Abstract
The Cre/loxP system is increasingly showing promise for investigating genes involved in neural function. Here, we demonstrate that in vivo modification of genes in the mouse brain can be accomplished in a spatial- and temporal-specific manner by targeted delivery of an adeno-associated virus (AAV) encoding a green fluorescent protein/Cre recombinase (GFP/Cre) fusion protein. By using a reporter mouse, in which Cre recombinase activates β-galactosidase expression, we demonstrate long-term recombination of neurons in the hippocampus, striatum, and septum as early as 7 days after stereotaxic injection of virus. Recombined cells were observed for at least 6 months postinjection without evidence of cell loss or neural damage. AAV-mediated delivery of GFP/Cre provides a valuable approach to alter the mouse genome, as AAV delivers genes efficiently to neurons with low toxicity. This approach will greatly facilitate the study of genetic modifications in the mouse brain.
Keywords: gene transfer‖Cre recombinase‖transgenic mouse‖loxP‖ROSA26
The analysis of gene function in vivo has progressed rapidly as a result of our ability to engineer specific genetic mutations in mice (1). Transgenic mouse models have allowed the elucidation of molecular and cellular mechanisms underlying neurodegenerative disease (2–4) and behavior (5, 6). The more recent demonstration that controlled, site-specific recombination by Cre recombinase is feasible in mammalian cells has led to the development of approaches for temporally and spatially controlled gene ablation in vivo (7). These genetic approaches have provided the possibility of exploring the role of genes in select brain regions without confounding interpretations of phenotype stemming from effects on other brain functions, altered development or embryonic lethality, as may occur in global knockouts.
This “second generation” knockout approach relies on the ability of Cre recombinase to excise DNA sequences located between loxP sites. The Cre/loxP system is a phage P1-derived, site-specific recombination system in which Cre recombinase catalyzes the recombination between 34-bp loxP recognition sequences (8–10). The loxP sites are engineered around crucial exons by gene targeting in such a way so as not to affect normal gene function (11). Upon Cre recombinase expression in selective tissue regions, excision of the DNA segment between the loxP sites occurs. This excision results in a loss of the crucial exon and subsequent loss or gain of gene function in that tissue (8, 11).
To date, approaches for site-specific recombination in the brain have depended on engineering transgenic mice so that Cre recombinase is placed under the control of a brain region-selective promoter (7, 12). Recently, mice have been engineered to allow Cre recombinase to be regulated spatially and temporally, by using a ligand-inducible system under the control of region-selective promoters (13–15). These mice expressing Cre recombinase were then mated to mice engineered with loxP sites to allow inducible gene ablation.
Although showing substantial promise, this approach has important limitations. First, promoters for selective expression in the brain region of interest are not always available. Second, expression of a construct integrated at different genomic locations often varies because of position variegation effects, necessitating the generation and analysis of multiple lines of transgenic founder lines. Third, it is necessary to cross the Cre recombinase-expressing mouse line with mice engineered with loxP sites, a process that is resource-intensive and can increase the variation in the genetic background of the resultant offspring, potentially confounding behavioral and other analyses. Finally, it is conceivable that inducible systems may be leaky, resulting in low-level Cre recombinase expression in the absence of ligand and allowing for gene knockout to occur without induction.
A potential solution to this problem is to use recombinant viruses to deliver Cre recombinase to the target region of the adult animal. Previous studies from other laboratories have shown the utility of adenovirus (16–18) and herpes virus (6, 19) to deliver functional Cre recombinase to the mouse in vivo. However, the potential for host immune response against the viral genes of these vectors poses substantial difficulties. Recently, a number of vectors have emerged that show great promise as gene delivery vehicles to the nervous system. Because lentiviral and adeno-associated viral (AAV) vectors are deleted of all viral genes, they are nontoxic and elicit a minimal immune response (20–22). Reports on AAV vectors have shown long-term gene expression in the central nervous system (21, 23–25). Here, we report that stereotaxic injections of AAV expressing a Cre recombinase fused with green fluorescent protein (GFP) can mediate extensive recombination in neural cells of defined brain regions, including the hippocampus, striatum, septum, and substantia nigra. This approach will prove useful for selective ablation of genes in the adult nervous system without necessitating the generation of transgenic mouse lines expressing inducible Cre recombinase.
Materials and Methods
Vector Construction, Viral Production, and Purification.
AAV-GFP/Cre vector was constructed by PCR of Cre306c with BspE1 and XhoI ends and cloning into pBluescript II vector containing GFP cloned into the Eco47III and XhoI sites, creating an N-terminal fusion of GFP (CLONTECH) to Cre recombinase. The AAV plasmid containing the inverted terminal repeats contained the human immediate early cytomegalovirus promoter (CMV) with a splice donor/acceptor sequence and polyadenylation signal from human β-globin gene with a multiple cloning site to insert transgenes. AAV-GFP vector was constructed by cloning the Eco47III and XhoI fragment from pBluescript II into the AAV multiple cloning site.
