Abstract
Approximately two billion people, mainly women and children, are iron deficient. Two studies examined the effects of iron deficiency and supplementation on rats. In study 1, mitochondrial functional parameters and mitochondrial DNA (mtDNA) damage were assayed in iron-deficient (≤5 μg/day) and iron-normal (800 μg/day) rats and in both groups after daily high-iron supplementation (8,000 μg/day) for 34 days. This dose is equivalent to the daily dose commonly given to iron-deficient humans. Iron-deficient rats had lower liver mitochondrial respiratory control ratios and increased levels of oxidants in polymorphonuclear-leukocytes, as assayed by dichlorofluorescein (P < 0.05). Rhodamine 123 fluorescence of polymorphonuclear-leukocytes also increased (P < 0.05). Lowered respiratory control ratios were found in daily high-iron-supplemented rats regardless of the previous iron status (P < 0.05). mtDNA damage was observed in both iron-deficient rats and rats receiving daily high-iron supplementation, compared with iron-normal rats (P < 0.05). Study 2 compared iron-deficient rats given high doses of iron (8,000 μg) either daily or every third day and found that rats given iron supplements every third day had less mtDNA damage on the second and third day after the last dose compared to daily high iron doses. Both inadequate and excessive iron (10 × nutritional need) cause significant mitochondrial malfunction. Although excess iron has been known to cause oxidative damage, the observation of oxidant-induced damage to mitochondria from iron deficiency has been unrecognized previously. Untreated iron deficiency, as well as excessive-iron supplementation, are deleterious and emphasize the importance of maintaining optimal iron intake.
Iron deficiency is the most common nutritional deficiency worldwide, affecting approximately two billion people, mostly women and children (1, 2). In the United States, an estimated nine million people are iron deficient (3). Iron deficiency is a significant public health concern (reviewed in ref. 4) that is associated with an increased risk of poor pregnancy outcomes (2) and impaired cognitive development in young children (5). Pregnant women in developing countries are commonly given daily supplements containing 120 mg of iron (6) to prevent and correct gestational iron deficiency. This dose of iron, which is 10 times the normal daily dietary iron intake, can cause gastrointestinal side effects (7).
We have found that equivalent doses of daily high-iron supplements in rats (i.e., 10 × normal intake, or 8,000 μg iron/day) results in an abnormal accumulation of intestinal mucosal (8) and hepatic nonheme iron and significant increases in lipid peroxidation (9). We unexpectedly observed that iron-deficient rats also had markedly increased lipid peroxidation (9), suggesting that both iron deficiency and iron excess promote oxidative stress.
The increased oxidative stress observed in both iron deficiency and excess may involve mitochondrial dysfunction, as has been observed also in aging and associated degenerative diseases (10). Mitochondria use 90% of inspired oxygen, produce a significant amount of cellular superoxide, and accumulate iron for heme and iron-sulfur cluster formation. Studies of severe iron overload modeling hemochromatosis have reported increased hepatic lipid peroxidation, nuclear DNA damage, and mitochondrial dysfunction (reviewed in ref. 11). The severe iron overload of mitochondria has been found to induce mitochondrial DNA (mtDNA) damage (12, 13). Damage to mtDNA correlates with the mitochondrial dysfunction associated with oxidant stress and aging (14).
Previous morphological and biochemical studies have shown that iron deficiency causes mitochondrial dysfunction in heart, skeletal muscle, liver, and blood cells. The heart and skeletal muscle were more severely affected than the liver (15). The mitochondria in the morphological studies appeared enlarged and rounded, and had fewer cristae, resulting in enhanced electron lucency (16, 17); the mitochondria also had decreased cytochrome concentration, respiratory control, and gluconeogenesis relative to iron normal controls (18, 19).
Although studies of iron deficiency previously have documented either impaired mitochondrial function (18) or increased oxidant stress (9, 20), no studies have related the two phenomena. Furthermore, no studies have investigated whether iron deficiency can cause mtDNA damage. The effects of iron deficiency and mild iron excess on mitochondrial function and oxidative stress are the focus of this report. Because supplementation with high iron either given daily or every 3 days are equally effective in correcting iron deficiency, and because there is less accumulation of iron when using an intermittent scheme (8, 21), we compared iron-deficient rats under both supplementation schemes with iron-normal rats. Iron deficiency disrupted normal liver mitochondrial function and induced an increase in oxidants in polymorphonuclear leukocytes (PMNs). The respiratory efficiency of isolated liver mitochondria was also lower in all high-iron-supplemented rats. Biomarkers of damage correlated with liver-iron stores (R2 ≈ 0.4 or higher, depending on the variable).
Materials and Methods
Materials.
All materials were of the highest quality available and came from the following vendors: rotenone, histopaque, ADP (Sigma); NaCl, succinate, mannitol, sucrose, Hepes (N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid]), EDTA, and succinate (Fisher Scientific); 2′7′-dichlorodihydrofluorescein diacetate (DCFH), rhodamine 123 (Rh123) (Molecular Probes); SDS, (Fluka); BSA (fraction V, fatty acid free), m-chlorocarbonyl cyanide phenylhydrazone (CCCP) (Calbiochem); and KH2PO4 (EM Science).
Animal Studies.
