Abstract
Acidic extracellular solution activates transient H+-gated currents in dorsal root ganglion (DRG) neurons. The biophysical properties of three degenerin/epithelial sodium (DEG/ENaC) channel subunits (BNC1, ASIC, and DRASIC), and their expression in DRG, suggest that they might underlie these H+-gated currents and function as sensory transducers. However, it is uncertain which of these DEG/ENaC subunits generate the currents, and whether they function as homomultimers or heteromultimers. We found that the biophysical properties of transient H+-gated currents from medium to large mouse DRG neurons differed from BNC1, ASIC, or DRASIC expressed individually, but were reproduced by coexpression of the subunits together. To test the contribution of each subunit, we studied DRG from three strains of mice, each bearing a targeted disruption of BNC1, ASIC, or DRASIC. Deletion of any one subunit did not abolish H+-gated currents, but altered currents in a manner consistent with heteromultimerization of the two remaining subunits. These data indicate that combinations of two or more DEG/ENaC subunits coassemble as heteromultimers to generate transient H+-gated currents in mouse DRG neurons.
Transient proton-activated currents in peripheral sensory neurons were first described more than 20 years ago (1). These currents were thought to mediate acid-induced pain sensation (2). However, their molecular basis remained uncertain until the recent discovery of degenerin/epithelial sodium channels (DEG/ENaC), which reconstitute several properties of these currents: H+ activation, Na+ selectivity, and amiloride block.
Within the DEG/ENaC family, three genes encode H+-gated channels: BNC1 (3), ASIC (4), and DRASIC (5).¶ BNC1 and ASIC each have alternative splice forms [BNC1a and BNC1b (3, 6) and ASICα and ASICβ (7), respectively], which differ in a segment including the N terminus and first membrane-spanning domain. mRNA corresponding to each of these subunits are present in mammalian dorsal root ganglion (DRG; refs. 7–10), and protein expressions for some of the subunits have been identified at sensory nerve terminals (9–11) where they may participate in the transduction of acid-mediated pain (8, 11) and mechanosensation (9, 11).
Expression of BNC1, ASIC, or DRASIC individually in heterologous cells generates transient H+-gated Na+ currents. Moreover, when coexpressed in combination, they associate (12, 13) and produce currents with unique functional properties (6, 12, 13), suggesting that they have the capacity to form heteromultimers. Previous data comparing the properties of native H+-gated currents with those of heterologously expressed DEG/ENaC subunits provide some insight into the in vivo channel composition. For example, the properties of DRASIC match the H+-gated current properties in rat cardiac sensory neurons (14). However, genetic data regarding the role of specific DEG/ENaC subunits is lacking, and the precise composition of the channels in vivo remains uncertain.
Our goal, therefore, was to determine the functional composition of the DEG/ENaC channels in DRG neurons. We used two strategies. First, we compared the properties of the currents in mouse DRG neurons with those of the cloned mouse channel subunits expressed individually or in combination. Second, we examined the properties of the currents in genetically altered mice lacking BNC1, ASIC, or DRASIC.
Materials and Methods
Culture of DRG Neurons.
Thoracic DRG neurons were collected and dissociated from 6- to 12-month-old mice as described (9). DRGs were dissociated with papain, collagenase, and dispase, plated on poly(D-lysine)/laminin-coated plastic, and stored at either room temperature in L15 medium supplemented with 50 nM nerve growth factor, or at 37°C in F12 medium supplemented with nerve growth factor. Neurons 20–35 μm in diameter were studied 18–48 h after plating.
cDNA Constructs.
Mouse BNC1a was cloned from mouse brain mRNA (CLONTECH) by reverse transcription–PCR by using the 5′ primer: 5′-CCA TCG ATG GAG CCA TGG ACC TCA AGG AGA GCC CCA GC-3′, and 3′ primer: 5′-G GGG TAC CCC TCA GCA GGC AAT CTC CTC CAG GGT GCC-3′. Mouse ASICα and ASICβ were cloned by reverse transcription–PCR from total RNA isolated from mouse brain and DRG, respectively, by using the primer pairs: ASICα (5′ primer: 5′-GCC ATC GAT ATG GAA CTG AAG ACC GAG GAG G-3′; 3′ primer: 5′-CGG GGT ACC TTA GCA GGT AAA GTC CTC AAA CG-3′), ASICβ (5′ primer: 5′-GCC ATC GAT ATG GAG CTG GAT GAG GGT GAC TC-3′; 3′ primer: 5′-CGG GGT ACC TTA GCA GGT AAA GTC CTC AAA CG-3′). Mouse DRASIC was cloned as described (15). The constructs were cloned into pMT3 for expression in COS-7 cells.