Human embryonic kidney 293 cells (American Type Culture Collection) were propagated as described (26). Recombinant AAV-2 encoding GFP or the GFP-Cre recombinase fusion was produced by calcium phosphate transient triple transfection of vector plasmid, pAAV/Ad8 helper plasmid, and pXX6 as previously described (27). All virus preparations were purified by two cesium chloride density gradients, dialyzed against Hepes-buffered saline, and heated at 56°C for 45 min to ensure purity and stability of the vector. Recombinant virus titers were determined as infectious titer per cell (multiplicity of infection) and found to be 1 × 109 infectious particles per milliliter. All viral stocks were tested and found to be free from contaminating adenovirus, as described (28).
In Vivo AAV Injections and Tissue Preparation.
Homozygous ROSA26 mice (18–20 weeks of age) were anesthetized by i.p. injection of the following mixture: 100 mg/kg ketamine and 10 mg/kg xylazine. All surgical procedures were performed in sterile conditions and in accordance with National Institutes of Health and Salk guidelines for animal care and use. Mice were positioned in a Kopf stereotaxic frame for injection into the hippocampus (−2.0 anterior–posterior, −1.5 mediolateral, and −2.0 dorsoventral), striatum (+0.5 anterior–posterior, +2.0 mediolateral, and −2.5 dorsoventral), and medial septum (+0.8 anterior–posterior, +0.1 mediolateral, and −3.8 to −3.6 dorsoventral). One microliter of AAV-GFP or AAV-GFP/Cre recombinase was injected per site at a rate of 0.50 μl/min by using a 5-μl Hamilton syringe with a 30-gauge beveled needle. Following virus delivery, the syringe was left in place for 1 min, raised 0.5 mm, and left in place an additional minute before being slowly withdrawn from the brain.
For histological analyses, animals were anesthetized and perfused through the ascending aorta with 50 ml of PBS followed by 150 ml of 4% paraformaldehyde in 0.1 M phosphate buffer. After perfusion, the brains were removed and incubated several additional hours in the same fixative, followed by phosphate-buffered 30% sucrose for cryoprotection. Serial 40-μm-thick coronal sections were cut on a freezing microtome and collected in 0.1 M Tris buffer.
Immunohistochemistry and Cell Counting.
Multiple immunofluorescent labeling used antibodies against β-galactosidase (β-gal; rabbit, 1:1,000 Cortex or mouse, 1:1,000 Promega), the neuron-specific nuclear protein NeuN (mouse, 1:50, R. Mullen, Univ. Utah), tyrosine hydroxylase (rabbit, 1:1,000, Chemicon), anti-glial fibrillary acidic protein (guinea pig, 1:1,000, Advanced Immunochemical), choline acetyltransferase (ChAT; goat, 1:250, Chemicon), anti-CD4 and CD8 (mouse, 1:1,000, PharMingen), and anti-GFP (rabbit, 1:500, CLONTECH) to enhance detection of the reporter gene. Donkey anti-species antibodies conjugated to biotin, FITC, Cy3, or Cy5 and streptavidin-FITC (1:250; all from Jackson ImmunoResearch) were used for detecting primary antibodies. 4′,6-diamidino-2-phenylindole (30 ng/ml, Molecular Probes) was used as a fluorescent DNA stain. Microscopy was performed by using a confocal microscope (Bio-Rad MRC1024UV). Quantification of β-gal-positive cells at each time point [2 days (n = 3), 7 days (n = 3), 14 days (n = 4), 28 days (n = 5), and 6 months (n = 2)] was performed on every sixth section by using the optical fractionator procedure (MicroBrightField, Colchester, VT) as previously described (29, 30). Statistical analysis was performed by multiway ANOVA followed by a Bonferroni post hoc analysis of means differences between groups (GraphPad, San Diego).
Results
Recombinant AAV Constructs Are Functional in Vitro.
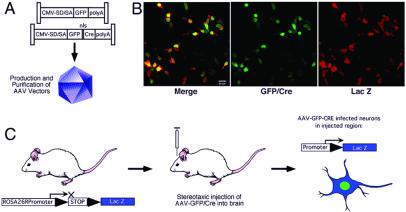
As shown in Fig. 1A, two different AAV constructs were designed. One contains a cDNA encoding GFP only (AAV-GFP). The other contains a cDNA encoding an N-terminal GFP, fused in frame to a nuclear localization signal from SV40 DNA and Cre recombinase (AAV-GFP/Cre). Gene expression in both of these constructs is driven by a CMV immediate early gene promoter.