The animal studies described here continue our studies of iron deficiency and daily high-iron supplementation reported in (9), where all details of the animal studies can be found. A brief description follows. Weanling male Sprague–Dawley rats (Bantin and Kingman, Fremont, CA) were housed in stainless steel, wire-bottomed cages in a room with light control. All rats consumed AIN-93G diet (Dyets, Bethlehem, PA) without iron twice daily, preceded by a premeal that provided no extra iron in the iron-deficient group, 800 μg/day in the iron-normal group, and 8,000 μg either every day or once every third day in the high-iron-supplemented groups. The iron supplement (crystalline ferrous sulfate) was mixed in 0.8 g of a premeal containing diet and sucrose in a 1:1 ratio. Rats receiving the high-iron-supplementation dose once daily or every third day (intermittent iron supplementation) were either iron-normal or first were made iron-deficient by consuming an iron-deficient diet for 12 days.
Two studies were performed. Study 1 had four groups of rats: an iron-normal group, an iron-deficient group, an iron-normal group that received a daily high-iron supplement, and an iron-deficient group that received a daily high-iron supplement for 34 days. Each group consisted of 12 rats in a 2 × 2 factorial design. Study 2 had three groups of rats: an iron-normal group (6 rats), a group that received a daily high-iron supplement (12 rats), and a group that received iron once every third day (18 rats). Rats were fed this regime for 34 days, except those in the group that received iron supplements every third day (see below for details). The rats then were killed, and tissue and blood samples were taken. To investigate the possibility that iron status and liver mitochondrial parameters could vary in the days after every-third-day iron dosing, the 18 rats were killed in groups of 6 rats 1, 2, and 3 days after having received their final iron dose, i.e., on days 35, 36, and 37, respectively. Rat liver nonheme iron levels were measured colorimetrically after acid digestion (22). The Animal Care and Use Committee of the University of California, Berkeley approved the experimental protocol.
Isolation of Mitochondria.
After anesthesia by Nembutal, the rats were killed by exsanguination through aortic puncture, and their livers were quickly removed. Rat-liver mitochondria were isolated by differential centrifugation (23) by using a buffer containing 210 mM mannitol, 70 mM sucrose, 5 mM Hepes (MSH) buffer, and 1 mM of EDTA to bind divalent metals, such as the excess iron), pH 7.5. The final two washes had 0.1% BSA and did not contain EDTA. Protein concentration was determined by the Biuret method, by using BSA as a standard.
Isolation and Analysis of mtDNA.
Damage to mtDNA induced by excess iron during the isolation was controlled by the inclusion of iron chelators. Twelve mg of freshly prepared mitochondrial protein was resusupended in MSH buffer plus 20 mM diethylenetriamine pentaacetic acid (DTPA). mtDNA was isolated with a gentle neutral lysis in 1.5% SDS plus 20 mM DTPA (12). Electrophoresis was performed as in ref. 12. For analysis of mtDNA, Southern blotting (24) with chemiluminescent detection was used. After agarose electrophoresis, mtDNA was transferred to nylon membranes by using alkaline transfer methods, and the membranes were hybridized by using an mtDNA probe. mtDNA probes for hybridization were made from intact form I mtDNA molecules (16.3 kb) that were isolated from agarose after gel purification.
Forms of mtDNA were determined by comparison against a supercoiled marker (New England Biolabs) for form I, comparison against λ DNA (cleaved with Hind III) for form II, and restriction enzyme analysis (ClaI) for form III.
After electrophoresis, a digital image of the agarose gel and Southern blot was obtained with an Alpha Imager CCD camera system (Alpha Innotech, San Leandro, CA). The amount of mtDNA in each band of the acquired digital image was determined with ALPHA IMAGER 2000 v.3.3b computer software. The estimation of mtDNA fragmentation was determined from the ratio of fragment density to supercoiled density of mtDNA, as described (25).
Flow Cytometry of White Blood Cells.
Heparinized blood was layered over histopaque for gravitational erythrocyte sedimentation to isolate PMNs, macrophages, and lymphocytes. Leukocytes were enriched further through the removal of contaminating red blood cells by a 10-min exposure to 150 mM ammonium chloride at 4°C and subsequent washing by centrifugation (700 × g) and resuspension in PBS. The resulting leukocytes were incubated for 30 min at 37°C with either 10 μM Rh123 or 25 μM DCFH. Cells then were subjected to fluorescent flow cytometry on a FACSort (Becton Dickinson). Flow cytometry was not performed on cells from rats that had been fed iron every 3 days.
Mitochondrial Respiration.
Freshly isolated mitochondria (2 mg protein/ml) were incubated in MSH buffer (without EDTA) plus 3 mM phosphate with stirring at 23°C. A Clarke electrode (YSI Life Sciences, Yellow Springs, OH) was used to monitor oxygen consumption. Measurements were initiated by the addition of 10 μl of succinate (2.5 mM), establishing maximal state 4 respiration. State 3 respiration was achieved with the addition of 2.5 μl of ADP (200 μM). The respiratory control ratio (RCR) was determined by comparing the ratio of state 3 to state 4 oxygen consumption. Uncoupled respiration was induced by the addition of 5 μl of CCCP (5 μM).
Statistics.
Data are reported as means ± SE by using at least 12 rats per group (or as otherwise indicated). Comparisons among groups were made by standard ANOVA analysis after natural-logarithmic (ln) transformation of each value, where P < 0.05 was considered significant. Further post tests were done by Tukey's analysis. Nonlinear analyses were performed by using second-order polynomial regression analyses.
Ln transformation of liver-iron values (independent variable) were used because of its skewed distribution to the right; this procedure yields a variable that is closer to being normally distributed. Ln transformations are a common parametric statistical procedure when there are a limited number of observations. The ln transformation of the six dependent variables achieves a more normal distribution and variances that are less dissimilar (homoscedasticity) for each variable (i.e., RCR among iron-deficient, normal, or supplemented groups), which is desirable when using parametric statistical procedures.
