that are proteases. The sequence of the phage cDNA that bound to the homing peptide, although very similar, was not identical with the published APaseP sequence, presumably because of instability of the cloned sequences in the phage. Alternatively, more than one APaseP may exist. Nonetheless, inhibition of phage homing with an anti-ApaseP and binding to the homing peptide of an APaseP recognized by this antiserum from tissues confirmed the identification of the receptor. The prevalence of proteases as receptors for vascular homing peptides may reflect a diversity of protease expression in various tissues. However, we cannot exclude the possibility that peptide library screening might favor proteases as targets, because proteases possess a peptide recognition site, whereas other types of molecules often recognize larger surfaces on proteins.
Our CPGPEGAGC peptide may mimic the natural substrates for APaseP, because the CPGPEGAGC peptide contains potential recognition sequences for APaseP. APaseP cleaves peptides at an N-terminal X-P-Z sequence (28–30). The peptide contains two X-P-Z sequences, although neither is N-terminal in the phage. This location of the recognition sequence, and the cyclic structure of the peptide, may explain why the phage stays bound to the enzyme; the enzyme may recognize the peptide, but is unable to cleave it. The active site of APaseP has not been identified, but our results suggest that the very N terminus of the mature protein may form at least part of the substrate recognition site of this enzyme.
APaseP is a GPI-linked membrane protein expressed on the surface of vascular endothelial cells in various tissues, on lymphoid cells, and on the brush-border membrane in the intestine and in kidney tubules (20). Potential physiological substrates for APaseP include interleukins and bradykinin (30). We found that APaseP is expressed at higher levels in mouse breast tissue than in other tissues (M.E. and E.R., unpublished work). However, this difference may not be sufficiently large to explain the highly selective homing of the CPGPEGAGC peptide to breast vasculature. Our immunoblotting results also suggest that individual tissues may express different isoforms of APaseP. Thus, the CPGPEGAGC peptide might specifically recognize a particular isoform of APaseP.
The breast-homing peptide we describe here may be useful in targeting therapies to breast tissue. A therapeutic benefit gained by coupling anticancer drugs to homing peptides has been demonstrated in preclinical models of cancer (7, 17–19). Therapeutics based on conjugates of the breast-homing peptide could provide a means of reducing breast tissue in individuals with high risks for breast cancer and may also be useful in enhancing the efficacy of anticancer drugs in early breast cancer.
Acknowledgments
We thank Drs. Jürgen Lasch, William Muller, and Robert Oshima for reagents, and Dr. Richard Hynes and Jason A. Hoffman for comments on the manuscript. This study was supported by Grants CA74238 and CA82715 and Cancer Center Support Grant CA 30199 from the National Cancer Institute, and Grant 99–3339 from the Komen Foundation. M.E. is supported by a fellowship from the Deutsche Forschungs Gemeinschaft.
Abbreviations
- APaseP
aminopeptidase P
- pfu
plaque-forming units
- MMTV
mouse mammary tumor virus
- PyMT
polynoma middle T antigen
References
- 1.Ruoslahti E, Rajotte D. Annu Rev Immunol. 2000;18:813–827. doi: 10.1146/annurev.immunol.18.1.813. [DOI] [PubMed] [Google Scholar]
- 2.Streeter P R, Berg E L, Rouse B T N, Bargatze R F, Butcher E C. Nature (London) 1988;331:41–46. doi: 10.1038/331041a0. [DOI] [PubMed] [Google Scholar]
- 3.Johnson R C, Augustin-Voss H G, Zhu D, Pauli B U. Cancer Res. 1991;51:394–399. [PubMed] [Google Scholar]