Figure 1.
The AAV-GFP/Cre loxP system for recombination. (A) Schematic representation of the AAV vectors used in this study (control AAV-GFP and AAV-GFP/Cre recombinase; nls, nuclear localization signal). Expression was driven by the CMV promoter with a splice donor/acceptor sequence (SD/SA) and flanked by the inverted terminal repeats. High titer, purified virus was prepared for stereotaxic injection into the adult ROSA26 reporter mouse at the desired time point. (B) In vitro validation of Cre-mediated recombination in HEK293 cells transfected with a loxP-flanked β-gal reporter construct that contains a stuffer sequence that stops gene transcription surrounded by loxP sites. Upon Cre/loxP-mediated recombination, the stuffer sequence is excised, allowing transcription. Infection of 1 × 105 cells with AAV-GFP/Cre at a multiplicity of infection of 1 demonstrates the high efficiency of the system for virus-mediated recombination and expression of β-gal 2 days following infection. (Scale bar = 20 μm.) (C) Schematic of in vivo AAV gene delivery and recombination in the ROSA26 reporter mice. Transcription of the ROSA26 reporter before recombination is blocked because of a stop (polyadenylation) sequence. Upon stereotaxic delivery of AAV-GFP/Cre to the brain, infected neurons undergo recombination and excise the stop sequence, allowing transcription of lacZ. (Scale bar = 20 μm.)
We tested the functionality of these AAV constructs by infecting HEK293 cells in vitro. Infected cells were identified by GFP fluorescence. In cells infected with AAV-GFP/Cre, GFP fluorescence was localized primarily to the nuclei, whereas cells infected with AAV expressing GFP alone demonstrated diffuse cytoplasmic and nuclear fluorescence. Functionality of GFP/Cre fusion was subsequently demonstrated in vitro by infecting HEK cells that contained a reporter plasmid that is nonfunctional until GFP/Cre-mediated recombination removes a loxP-flanked stuffer sequence, allowing β-gal expression to occur (similar to the reporter construct used in the ROSA26 reporter mouse) (31). By counting fluorescent cells, we calculated the titer to be 1 × 109 infectious particles per ml. In essentially all cells in vitro that were GFP-fluorescent, β-gal expression was observed, indicating that GFP/Cre efficiently induces loxP-mediated recombination (Fig. 1B).
Recombinant AAV Vectors Are Functional in the Mouse Brain in Vivo.
We used a reporter mouse strain generated by Soriano as a model system to study the functionality of AAV-GFP/Cre in vivo (31). In this mouse, a β-gal reporter gene is genetically engineered into the ubiquitously active ROSA26 locus in such a way that a loxP-flanked stuffer sequence prevents β-gal expression. When GFP/Cre is expressed in any cell, it mediates recombination between the loxP sites to splice out the stuffer sequence, thereby allowing β-gal expression in that cell (Fig. 1C).
To characterize the potential for AAV-GFP/Cre to mediate recombination between loxP sites when delivered to the adult brain, the reporter mice were stereotaxically injected with 1 × 106 infectious particles of AAV-GFP or AAV-GFP/Cre in the hippocampus, striatum, septum, or substantia nigra. These regions were selected because temporally and spatially selective knockout of loxP-flanked genes in these regions is of substantial interest for testing theories regarding the involvement of genes in learning and memory and in neurodegenerative disorders.
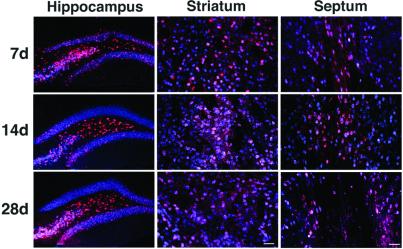
To determine the time dependence and distribution of AAV-GFP/Cre-mediated recombination in vivo, we performed immunohistochemistry by using an antibody against β-gal on brain sections taken from animals 2, 7, 14, and 28 days after stereotaxic injection of AAV-GFP/Cre (Fig. 2). Two days following intrahippocampal viral injection, several β-gal staining cells were observed in the hippocampus, indicating recombination had occurred. Very little β-gal staining was observed at the striatal or septal injection sites at this time point (data not shown). However, the number of cells expressing β-gal increased over time, indicating a time dependence of functional GFP/Cre activity. By 14 days, the area of recombination routinely extended up to 2.5 mm along the rostral–caudal axis, 1.2 mm along the medial–lateral axis, and up to 3.1 mm along the dorsal–ventral axis. No cells expressing β-gal were found at any of the time points in the control AAV-GFP-infected animals (data not shown).