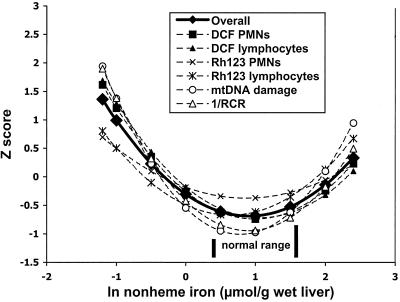
After comparisons among groups were made by standard ANOVA, each of the six ln-transformed dependent variables analyzed by nonlinear regression was transformed to a Z score and modeled as a quadratic function of the ln of liver iron as the independent variable. The equation for the RCR ratio's Z score was obtained from inverted RCR values, which resulted in the normal rats having lower instead of higher values. These six equations were submitted to a generalized F test, which compared a nested model that included the six quadratic functions with a single combined model for all equations (see Fig. 3). The units of the dependent variables are very different; the Z transformation of their ln-transformed values converted them to unit-less variables. This result allowed comparisons of response patterns to be made as a function of ln-liver iron.
Figure 3.
Analysis of nonlinear regression models: comparison of an overall model and individual models of Z-transformed values of the six dependent variables vs. liver ln-nonheme iron. Each of the six dependent variables (five were analyzed by nonlinear regression, as shown in Figs. 1 and 2) was transformed to a Z score and modeled as a quadratic function, using ln-liver nonheme iron as the independent variable. The equation for the RCR ratio's Z score was obtained from inverted RCR values (1/RCR, a measure of inefficiency), resulting in normal rats having lower rather than higher values. Each model line was obtained from nine values of liver iron. All statistics were performed as described in Materials and Methods.
Results
Study 1: Daily Iron Supplementation
Mitochondrial respiration.
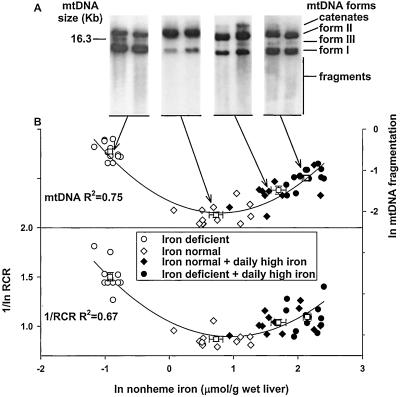
Mitochondria from iron-deficient rats had only 58% of the RCR that iron-normal rats had and 76% of the RCR that iron-deficient daily high-iron-supplemented rats had (P < 0.05, Table 1), as assayed by using an oxygen electrode. The lower RCRs were caused primarily by increases in state 4 respiration (Table 1). There was no significant change in state 3 (Table 1) or in uncoupled respiration (data not shown) in the different animal groups. Ln values of inverse (inefficient) mitochondrial respiratory control capacity (1/RCR), showed a strong nonlinear (“U” shape) correlation to ln values of liver nonheme-iron stores (R2 = 0.67; Fig. 1B Lower).
Table 1.
Effect of iron deficiency and daily supplementation with high iron on mitochondrial respiration and DNA damage in rat liver
| Treatment | State 4 respiration, nmol O2/min/mg | State 3 respiration, nmol O2/min/mg | RCR state 3/state 4 | mtDNA damage, % linear mtDNA | mtDNA damage Fragmented DNA supercoiled DNA |
|---|---|---|---|---|---|
| Iron deficient | 15.74 ± 1.23 (a) | 30.49 ± 2.63 (a) | 1.9 ± 0.06 (a) | 14.7 ± 1.35 (c) | 0.58 ± 0.04 (d) |
| Iron normal | 9.45 ± 0.93 (b) | 30.87 ± 3.06 (a) | 3.3 ± 0.12 (b) | 4.84 ± 0.51 (a) | 0.13 ± 0.01 (a) |
| Iron normal + daily high iron | 9.25 ± 0.79 (b) | 24.65 ± 2.57 (a) | 2.7 ± 0.12 (c) | 7.02 ± 0.66 (b) | 0.28 ± 0.04 (b) |
| Iron deficient + daily high iron | 12.24 ± 0.99 (a,b) | 31.08 ± 2.77 (a) | 2.5 ± 0.10 (c) | 10.9 ± 0.72 (c) | 0.31 ± 0.02 (c) |
Mitochondrial respiration and DNA damage were measured as in Materials and Methods. Values are means ± SEM; number of samples analyzed = 12 in each treatment group. Within a column mean values not sharing the same letter (a, b, c, or d) are significantly different (P < 0.05). Data were ln-transformed prior to statistical analysis.
Figure 1.
Relationship between mitochondrial 1/RCR and mtDNA fragmentation and liver nonheme-iron levels. (A) Representative Southern blots from the four groups showing mtDNA forms and fragments. mtDNA fragmentation was determined by using densitometry of digitized images from Southern blots of agarose gels, as described in Materials and Methods. (B) Nonlinear correlation between 1/ln-RCR (left ordinate), ln-mtDNA fragmentation normalized to supercoiled mtDNA (right ordinate) and ln-nonheme liver-iron stores. Boxes (□) represent averages from 12 animals ± SE; regression analysis was performed using all data points (n = 48). Mitochondria and mtDNA were isolated from iron-deficient rats (○), iron-normal control rats (⋄), daily high-iron-supplemented iron-normal rats (♦), and daily high-iron-supplemented iron-deficient rats (●), as described in Materials and Methods. RCR was determined by measuring the ratio of state 3 to state 4 oxygen consumption by using a Clark-type oxygen electrode, as described in Materials and Methods.
mtDNA fragmentation in liver.
mtDNA fragmentation increased in the iron-deficient and iron-supplemented rats 3.5- and 1.4-fold, respectively, compared with the iron-normal rats (P < 0.05, Table 1), as measured by densitometric analysis of the Southern blots. The ln-mtDNA fragmentation pattern also correlated nonlinearly with the ln values of liver iron (R2 = 0.75, Fig. 1A, B Upper). Iron-deficient rats and iron-deficient rats supplemented with daily high doses of iron had more linear mtDNA (indicative of double-strand breaks) in liver mitochondria than did iron-normal rats. (Table 1, P < 0.001).