- 4.Pasqualini R, Ruoslahti E. Nature (London) 1996;380:360–364. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 5.Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. J Clin Invest. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samoylova T I, Smith B F. Muscle Nerve. 1999;22:460–466. doi: 10.1002/(sici)1097-4598(199904)22:4<460::aid-mus6>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Arap W, Pasqualini R, Ruoslahti E. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 8.Houston P, Goodman J, Lewis A, Campbell C J, Braddock M. FEBS Lett. 2001;492:73–77. doi: 10.1016/s0014-5793(01)02191-3. [DOI] [PubMed] [Google Scholar]
- 9.Brooks P C, Montgomery A M, Rosenfeld M, Reisfeld R A, Hu T, Klier G, Cheresh D A. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 10.Gross J, Azizkhan R G, Biswas C, Bruns R R, Hsieh D S, Folkman J. Proc Natl Acad Sci USA. 1981;78:1176–1180. doi: 10.1073/pnas.78.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coussens L M, Tinkle C L, Hanahan D, Werb Z. Cell. 2000;103:481–490. doi: 10.1016/s0092-8674(00)00139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koivunen E, Arap W, Valtanen H, Rainisalo A, Medina O P, Heikkilä P, Kantor C, Gahmberg C G, Salo T, Konttinen Y T, et al. Nat Biotechnol. 1999;17:768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- 13.Schlingemann R O, Rietveld F J R, de Waal R M W, Ferrone S, Ruiter D J. Am J Pathol. 1990;136:1393–1405. [PMC free article] [PubMed] [Google Scholar]
- 14.Burg M A, Pasqualini R, Arap W, Ruoslahti E, Stallcup W B. Cancer Res. 1999;59:2869–2874. [PubMed] [Google Scholar]
- 15.Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun R A, Shapiro L, Arap W, Ruoslahti E. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- 16.Rajotte D, Ruoslahti E. J Biol Chem. 1999;274:11593–11598. doi: 10.1074/jbc.274.17.11593. [DOI] [PubMed] [Google Scholar]
- 17.Ellerby H M, Arap W, Ellerby L M, Kain R, Andrusiak R, Del Rio G, Krajewski S, Lombardo C R, Rao R, Ruoslahti E, Pasqualini R. Nat Med. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 18.Curnis F, Sacchi A, Borgna L, Magni F, Gasparri A, Corti A. Nat Biotechnol. 2000;18:1185–1190. doi: 10.1038/81183. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Xu X, Hong S, Chen J, Liu N, Underhill C B, Creswell K, Zhang L. Cancer Res. 2001;61:2434–2438. [PubMed] [Google Scholar]
- 20.Lasch J, Moschner S, Sann H, Zellmer S, Koelsch R. Biol Chem. 1998;379:705–709. doi: 10.1515/bchm.1998.379.6.705. [DOI] [PubMed] [Google Scholar]
- 21.Siegel P M, Tyan E D, Cardiff R D, Muller W J. EMBO J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman J A, Laakkonen P, Porkka K, Ruoslahti E. In: Phage Display: A Practical Approach. Clarkson T, Lowman H, editors. Oxford: Oxford Univ. Press; 2002. , in press. [Google Scholar]
- 23.Sche P, McKenzie K M, White J D, Austin D J. Chem Biol. 1999;6:707–716. doi: 10.1016/s1074-5521(00)80018-6. [DOI] [PubMed] [Google Scholar]
- 24.Sidhu S S. Curr Opin Biotechnol. 2000;11:610–616. doi: 10.1016/s0958-1669(00)00152-x. [DOI] [PubMed] [Google Scholar]
- 25.Cochrane D, Webster C, Masih G, McCafferty J. J Mol Biol. 2000;297:89–97. doi: 10.1006/jmbi.2000.3561. [DOI] [PubMed] [Google Scholar]
- 26.Pytela R, Pierschbacher M D, Ruoslahti E. Cell. 1985;40:191–198. doi: 10.1016/0092-8674(85)90322-8. [DOI] [PubMed] [Google Scholar]
- 27.Hovey R C, MacKenzie D D, McFadden T B. In Vitro Cell Dev Biol Anim. 1998;34:385–392. doi: 10.1007/s11626-998-0020-2. [DOI] [PubMed] [Google Scholar]
- 28.Vanhoof G, de-Mester I, van-Sande M, Scharpe S, Yaron A. Eur J Clin Chem Clin Biochem. 1992;30:333–338. doi: 10.1515/cclm.1992.30.6.333. [DOI] [PubMed] [Google Scholar]
- 29.Cottrell G S, Hyde R J, Lim J, Parsons M R, Hooper N M, Turner A J. Biochemistry. 2000;39:15129–15135. doi: 10.1021/bi0015865. [DOI] [PubMed] [Google Scholar]
- 30.Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpe S. FASEB J. 1995;9:736–744. [PubMed] [Google Scholar]