Figure 2.
Targeted recombination of cells in the brain of ROSA26 mice. Unilateral injections of AAV-GFP/Cre into the hippocampus, striatum, and septum were assessed at three time points. Brains were evaluated by immunohistochemistry for NeuN (blue) and β-gal (red) at 7 (n = 3), 14 (n = 4), or 28 (n = 5) days post-AAV delivery for each structure. Coexpression of the neuron-specific marker NeuN (blue) and β-gal (red) demonstrates widespread targeted neurotropic transduction and GFP/Cre-mediated recombination at all time points. (Scale bars = 40 μm.)
To quantify the AAV-GFP/Cre-mediated recombination, we analyzed the number of cells that showed β-gal staining at each time point postinjection (Table 1). In all regions injected, there was a statistically significant increase in the number of cells between 2 and 7 days postinjection (P < 0.001). No statistically significant change in the number of recombined cells was observed in the hippocampus or septum after 7 days (P > 0.05). This finding indicates that recombination was mostly complete at the 7-day time point in these regions. In the striatum, however, the number of cells expressing β-gal increased significantly up to 14 days (P < 0.001) but did not vary significantly after that (P > 0.05). The time dependence of recombination may reflect several factors, including the slow conversion of the AAV vector genome to a transcriptionally active, double-stranded form. Recently, it has been shown that AAV vectors have a slow conversion rate in vivo that requires several days to weeks for maximal gene expression to occur (32, 33).
Table 1.
Quantification of β-gal-positive recombined cells
| Days | Hippocampus
|
Striatum
|
Septum
|
|||
|---|---|---|---|---|---|---|
| No. β-gal cells | % neurons recombined | No. β-gal cells | % neurons recombined | No. LacZ cells | % neurons recombined | |
| 2 | 153 (35) | ND | NS | ND | NS | ND |
| 7 | 9,217 (473) | 95 | 19,242 (631) | 45 | 6,089 (980) | 12 |
| 14 | 9,956 (175) | 98 | 28,144 (1,336) | 67 | 6,117 (473) | 18 |
| 28 | 10,326 (1,021) | 99 | 30,421 (2,143) | 82 | 5,599 (793) | 28 |
Stereological quantification of total recombined cells in the hippocampus, striatum, and septum based on β-gal immunohistochemistry. Standard deviations are provided in parentheses. The percentage neurons that expressed β-gal within each injection site was quantified and expressed as % neurons recombined. Statistics were performed on the total number of β-gal positive cells at each time point for animals evaluated at 2 (n = 3), 7 (n = 3), 14 (n = 4), and 28 days (n = 5) postinjections. ND, not determined; NS, not a significant number of recombined cells.
AAV-GFP/Cre-Mediated Recombination Occurs Selectively in Neurons.
To assess the cellular selectivity of AAV-GFP/Cre, we quantified the percentage of cells expressing the neuronal marker NeuN that also expressed β-gal (Figs. 2 and 3 A–D). Consistent with the previously reported tropism of AAV toward neurons (34), all recombined cells were NeuN positive, indicating that AAV-GFP/Cre was effective only in neurons. No glial cells were ever found to be recombined at any time point studied (Fig. 4 A–F). Similarly, following AAV-GFP infection, GFP expression was observed only in NeuN-positive cells (data not shown).
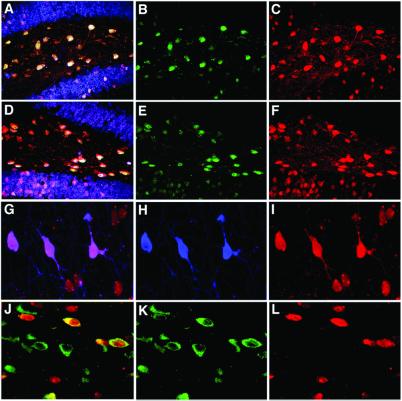
Figure 3.