Flow cytometry analysis of WBC oxidant production using 2′7′-dichlorofluorescein hydrochloride (DCF).
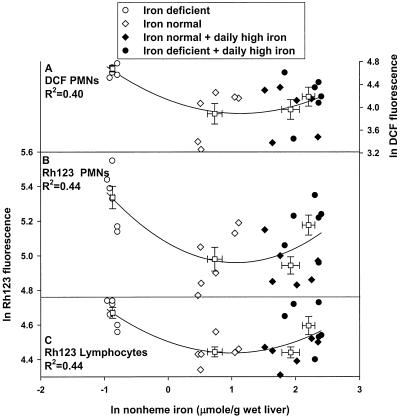
When oxidized, the nonfluorescent probe DCFH produces the fluorescent product DCF, which can be monitored by using flow cytometry (26). PMNs from the iron-deficient group had twice the DCF fluorescence (P < 0.05), indicative of increased oxidants relative to the iron-normal group. The normal group was not different from both iron supplement groups (Table 2). The relationship between ln values of DCF fluorescence in PMNs correlates with ln values of liver iron as a second-order polynomial regression (R2 = 0.4; Fig. 2A). Isolated lymphocytes from the iron-deficient iron-supplemented rat groups also demonstrated an increased trend in DCF fluorescence that was consistent with that in PMNs; however, it did not reach significance (Table 2).
Table 2.
Effect of iron deficiency and daily high-iron supplementation on the fluorescent intensity of DCF or Rh123 during flow cytometry of isolated white blood cells from rats
| Treatment | DCF fluorescence (mean channel no.)
|
Rh123 fluorescence (mean channel no.)
|
||
|---|---|---|---|---|
| PMNs | Lymphocytes | PMNs | Lymphocytes | |
| Iron deficient | 108.5 ± 6.84 (a) | 51.8 ± 5.36 (a) | 210.1 ± 13.4 (b) | 106.7 ± 3.27 (b) |
| Iron normal | 52.4 ± 7.89 (a) | 35.3 ± 6.41 (a) | 147.3 ± 10.1 (a) | 85.3 ± 2.43 (a) |
| Iron normal + daily high iron | 56.2 ± 8.40 (a) | 41.2 ± 6.54 (a) | 141.4 ± 7.4 (a) | 85.0 ± 2.58 (a) |
| Iron deficient + daily high iron | 69.8 ± 9.64 (a, b) | 49.6 ± 10.5 (a) | 178.3 ± 10.1 (a, b) | 99.7 ± 5.08 (b) |
Fluorescent intensity of DCF or Rh123 as measured by flow cytometry of isolated PMNs or lymphocytes. Values are means ± SEM; number of samples analyzed = 6. In each treatment group within a column, mean values not sharing the same letter (a or b) are significantly different (P < 0.05). Data were ln-transformed prior to statistical analysis.
Figure 2.
Relationship between liver nonheme iron and DCF fluorescence (A) and Rh123 fluorescence (B and C) in PMNs and lymphocytes in rats. PMNs and lymphocytes were isolated from the different iron treatment groups (as in Fig. 1) as described in Materials and Methods. Boxes (□) represent averages of ln-DCF (right ordinate) or ln-Rh123 fluorescence (left ordinate, mean channel number) from six animals ± SE from each iron-treatment group. Cells were collected immediately after the blood draw and subjected to flow cytometry, as described in Materials and Methods. Nonlinear regression analysis was performed using all data points (24 animals).
Flow cytometry analysis of mitochondria using Rh123.
Rh123 is a lipophilic cationic probe that is readily sequestered by active mitochondria and that produces a fluorescent signal (27). The Rh123 florescence of PMNs was at least 40% higher in the iron-deficient group and the iron-normal group that had received high-iron supplementation compared with the iron-normal group (P < 0.05, Table 2). Lymphocytes from rats in both the iron-deficient group and the iron-deficient group that had received daily high-iron doses had approximately a 20% (P < 0.05) elevation of Rh123 fluorescence relative to the iron-normal group and the iron-normal group that received high-iron supplementation (Table 2). The ln values of Rh123 fluorescence of PMNs and lymphocytes (Fig. 2 B and C) also showed a nonlinear correlation to ln-liver nonheme iron (R2 = 0.44).
Analysis of nonlinear regression models: An overall model and individual models compared.
Z scores of the six dependent variables that were modeled as quadratic functions of the natural logarithm (ln) using liver-iron values as the independent variable (as described in Material and Methods) were compared with a single combined model in Figure 3. This analysis indicated that all variables behaved similarly (Fig. 3, P = 0.356). The quadratic functions obtained from all data points were:
Single combined model: Y = −0.306 − 0.838X + 0.460X2.
Variable 1 (1/RCR): Y = −0.421 − 1.162X + 0.643X2.
Variable 2 (DCF PMNs) Y = −0.257 − 0.973X + 0.490X2.
Variable 3 ( DCF Lymphocytes) Y = −0.179 − 1.000X + 0.465X2.