Heterologous Expression of cDNA in COS-7 Cells.
cDNAs were expressed by electroporation (950 μF, 320 V, GenePulser II; Bio-Rad) in COS-7 cells that had less than 150 pA of endogenous pH-evoked current. cDNA for green fluorescent protein (3 μg/0.4 ml) was expressed with DEG/ENaC subunits (15 μg/0.4 ml) to facilitate detection of transfected cells by epifluorescence. For DEG/ENaC coexpression experiments, cDNAs were transfected at equal concentrations for a total of 15 μg/0.4 ml. Cells were cultured in DMEM with 10% FBS and 1% Pen/Strep at 37°C and were studied 48–72 h after transfection.
Electrophysiology.
Whole-cell patch-clamp recordings (at −70 mV) from DRG neurons and COS-7 cells were performed with an Axopatch 200B amplifier (Axon Instruments, Foster City, CA) and acquired and analyzed with PULSE/PULSEFIT 8.30 (HEKA Electronics, Lambrecht, Germany) and igor pro 3.16 (WaveMetrics, Lake Oswego, OR) software. Currents were filtered at 5 kHz and sampled at 2 or 0.2 kHz. Micropipettes (2–5 MΩ) were filled with internal solution: 100 mM KCl, 10 mM EGTA, 40 mM Hepes, and 5 mM MgCl2, pH 7.4 with KOH. External solution contained: 120 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, 10 mM Mes, pH adjusted with tetramethylammonium hydroxide, and osmolarity adjusted with tetramethylammonium chloride. Extracellular solutions were changed within 20 msec by using a computer-driven solenoid valve system (16).
We focused on the transient H+-activated currents because they are believed to be carried solely by DEG/ENaC channels (8), whereas the sustained transient H+-activated currents are probably the result of multiple channel types, including the vanilloid receptors (17, 18). Data are means ±SEM unless otherwise stated. Kinetics of desensitization and recovery from desensitization curves were fit with single exponential equations and time constants (τ) reported. pH activation curves were fit by using the Hill equation: Fraction of open channels = 1/{1 + (pH10/K0.510)n}, where K0.5 is the pH at which half of the channels are opened.
Generation of ASICα and BNC1 Knockout Mice.
The generation and description of the BNC- and DRASIC-null mice have been reported (9, 11). Mice with disruption of the gene for ASIC (J.A.W. and M.J.W., unpublished work) were generated by similar methods. All mice were F2 generation from 129/Sv:C57BL-6 crosses. Wild-type littermates from BNC1 and ASIC knockout mice were analyzed separately, however, no differences were observed between the two groups and so their data were pooled.
Results
Individual DEG/ENaC Subunits Do Not Reproduce the H+-Gated Currents of DRG Neurons.
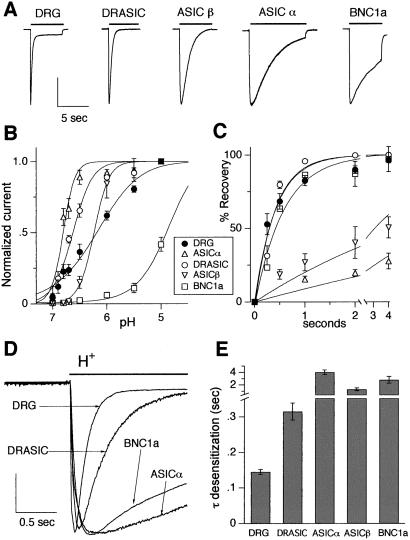
To determine which subunits generate transient acid-evoked currents in DRG neurons, we compared currents in mouse DRG with those of mouse H+-gated DEG/ENaC subunits expressed in COS-7 cells; Fig. 1A shows examples (BNC1b does not form homomeric H+-gated channels; ref. 6). We focused on three properties that distinguish between individual subunits (14). First, the DEG/ENaC subunits form channels with differing sensitivities to pH. We found that the pH sensitivity of DRG neurons did not match that of DRASIC, ASICα, or BNC1a (Fig. 1B). Although DRG neurons and ASICβ showed similar half-maximal activation, the slopes of their pH-activation curves differed markedly. Thus, a single H+-gated DEG/ENaC subunit cannot reconstitute the pH sensitivity of DRG neurons.
Figure 1.