Characterization of AAV-GFP/Cre-mediated recombination in the brain. (A–C) Analysis of brains 28 days following intrahippocampal injection indicates AAV-GFP/Cre mediates efficient recombination in neurons within the hilus and dentate gyrus. (A) Coexpression of neuron-specific marker NeuN (blue) with β-gal (red) and GFP (green) demonstrates neurotropic transduction and recombination with GFP/Cre protein found in recombined cells. (B) GFP immunohistochemistry (green) demonstrates the localization of the GFP/Cre protein predominantly to the nucleus of infected cells. (C) β-gal expression (red) fills neuronal cell bodies and processes. (D–F) Immunohistochemistry also reveals that Cre protein is also localized predominantly to the nucleus of infected cells at 28 days postinjection in the hippocampus. (D) Merged images of neuron-specific staining (NeuN, blue) with β-gal (red) and Cre (green) demonstrates neurotropic recombination with GFP/Cre protein found in recombined cells. (E) Cre immunohistochemistry (green) also demonstrates nuclear localization. (F) β-gal expression (red) fills neuronal cell bodies and processes. (G–I) Analysis of septum 28 days post-AAV-GFP/Cre injection reveals recombination in ChAT-positive neurons. (H) Immunohistochemistry for ChAT (blue) shows several positive cholinergic cells in the septum. (I) β-gal expression (red) shows numerous recombined cells within the septum. (G) Merging ChAT (blue) with β-gal (red) demonstrates β-gal colocalization of the ChAT neurons. (J–L) Analysis of substantia nigra 14 days postinjection indicates recombination in tyrosine hydroxylase-staining (dopaminergic) neurons. (K) Tyrosine hydroxylase immunohistochemistry (green) defines the substantia nigra compacta dopaminergic cells. (L) Cells positive for β-gal expression (red) indicate recombination occurred. (J) β-gal expression (red) and tyrosine hydroxylase (green) colocalization demonstrate a significant number of recombined cells are dopaminergic neurons.
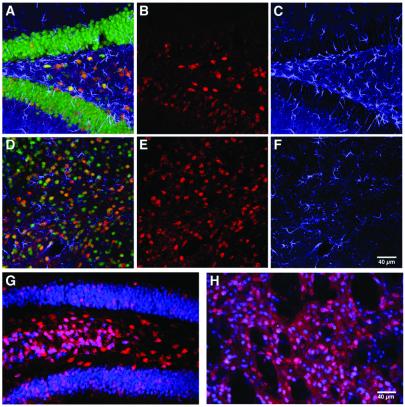
Figure 4.
Lack of significant tissue damage and inflammatory responses after AAV-GFP/Cre delivery allowing for long-term genetic modification of AAV-GFP/Cre-recombined cells. (A–F) Analysis for gliosis or recombined glial fibrillary acidic protein-positive cells 1 week postinjection of 1 × 106 infectious particles into the hippocampus (A–C) or striatum (D–F). Colocalization of β-gal (red; B and E) with glial fibrillary acidic protein staining (blue; C and F) was never detected, whereas NeuN staining (green) showed colocalization with β-gal (A and D merges). There was no evidence for gliosis occurring in either hippocampus (A and C) or striatum (D and F) in response to AAV-GFP/Cre transduction. Long-term expression of β-gal (red) was assessed at 6 months (n = 2) postinjection in the hippocampus (G) and striatum (H). Expression of β-gal (red) was colocalized with NeuN (blue). At 6 months, there was no evidence of any cell loss, vascular cuffing, or infiltrating cells indicative of toxicity.
Additionally, to show the degree of neuronal infectivity and recombination potential within an injection site, we quantified the total percentage of neurons undergoing recombination and expressing β-gal within the injection site. Within the hilus, nearly every neuron expressed β-gal (Fig 3 A–F and Table 1). Regions of the dentate granule cell layer showed significant recombination efficiency; however, given the structure of the region and stereotaxic injections targeted to the hilus, we did not quantify this subregion of the hippocampus. The striatum also showed a high degree of recombination in nearly 50% of the neurons within the injection site at 7 days. The total percentage of β-gal-positive neurons in this region continued to increase at the 14- and 28-day time points, with neuronal recombination efficiency up to 67% and 82%, respectively. Recombination efficiency in the septum was lower, with only 28% of the total neurons within the transduced volume expressing β-gal. These results demonstrate AAV-GFP/Cre is capable of inducing recombination at high efficiency in the majority of neurons within the injection area (Table 1). Further increases in total area of recombination and increased percentage of recombined neurons are likely achievable by multiple-overlapping injections and increasing the total virus injected.
GFP Immunofluorescence as an Indicator of Functional GFP/Cre Activity.
To assess the validity of using GFP staining alone as an indicator of cells that have undergone Cre/loxP recombination following AAV-GFP/Cre infection, we determined the number of recombined, β-gal-positive cells that also showed GFP staining. Brain slices were analyzed for GFP expression 28 days after AAV-GFP/Cre injection. Whereas GFP was readily detected in AAV-GFP-expressing animals (data not shown), GFP fluorescence was difficult to observe directly in the AAV-GFP/Cre-injected animals. To overcome this difficulty, we performed immunohistochemistry with amplification to detect GFP protein in the hippocampus and counted cells that showed both GFP and β-gal colabeling, compared with either alone. GFP expression was observed in most cells that also showed β-gal staining (Fig. 3 A–C). In rare cases, GFP-positive cells did not show β-gal staining, indicating a very small percentage of cells expressing GFP/Cre did not undergo GFP/Cre-mediated recombination. To demonstrate that Cre and β-gal were also colocalized following infection of ROSA26 mice with AAV-GFP/Cre, we performed immunostaining with an anti-Cre antibody. As expected, all cells that showed β-gal expression were also Cre positive, indicating that the GFP/Cre fusion construct expressed the GFP/Cre fusion product (Fig. 3 D–F). Furthermore, the GFP and Cre signals were, as expected, predominantly localized to the nucleus (Fig. 3 A–F).