Variable 4 (Rh123 PMNs) Y = −0.186 − 0.434X + 0.250X2.
Variable 5 ( Rh123 Lymphocytes) Y = −0.492 − 0.559X + 0.435X2.
Variable 6 ( Liver mtDNA fragments) Y = −0.547 − 1.175X + 0.749X2.
The function of the inverse RCRs showed the greatest changes between the liver iron from iron-deficient and iron-deficient supplemented rats. Consequently, although the generalized F test among all of the functions showed no differences, the ln values of the 1/RCR quadratic function was tested against the ln values of the other five to determine whether its behavior approached a significant difference; it did not (P = 0.153). Thus, all of the variables show the same response to variation in liver nonheme-iron levels; this result indicates that iron deficiency or excess changes these biochemical parameters in a similar manner and magnitude.
Study 2: Comparison of Daily and Every-Third-Day Iron Supplementation.
Mitochondrial respiration.
Compared with normal rats (RCR = 2.0 ± 0.06, n = 4), liver mitochondrial RCRs were significantly lower in pooled data from rats killed on days 1, 2, and 3 after the last intermittent supplemental iron dose (RCR = 1.7 ± 0.1, n = 12), as well as iron-deficient rats that received daily high-iron supplements (RCR = 1.5 ± 0.12, n = 8, P < 0.01). The difference in mean RCRs between the high-iron-supplemented groups was not significant.
mtDNA fragmentation.
Densitometry of Southern blots showed significant liver mtDNA damage in iron-deficient rats supplemented daily and only in mitochondria isolated one day after the last intermittent dose given to iron-deficient rats (Table 3). mtDNA damage was progressively less 2 and 3 days after the last intermittent iron dose and did not significantly differ anymore from that in iron-normal rats.
Table 3.
Effect of daily and every-three-day high-iron supplementation on mtDNA damage of iron-deficient rats
| Treatment and timing of sampling | mtDNA damage*Fragmented DNA supercoiled DNA |
|---|---|
| Iron normal | 0.17 ± 0.01 (n = 6) (a) |
| Iron deficient + daily high iron | 0.36 ± 0.03 (n = 12) (b) |
| Iron deficient + iron every 3 days (1 day post) | 0.31 ± 0.02 (n = 6) (b, c) |
| Iron deficient + iron every 3 days (2 days post) | 0.26 ± 0.06 (n = 6) (a, b, c) |
| Iron deficient + iron every 3 days (3 days post) | 0.22 ± 0.03 (n = 6) (a, b) |
Mitochondrial DNA damage was measured as in Materials and Methods. Values are means ± SEM. Within a column, mean values not sharing the same letter (a, b, or c) are significantly different (P < 0.05).
Data were ln-transformed prior to statistical analysis.
Discussion
Severely anemic, iron-deficient rats, as well as rats supplemented daily with high iron, were shown previously to have increased lipid peroxidation, as indicated by elevated levels of malondialdehyde in liver and kidney and ethane in breath (9). The current study shows that iron deficiency increased oxidant levels in PMNs and mitochondrial fluorescence of Rh123 (in PMNs and lymphocytes) and liver mtDNA damage. Iron deficiency also decreased liver mitochondrial RCR, a measure of respiratory efficiency.
The statistical relationship of all these mitochondrial biomarkers to liver nonheme iron content has a U shape (nonlinear polynomial correlation, Fig. 3); plotting 1/RCR (inefficiency) also shows a U shape. The data show that a range of liver nonheme iron levels is associated with normal function, and that both iron deficiency and moderate iron excess induce functional deterioration of mitochondria. This response to trace elements (first observed in plant mineral nutrition) is known as Bertrand's Rule (28). Mertz (29) has proposed that Bertrand's Rule also is relevant to human nutritional needs. The uniformity of our results obtained from the nonlinear regressions of Z-transformed ln values of mitochondrial-dependent variables against the ln values of liver nonheme iron (Fig. 3) provides evidence that mitochondrial dysfunction and mtDNA damage can be described by a single model that quantifies the effects of both iron deficiency and moderate excess.
Liver Mitochondria.
The iron-deficient rats and those that received daily high iron had impaired liver mitochondrial RCR relative to the iron-normal rats as well as higher levels of oxidants and lipid peroxidation (9). The low RCRs indicate an uncoupling of electron transport to oxidative phosphorylation. The increased level of oxidants and lipid peroxidation in both these groups may have damaged the integrity of the mitochondrial membrane, which resulted in the uncoupling.
The higher level of oxidants found in the iron-deficient group could be caused by decreased heme levels and complex IV activity (ref. 30; see below); iron excess could result in more reactive oxidants because of increased Fenton chemistry. The increased rate of state 4 respiration in iron-deficient relative to iron-normal animals also was found by Masini et al. (18), who suggested this could be caused by fatty acid-induced uncoupling. Masini et al. reached this conclusion because of the increased serum total lipid and triglycerides associated with iron deficiency (9, 31), and because inclusion of BSA in their mitochondrial respiration studies prevented the loss of respiratory control in iron-deficient animals (18). The mitochondrial preparation used in the present work included BSA in the final washes, thus making uncoupling because of free fatty acids unlikely.
The lower RCRs of both iron-supplemented groups of rats relative to control animals supports past research that iron overload impairs mitochondrial function (11). Liver-iron loading reported in those studies was extremely high (15- to 100-fold higher than normal) to model human hemochromatosis. In the present study, adverse effects were seen when daily-iron-supplemented rats had mean liver nonheme iron levels that were only 2.7-fold higher than normal.