Properties of acid-evoked currents in DRG neurons are not matched by any one DEG/ENaC subunit. (A) Representative acid-evoked currents in a wild-type DRG neuron and COS-7 cells expressing the indicated DEG/ENaC subunits. The bar above each current represents a fast solution change from pH 7.4 to pH 6, except for BNC1a, which is evoked by pH 5 (because of the relative pH insensitivity of BNC1a channels). Vertical scale bar: 2.71 nA DRG, 1.23 nA DRASIC, 1.78 nA ASICβ, 0.943 nA ASICα, 3 nA BNC1a. (B) Effect of pH on currents from DRG and COS cells expressing the indicated subunits from a holding pH of 8. Currents were normalized to those evoked by pH 5 (except BNC1a, which was normalized to pH 4 current). Lines are fit of Hill equation. n = at least four cells for all data points. (C) Time course of recovery from desensitization. Current was completely desensitized by a prolonged pulse to pH 6 (pH 4 for BNC1a). Cells then were bathed in pH 7.4 solution for the indicated times before they were stimulated again with a pH 6 (pH 4) solution. Recovery is percentage of current evoked by the second pulse compared with the first. Lines are fit of single exponentials. n = at least three cells for each data point. (D) Superimposed currents evoked by a solution change from pH 7.4 to pH 6 (pH 4 for BNC1a). Vertical scale bar: 1.27 nA DRG, 0.36 nA DRASIC, 1 nA ASICα, 4.95 nA BNC1a. (E) Mean time constants of desensitization as measured from single exponential fits to the falling phase of the currents evoked by pH 6 (pH 4 for BNC1a) application to wild-type DRG neurons (n = 29) and COS-7 cells expressing indicated subunits (n = at least five cells).
Second, we measured recovery from desensitization. After prolonged exposure to acidic pH, DRG currents recovered by ≈50% in 300 ms (Fig. 1C). Expression of DRASIC or BNC1a generated currents with similar recovery times, but ASICα and ASICβ currents recovered more slowly. These data indicate that neither ASICα nor ASICβ alone account for H+-gated currents in DRG neurons. However, the rate of recovery from desensitization does not exclude channels composed of DRASIC or BNC1a subunits.
Third, we examined desensitization kinetics during an acid stimulus. DRASIC desensitized faster than BNC1a, ASICα, or ASICβ (Fig. 1 D and E). Currents from mouse DRG neurons desensitized even faster than any of the individual subunits.
These data (summarized in Table 1) indicate that no single DEG/ENaC subunit generates the acid-evoked currents observed in DRG neurons; none of the individual subunits reproduced the pH sensitivity, recovery from desensitization, and desensitization kinetics, suggesting the currents result from coexpression of two or more DEG/ENaC subunits in the same cell. Subunits could assemble as homomultimers, with total native current equaling the sum of current from the various homomultimers. However, this arrangement could not explain our finding that acid-evoked DRG current desensitized faster than any individual subunit. Alternatively, subunits could assemble into heteromultimers containing two or more of the DEG/ENaC subunits. Such a model could explain the data if coassembly generated a channel with faster kinetics desensitization.
Table 1.
Functional properties of mouse DRG neurons and DEG/ENaC subunits expressed individually and in combination (in COS-7 cells)
| Amplitude (nA at pH 5) | Half-maximum pH activation | τ desensitization (sec) | τ recovery (sec, at pH 7.4) | |
|---|---|---|---|---|
| DRG neurons | ||||
| Wild-type | 4.1 ± 0.45 | 6.2 | 0.15 ± 0.01 | 0.42 |
| ASIC null | 1.4 ± 0.18* | 6.2 | 0.20 ± 0.02 | 0.20 |
| BNC1 null | 3.9 ± 1.00 | 6.6 | 0.15 ± 0.01 | 0.68 |
| DRASIC null | 2.66 ± 0.40* | |||
| DEG/ENaC subunits expressed individually | ||||
| DRASIC | 6.6 | 0.32 ± 0.02* | 0.40 | |
| ASICα | 6.8 | 4.0 ± 0.35* | 11 | |
| ASICβ | 6.2 | 1.3 ± 0.20* | 4.4 | |
| BNC1a | 4.9 | 2.8 ± 0.55* | 0.60 | |
| Coexpressed DEG/ENaC subunits | ||||
| DRASIC + ASICα | 6.6 | 0.14 ± 0.01 | 0.64 | |
| DRASIC + BNC1a | 6.1 | 0.26 ± 0.02* | 0.08 | |
| ASICα + BNC1a | 6.1 | 0.90 ± 0.20* | 0.63 | |
| DRASIC + ASICα + BNC1a | 6.4 | 0.18 ± 0.02 | 0.32 |
Values without standard errors are derived from curve fits of the means (n = 3–14 for each data point on the curves). Current amplitude at pH 5 (n = 22–65); τ desensitization at pH 6, except BNC1a is at pH 4 (n = 5–29). Asterisks denote statistical differences from wild-type DRG for data with standard errors of the mean (P < 0.01, two-tailed t test).