AAV-GFP/Cre-Mediated Recombination Occurs in Several Neuronal Subtypes.
We investigated whether AAV-GFP/Cre is able to mediate loxP recombination of specific cell types in each region. In the hippocampus, consistent with earlier reports on AAV infectivity (23, 34), we found the greatest number of β-gal expressing recombined cells in the dentate hilus. However, a large number of dentate granule neurons also recombined and expressed β-gal. β-gal expression was robust and filled the cytoplasm, nucleus, and processes of these cells.
ChAT-containing neurons synthesize acetylcholine and project to the hippocampus, and their degeneration has been implicated in the etiology of Alzheimer's disease (35). Selective ablation or activation of genes in ChAT neurons would provide a valuable approach to study the role of selective genes implicated in this disease. We were therefore specifically interested to determine whether ChAT-containing neurons could be targeted for Cre/loxP recombination by AAV-GFP/Cre. Colabeling for β-gal and ChAT revealed that up to 60% of ChAT neurons within the injection site of the septum showed β-gal expression 28 days after injection, indicating the potential to target ChAT-positive cells (Fig. 3 G–I).
Similarly, we determined whether tyrosine hydroxylase dopaminergic neurons expressed β-gal following injection. AAV-GFP/Cre ablation of genes in dopaminergic neurons would be of interest for studying genes relevant to Parkinson's disease. In the substantia nigra pars compacta, over 50% of tyrosine hydroxylase dopaminergic neurons expressed β-gal (Fig. 3 J–L). These results taken together suggest that delivery of AAV-Cre can efficiently target specific populations of neurons within the brain. As noted above, we expect that multiple injections to these sites would enhance the number of infected cells.
AAV-GFP/Cre Yields Long-Term Recombination Without Associated Toxicity.
A number of studies have demonstrated that Cre recombinase expressed at high levels can have cytotoxic effects in vitro (36, 37) and in vivo (38, 39), whereas more moderate expression of Cre is not cytotoxic (36). The utility of AAV-GFP/Cre for the study of long-term effects of gene modification would require that AAV-GFP/Cre does not affect cells adversely. We therefore carefully determined whether any pathological signs of toxicity to the brain arose following delivery of AAV-GFP/Cre. If AAV-GFP/Cre resulted in neuronal loss, decreases in numbers of β-gal-positive cells would be observed over time after injection. As noted in Table 1, however, there were no decreases in the number of cells expressing β-gal up to 28 days following AAV-GFP/Cre infection. This finding suggests that no cell loss had occurred over this time period. Additionally, the numbers of recombined cells at the 6-month time point were similar with 10,121 and 31,028 recombined cells in the hippocampus and striatum, respectively (Fig 4 G–I). No statistics were performed at 6 months postinjection as only two animals were evaluated at this time point. Furthermore, we found no indication of cytotoxicity in a series of studies that investigated neuronal clearance, vascular cuffing, and glial cell infiltration (as evidenced by glial fibrillary acidic protein staining). We found no evidence of cytotoxicity in brain sections of AAV-GFP/Cre-infected mice at any postinjection time point investigated (Figs. 2 and 4 A–F). Additionally, immune markers were evaluated at each time point by investigating CD4 and CD8 cell infiltration. There were very few CD4 or CD8 cells at any time point and no detectable difference between the AAV-GFP and the AAV-GFP/Cre groups (data not shown).
Discussion
We describe the use of recombinant AAV to deliver GFP/Cre as a new approach for achieving temporally and spatially controlled gene modification in the adult nervous system. Stereotaxic injection of AAV-GFP/Cre in the adult mouse brain resulted in recombination between two loxP sites engineered into the ROSA26 locus (31). A significantly large number of neurons underwent recombination after a single stereotaxic injection of 1 × 106 infectious AAV-GFP/Cre particles. The maximum number of recombined cells was achieved and sustained as early as 7 days following injection into the hippocampus or septum and by 14 days in the striatum. The method is highly efficient because in nearly all neurons where GFP/Cre was detected, β-gal expression was observed, indicating high recombination efficiency at the ROSA26 locus. Although not all cells within the injection area were transduced, increasing the amount of virus injected may improve efficacy.