Although iron deficiency has never been shown previously to induce mtDNA damage, excess iron-induced damage to mtDNA has been reported (12, 13), as have double-strand breaks in isolated mitochondria subjected to mM concentrations of Fe2+ (32). Iron deficiency can induce nuclear DNA base damage (33). Whereas every-third-day-supplemented rats had levels of mtDNA damage that were not significantly higher than iron-normal rats on the second and third day after treatment, daily high-iron-supplemented rats had damage that was significantly different from iron-normal rats. This finding and the progressive decline in DNA damage 2 and 3 days after the last dose suggest either that there was repair of mtDNA in the 3 days after a high-iron dose, or that this feeding regimen was less severe.
WBCs.
The 2-fold increase in DCF fluorescence of PMNs isolated from iron-deficient rats indicates that oxidants are increased, possibly because of mitochondrial oxidant production (34). A plausible mechanism is that the lack of heme-containing electron-transport proteins (18) such as cytochrome c oxidase (complex IV) and/or iron sulfur clusters in the mitochondria promotes oxidant leakage (30) (see below).
Rh123 fluorescence increased in both PMNs and lymphocytes both in iron deficiency and excess. In parallel with our findings, studies have shown increased Rh123 fluorescence during oxidant stress (35) and aging (36, 37). It was suggested in these studies that the increase in fluorescence was caused by changes in mitochondrial morphology. Our finding of an increase in Rh123 fluorescence could be because of an augmentation of membrane potential (27), an increase in Rh123 uptake (38), or a decreased efflux, as has been proposed for other probes in apoptosis (39). We think the increased fluorescence was caused by changes in mitochondrial morphology because of the swelling associated with iron deficiency (16) or excess (40). Furthermore, apoptosis, which can follow the mitochondrial swelling associated with permeability transition, is often accompanied by a transient increase in R123 fluorescence (38). Previous research has shown that iron deficiency and excess can also impair immune function (41, 42) and neutrophil activation and metabolism (43, 44). These findings strongly suggest that both iron deficiency and moderate excess are detrimental to mitochondria.
Further Mechanisms by Which Iron Deficiency Can Cause Damage to Mitochondria and Its DNA.
We suggest four possible mechanisms:
(i) The uncoupling of mitochondria (low RCRs) can increase mitochondrial superoxide release (45), probably due to decreased heme availability (30, 46). Decreased cytochrome concentration (18) of the electron-transport chain could also possibly contribute to increased superoxide levels. The increased superoxide released from these uncoupled mitochondria could account for the observed elevation of liver MDA, mtDNA damage, and increased DCF fluorescence in PMNs.
(ii) Gastrointestinal up-regulation of iron absorption during iron deficiency increases copper absorption (47) and hepatic copper accumulation in rats (48). The increased copper absorption may be mediated by divalent metal transporter 1 (DMT1), which is dramatically up-regulated in iron-deficient duodenum (49). Copper can participate in Fenton chemistry with H2O2 generating reactive hydroxyl radicals that can damage lipids and DNA (50). Moreover, excess copper can cause severe mitochondrial dysfunction and mtDNA damage, as has been reported in the livers of patients with copper overload caused by Wilson's disease (51, 52).
(iii) Iron deficiency causes loss of activity of some important iron-containing repair enzymes. For example, the activity of ribonucleotide reductase is decreased in iron-deficient cell cultures (53); this could lead to decreased availability of deoxyribonucleotides for DNA repair. Treatment of cell cultures with desferrioxamine lowered iron levels as well as dNTPs (54) and DNA synthesis (53).
(iv) Iron deficiency also induces changes in the cellular iron homeostasis system. First, dietary iron deficiency causes an increase in liver IRP1 activity, which increases transferring receptor synthesis and decreases mitochondrial-aconitase activity (55). The decrease in mitochondrial-aconitase may prevent further mitochondrial release of oxidants by diminishing the supply of reducing equivalents to the electron-transport chain (55). The reduced electron flow may be one way by which the cell protects itself from oxidant stress in iron deficiency (55). However, after 3 weeks of iron deficiency, IRP1 activity is increased, and there is a marked decrease in ferritin synthesis (55), thus decreasing its potential as an antioxidant protein. Up-regulation of transferrin receptor (TFR) first maintains or increases cytosolic iron availability in the face of the initial iron deficiency. However, with TFR synthesis, there is down-regulation of liver DMT1 (56), shutting down a nontransferrin-dependent iron-uptake pathway. At the same time, decreasing iron supply may induce an increase in intracellular iron trafficking from iron stores in ferritin and hemosiderin. These two effects may account for some early increase in the potential for iron availability to catalyze some of the oxidant-induced damage. Last, ferritin sequesters Fe3+ and, through its ferroxidase activity, converts Fe2+ to Fe3+, both of which protect against oxidant-induced damage. Clearly, ferritin synthesis is decreased in iron deficiency.
In conclusion, these results extend our previous findings (9) and demonstrate that both iron deficiency and sustained moderate iron excess from daily supplementation are undesirable. Intermittent supplementation may mitigate the undesirable consequences and deserves consideration as a preventive measure for iron deficiency in at-risk populations, given its proven efficacy in human studies (57, 58). Iron deficiency is potentially more damaging than previously suspected; given its high prevalence worldwide, the prevention and safe treatment of iron deficiency deserve urgent attention.
Acknowledgments
We thank Tory Hagen and Hani Atamna for their helpful discussions and suggestions and Mark Hudes for his help with statistics. We also thank Benjamin Blount, Jiankang Liu, and Carol Wehr for help in experimental procedures and Mark Shigenaga, David Killilea, John Nides, Stuart Linn, and Elizabeth Theil for suggestions on the manuscript. This work was supported by the Wheeler Fund of the University of California, National Foundation for Cancer Research Grant 00CHORI, Ellison Medical Foundation Grant SS-0422–99, Department Of Energy Grant DE-FG03–00ER6Z943, and National Institute of Environmental Health Sciences Center Grant P30 ES01896 (to B.N.A.).