Whereas the ROSA26 reporter gene is functional in all cell types, including glial cells, AAV-GFP/Cre infection resulted in β-gal expression only in neurons, indicating that AAV-GFP/Cre is selective toward neurons in vivo. This finding is consistent with the known neuronal selectivity of AAV vectors in vivo (34). The neural selectivity of AAV-GFP/Cre may be further enhanced by the CMV promoter, which functions preferentially in neurons. Thus, AAV-GFP/Cre will be useful for studying regional selective ablation of loxP-flanked genes in neurons, without recombination in glial cells to confound interpretations. In contrast, infection by lentivirus, adenovirus, and other viral vectors is not selective for neurons.
Recently, in vitro and in vivo toxicity from Cre recombinase has been reported (36–39). The toxicity seems to be because of aberrant activity of Cre at “pseudo” loxP sites. An elegant study from the Berns' group (36) demonstrated that this Cre toxicity depends on expression levels, as Cre was not toxic at low levels. The results of our work, and of others, support this notion. In a previous study from our laboratories, lentivirus-mediated delivery of Cre had cytotoxic effects in vivo, which were overcome by use of a self-inactivating Cre recombinase (39). The lentivirus was designed for high expression levels, in which the vector contained a CMV promoter, a central ppt of HIV-1 pol, and a posttranscriptional element from the woodchuck hepatitis virus to further augment expression of the Cre transgene. Furthermore, the lentiviral Cre study injected 60-fold more infectious particles into the brain than the current AAV study, which likely led to multiple infections of individual cells (39). These factors combined would yield higher levels of Cre in individual cells, and thus toxicity. By contrast, we have not detected in vivo toxicity of a lentivirus expressing the GFP/Cre solely from a CMV promoter lacking the ppt and woodchuck hepatitis virus elements. By using a lower titer of virus (1 × 106 infectious particles), β-gal expressing cells persisted with no obvious signs of toxicity 1-year postinjection (unpublished observation). Similarly, GFP/Cre in our recombinant AAV vector was driven solely by the CMV promoter, and we did not observe any evidence of toxicity following delivery to the brain. Notably, in transgenic mice expressing Cre recombinase from specific cellular promoters, no gross cell loss is evident (7, 12). In these latter cases, expression levels of Cre are expected to be lower than in the lentiviral system that was originally designed to maximize expression of Cre (39).
A number of different approaches for achieving temporal and spatially regulated gene modification in the nervous system have been developed. However, AAV-GFP/Cre has several distinct advantages. It eliminates the time-consuming and expensive breeding strategies as well as eliminating potentially complex genetic backgrounds of transgenic approaches. In fact, it allows direct comparison of littermates homozygous for a loxP-flanked allele following injection or different sides of the brain of the same animal after unilateral injections. Further, direct delivery of virus at any time point postnatally overcomes many of the problems associated with inducible systems. A significant advantage is the AAV selectivity for neurons and its ability to induce recombination in nondividing cells with no vector toxicity. Finally, other recombinant viruses, with the exception of lentivirus, are not particularly suited for long-term studies, as they tend to invoke an immune response that results in subsequent damage or loss of infected cells.
We have shown that AAV-GFP/Cre will allow targeted recombination of loxP-flanked genes in the hippocampus, striatum, substantia nigra, and septum. Selective modification including ablation of genes in the hippocampus will facilitate study of their involvement in learning, memory, and neurodegeneration. Ablation of genes in the striatum and tyrosine hydroxylase-containing neurons of the substantia nigra is of interest for studies of motor coordination and movement disorders such as Huntington's disease and Parkinson's disease. Ablation of genes in ChAT-containing neurons of the septum will be of interest for examining their roles in cognition and neurodegenerative diseases. Moreover, recombination seems to be complete and stable 7–14 days after stereotaxic injection in the adult, allowing for a wide window of time in which to study gene function in these regions. Thus, AAV-GFP/Cre will be a powerful tool for ablating or activating loxP-modified genes in specific brain regions to test their roles in neural function and pathology.
Acknowledgments
We are grateful for the expertise of Linda Kitayabashi, and we thank Jamie Simon for imaging and illustration and Mary Lynn Gage for critical reading of the manuscript. The ROSA26 Cre reporter mice were kindly provided by Philippe Soriano, Fred Hutchinson Cancer Research Center, Seattle; the nls-Cre 306c was kindly provided by Steve O'Gorman. This work was supported by National Institutes of Health Grants P01 AG10435 and 5T32 AG00216, The Pasarow Foundation, and The Chapman Trust. B.V. was supported by The Hereditary Disease Foundation Leiberman Award.
Abbreviations
- AAV
adeno-associated virus
- GFP
green fluorescent protein
- β-gal
β-galactosidase
- CMV
cytomegalovirus
- ChAT
choline acetyltransferase
References
- 1.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 2.Price D L, Sisodia S S, Borchelt D R. Science. 1998;282:1079–1083. doi: 10.1126/science.282.5391.1079. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y, Romero M I, Ghosh P, Ye Z, Charnay P, Rushing E J, Marth J D, Parada L F. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dragatsis I, Levine M S, Zeitlin S. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Tonegawa S. Annu Rev Neurosci. 1997;20:157–184. doi: 10.1146/annurev.neuro.20.1.157. [DOI] [PubMed] [Google Scholar]
- 6.Brooks A I, Muhkerjee B, Panahian N, Cory-Slechta D, Federoff H J. Nat Biotechnol. 1997;15:57–62. doi: 10.1038/nbt0197-57. [DOI] [PubMed] [Google Scholar]
- 7.Tsien J Z, Chen D F, Gerber D, Tom C, Mercer E H, Anderson D J, Mayford M, Kandel E R, Tonegawa S. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 8.Sauer B, Henderson N. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sternberg N, Hamilton D, Hoess R. J Mol Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- 10.Sternberg N, Hamilton D. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 11.Sauer B. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 12.Tsien J Z, Huerta P T, Tonegawa S. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 13.Sauer B. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 14.Vooijs M, Jonkers J, Berns A. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 16.Anton M, Graham F L. J Virol. 1995;69:4600–4606. doi: 10.1128/jvi.69.8.4600-4606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohlmann A, Gotthardt M, Willnow T E, Hammer R E, Herz J. Nat Biotechnol. 1996;14:1562–1565. doi: 10.1038/nbt1196-1562. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Krushel L A, Edelman G M. Proc Natl Acad Sci USA. 1996;93:3932–3936. doi: 10.1073/pnas.93.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks A I, Cory-Slechta D A, Federoff H J. Proc Natl Acad Sci USA. 2000;97:13378–13383. doi: 10.1073/pnas.230169397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blomer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplitt M G, Leone P, Samulski R J, Xiao X, Pfaff D W, O'Malley K L, During M J. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 22.Kaspar B, Erickson D, Schafer D, Hinh L, Gage F, Peterson D. Mol Ther. 2002;5:50–56. doi: 10.1006/mthe.2001.0520. [DOI] [PubMed] [Google Scholar]
- 23.McCown T J, Xiao X, Li J, Breese G R, Samulski R J. Brain Res. 1996;713:99–107. doi: 10.1016/0006-8993(95)01488-8. [DOI] [PubMed] [Google Scholar]
- 24.Lo W D, Qu G, Sferra T J, Clark R, Chen R, Johnson P R. Hum Gene Ther. 1999;10:201–213. doi: 10.1089/10430349950018995. [DOI] [PubMed] [Google Scholar]
- 25.Bjorklund A, Kirik D, Rosenblad C, Georgievska B, Lundberg C, Mandel R J. Brain Res. 2000;886:82–98. doi: 10.1016/s0006-8993(00)02915-2. [DOI] [PubMed] [Google Scholar]
- 26.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 27.Xiao X, Li J, Samulski R J. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snyder R O S S, Lagarde C, Bohl D, Kaspar B, Sloan B, Cohen L K, Danos O. Hum Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- 29.Peterson D A, Lucidi-Phillipi C A, Eagle K L, Gage F H. J Neurosci. 1994;14:6872–6885. doi: 10.1523/JNEUROSCI.14-11-06872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sterio D C. J Microsc. 1984;134:127–136. doi: 10.1111/j.1365-2818.1984.tb02501.x. [DOI] [PubMed] [Google Scholar]
- 31.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 32.Ferrari F K, Samulski T, Shenk T, Samulski R J. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher K J, Gao G P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartlett J S, Samulski R J, McCown T J. Hum Gene Ther. 1998;9:1181–1186. doi: 10.1089/hum.1998.9.8-1181. [DOI] [PubMed] [Google Scholar]
- 35.Fisher L J, Raymon H K, Gage F H. Ann NY Acad Sci. 1993;695:278–284. doi: 10.1111/j.1749-6632.1993.tb23067.x. [DOI] [PubMed] [Google Scholar]
- 36.Loonstra A, Vooijs M, Beverloo H B, Allak B A, van Drunen E, Kanaar R, Berns A, Jonkers J. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silver D P, Livingston D M. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt E E, Taylor D S, Prigge J R, Barnett S, Capecchi M R. Proc Natl Acad Sci USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeifer A, Brandon E P, Kootstra N, Gage F H, Verma I M. Proc Natl Acad Sci USA. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]