Abbreviations
- mtDNA
mitochondrial DNA
- PMNs
polymorphonuclear-leukocytes
- RCR
respiratory control ratio
- Rh123
rhodamine 123
- DCF
2′7′-dichlorofluorescein hydrochloride
- DCFH
2′7′-dichlorodihydrofluorescein diacetate
References
- 1.World Health Organization. Maternal Health And Safe Motherhood Program, Nutrition Program. Geneva: W.H.O.; 1992. p. 100. [Google Scholar]
- 2.Viteri F E. Nutr Rev. 1997;55:195–209. doi: 10.1111/j.1753-4887.1997.tb01607.x. [DOI] [PubMed] [Google Scholar]
- 3.Looker A C, Dallman P R, Carroll M D, Gunter E W, Johnson C L. J Am Med Assoc. 1997;277:973–976. doi: 10.1001/jama.1997.03540360041028. [DOI] [PubMed] [Google Scholar]
- 4.Yip R. In: Present Knowledge in Nutrition. Bowman B A, Russell R M, editors. Washington, DC: International Life Sciences Institute; 2001. pp. 311–328. [Google Scholar]
- 5.Pollitt E. Annu Rev Nutr. 1993;13:521–537. doi: 10.1146/annurev.nu.13.070193.002513. [DOI] [PubMed] [Google Scholar]
- 6.Stoltzfus R J, Dreyfuss M L. A Report of the International Nutritional Anemia Consultative Group. Washington, DC: The Nutrition Foundation; 1999. [Google Scholar]
- 7.Hollan S, Johansen K S. Haematologia. 1993;25:69–84. [PubMed] [Google Scholar]
- 8.Viteri F E, Liu X, Tolomei K, Martin A. J Nutr. 1995;125:82–91. doi: 10.1093/jn/125.1.82. [DOI] [PubMed] [Google Scholar]
- 9.Knutson M D, Walter P B, Ames B N, Viteri F E. J Nutr. 2000;130:621–628. doi: 10.1093/jn/130.3.621. [DOI] [PubMed] [Google Scholar]
- 10.Shigenaga M K, Hagen T M, Ames B N. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britton R S, Ramm G A, Olynyk J, Singh R, O'Neill R, Bacon B R. Adv Exp Med Biol. 1994;356:239–253. doi: 10.1007/978-1-4615-2554-7_26. [DOI] [PubMed] [Google Scholar]
- 12.Yaffee M, Walter P, Richter C, Muller M. Proc Natl Acad Sci USA. 1996;93:5341–5346. doi: 10.1073/pnas.93.11.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsuhashi N, Miki T, Senbongi H, Yokoi N, Yano H, Miyazaki M, Nakajima N, Iwanaga T, Yokoyama Y, Shibata T, Seino S. J Biol Chem. 2000;275:17536–17540. doi: 10.1074/jbc.275.23.17536. [DOI] [PubMed] [Google Scholar]
- 14.Wallace D C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 15.Dallman P R. Annu Rev Nutr. 1986;6:13–40. doi: 10.1146/annurev.nu.06.070186.000305. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis J H, Jacobs A. J Clin Pathol. 1974;27:973–979. doi: 10.1136/jcp.27.12.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallman P R, Goodman J R. J Cell Biol. 1971;48:79–90. doi: 10.1083/jcb.48.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masini A, Salvioli G, Cremonesi P, Botti B, Gallesi D, Ceccarelli D. Biochim Biophys Acta. 1994;1188:46–52. doi: 10.1016/0005-2728(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 19.Klempa K L, Willis W T, Chengson R, Dallman P R, Brooks G A. J Appl Physiol. 1989;67:1868–1872. doi: 10.1152/jappl.1989.67.5.1868. [DOI] [PubMed] [Google Scholar]
- 20.Jansson L T, Perkkio M V, Willis W T, Refino C J, Dallman P R. Acta Haematol. 1985;74:218–221. doi: 10.1159/000206222. [DOI] [PubMed] [Google Scholar]
- 21.Tapiero H, Gate L, Tew K D. Biomed Pharmacother. 2001;55:324–332. doi: 10.1016/s0753-3322(01)00067-1. [DOI] [PubMed] [Google Scholar]
- 22.Torrance J D, Bothwell T H. S Afr Med J. 1968;33:9–11. [PubMed] [Google Scholar]
- 23.Klingenberg M, Slenczka W. Biochem Z. 1959;331:486–517. [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Enzmann H, Wiemann C, Ahr H J, Schluter G. Mutat Res. 1999;425:213–224. doi: 10.1016/s0027-5107(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 26.Robinson J P, Bruner L H, Bassoe C F, Hudson J L, Ward P A, Phan S H. J Leukocyte Biol. 1988;43:304–310. doi: 10.1002/jlb.43.4.304. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro H M. Methods. 2000;21:271–279. doi: 10.1006/meth.2000.1007. [DOI] [PubMed] [Google Scholar]
- 28.Bertrand G. Eighth Int Congr Appl Chem. 1912;28:30–40. [Google Scholar]
- 29.Mertz W. Science. 1981;213:1332–1338. doi: 10.1126/science.7022654. [DOI] [PubMed] [Google Scholar]
- 30.Atamna H, Liu J, Ames B N. J Biol Chem. 2001;276:48410–48416. doi: 10.1074/jbc.M108362200. [DOI] [PubMed] [Google Scholar]
- 31.Masini A, Trenti T, Caramazza I, Predieri G, Gallesi D, Ceccarelli D. Biochim Biophys Acta. 1994;1188:53–57. doi: 10.1016/0005-2728(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 32.Asin J, Perez-Martos A, Fernandez-Silva P, Montoya J, Andreu A L. FEBS Lett. 2000;480:161–164. doi: 10.1016/s0014-5793(00)01768-3. [DOI] [PubMed] [Google Scholar]
- 33.Adachi S, Takemoto K, Hirosue T, Hosogai Y. Carcinogenesis. 1993;14:265–268. doi: 10.1093/carcin/14.2.265. [DOI] [PubMed] [Google Scholar]
- 34.Swift L M, Sarvazyan N. Am J Physiol. 2000;278:H982–H990. doi: 10.1152/ajpheart.2000.278.3.H982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soltys B J, Gupta R S. J Cell Physiol. 1994;159:281–294. doi: 10.1002/jcp.1041590212. [DOI] [PubMed] [Google Scholar]
- 36.Atamna H, Paler-Martinez A, Ames B N. J Biol Chem. 2000;275:6741–6748. doi: 10.1074/jbc.275.10.6741. [DOI] [PubMed] [Google Scholar]
- 37.Martinez A O, Vara C, Castro J. Mech Ageing Dev. 1987;39:1–9. doi: 10.1016/0047-6374(87)90081-9. [DOI] [PubMed] [Google Scholar]
- 38.Darzynkiewicz Z, Traganos F, Staiano-Coico L, Kapuscinski J, Melamed M R. Cancer Res. 1982;42:799–806. [PubMed] [Google Scholar]
- 39.Ferlini C, Di Cesare S, Rainaldi G, Malorni W, Samoggia P, Biselli R, Fattorossi A. Cytometry. 1996;24:106–115. doi: 10.1002/(SICI)1097-0320(19960601)24:2<106::AID-CYTO2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 40.Thakerngpol K, Fucharoen S, Boonyaphipat P, Srisook K, Sahaphong S, Vathanophas V, Stitnimankarn T. Biometals. 1996;9:177–183. doi: 10.1007/BF00144623. [DOI] [PubMed] [Google Scholar]
- 41.Strauss R G. Am J Clin Nutr. 1978;31:660–666. doi: 10.1093/ajcn/31.4.660. [DOI] [PubMed] [Google Scholar]
- 42.Walker E M, Jr, Walker S M. Ann Clin Lab Sci. 2000;30:354–365. [PubMed] [Google Scholar]
- 43.Mackler B, Person R, Ochs H, Finch C A. Pediatr Res. 1984;18:549–551. doi: 10.1203/00006450-198406000-00015. [DOI] [PubMed] [Google Scholar]
- 44.Olynyk J K, Clarke S L. J Gastroenterol Hepatol. 2001;16:438–444. doi: 10.1046/j.1440-1746.2001.02456.x. [DOI] [PubMed] [Google Scholar]
- 45.Cadenas E, Davies K J. Free Radical Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 46.Atamna H, Walter P B, Ames B N. Arch Biochem Biophys. 2002;397:345–353. doi: 10.1006/abbi.2001.2671. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez-Matas M C, Lisbona F, Gomez-Ayala A E, Lopez-Aliaga T, Campos M S. Lab Anim. 1998;32:298–306. doi: 10.1258/002367798780559248. [DOI] [PubMed] [Google Scholar]
- 48.Sherman A R, Moran P E. J Nutr. 1984;114:298–306. doi: 10.1093/jn/114.2.298. [DOI] [PubMed] [Google Scholar]
- 49.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Nature (London) 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 50.Bremner I. Am J Clin Nutr. 1998;67:1069S–1073S. doi: 10.1093/ajcn/67.5.1069S. [DOI] [PubMed] [Google Scholar]
- 51.Gu M, Cooper J M, Butler P, Walker A P, Mistry P K, Dooley J S, Schapira A H. Lancet. 2000;356:469–474. doi: 10.1016/s0140-6736(00)02556-3. [DOI] [PubMed] [Google Scholar]
- 52.Mansouri A, Gaou I, Fromenty B, Berson A, Letteron P, Degott C, Erlinger S, Pessayre D. Gastroenterology. 1997;113:599–605. doi: 10.1053/gast.1997.v113.pm9247482. [DOI] [PubMed] [Google Scholar]
- 53.Furukawa T, Naitoh Y, Kohno H, Tokunaga R, Taketani S. Life Sci. 1992;50:2059–2065. doi: 10.1016/0024-3205(92)90572-7. [DOI] [PubMed] [Google Scholar]
- 54.Lederman H M, Cohen A, Lee J W W, Freedman M H, Gelfand E W. Blood. 1984;64:748–753. [PubMed] [Google Scholar]
- 55.Chen O S, Schalinske K L, Eisenstein R S. J Nutr. 1997;127:238–248. doi: 10.1093/jn/127.2.238. [DOI] [PubMed] [Google Scholar]
- 56.Trinder D, Oates P S, Thomas C, Sadleir J, Morgan E H. Gut. 2000;46:270–276. doi: 10.1136/gut.46.2.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Administrative Committee on Coordination/Sub-Committee on Nutrition. Fourth Report of the World Nutrition Situation. Geneva, Switzerland: ACC/SCN; 2000. p. 121. [Google Scholar]
- 58.Viteri F E. Arch Latinoam Nutr. 1999;49:15S–22S. [PubMed] [Google Scholar]