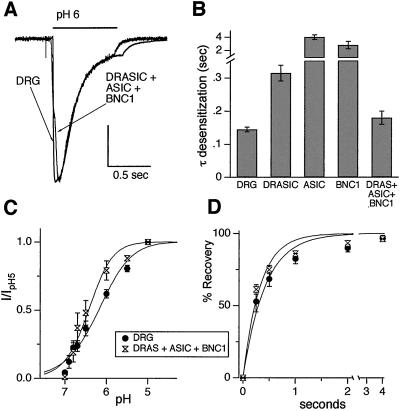
Coexpression of BNC1a, ASICα, and DRASIC Mimics the pH-Evoked Current in DRG Neurons.
To test the hypothesis that multiple DEG/ENaC subunits coassemble to generate DRG H+-gated currents, we coexpressed BNC1a, ASICα, and DRASIC. Coexpression reproduced the fast DRG desensitization kinetics; current desensitized faster than any individual subunit (Fig. 2 A and B). Coexpression also reproduced the DRG pH sensitivity (Fig. 2C) and recovery from desensitization (Fig. 2D) reasonably well. Thus, BNC1, ASIC, and DRASIC subunits assemble in some combination as heteromultimeric channels.
Figure 2.
Coexpression of DRASIC, ASICα, and BNC1a mimic the properties of the pH-evoked current in wild-type DRG neurons. (A) Currents evoked by application of pH 6 from pH 7.4 solutions in a DRG neuron, and a COS-7 cell coexpressing DRASIC, ASICα, and BNC1a. Vertical scale bar: 1.32 nA DRG, 0.5 nA COS-7. (B) Mean time constant of desensitization of the current evoked by pH 6 application to COS-7 cells expressing all three subunits (n = 6; other bars represent data collected in Fig. 1 for comparison). (C) pH dose-response data for currents evoked from COS-7 cells expressing all three subunits (n = at least 4 for all data points) compared with wild-type DRG neurons. (D) Recovery from desensitization data from COS-7 cells expressing all three subunits (n = at least six cells for all data points) compared with wild-type DRG neurons. All data were collected and fit as per Fig. 1.
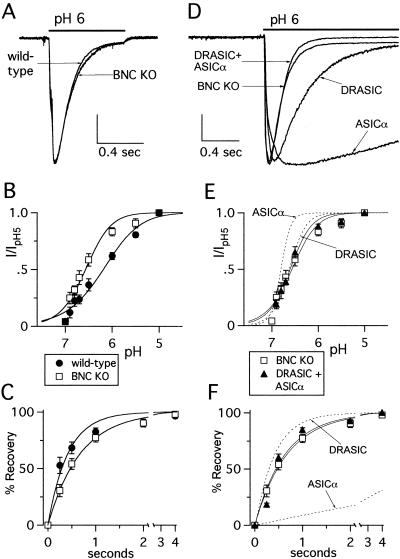
pH-Evoked Current in DRG Neurons from BNC1-Null Mice.
To test directly which subunits contribute to DRG proton-gated currents, we studied neurons from mice with targeted deletions in BNC1, ASIC, or DRASIC. Deletion of BNC1 did not abolish acid-evoked currents (Fig. 3A) or alter the peak amplitude (Table 1), but dramatically altered current properties. Currents in BNC1-null mice were more pH sensitive (Fig. 3B) (pH0.5 = 6.6, BNC null; 6.2, wild type), and recovered more slowly from desensitization (Fig. 3C) (τ = 0.68 sec, BNC null; 0.42 sec, wild type), than neurons from wild-type mice. Thus, BNC1 contributes to H+-gated currents in wild-type DRG neurons. However, deletion of BNC1 did not slow current desensitization (τ = 0.15 ± 0.01 sec; Fig. 3A), and because it was faster than any individual subunit, this finding suggests that more than one DEG/ENaC subunit coassembles to generate currents in BNC1-null neurons.
Figure 3.
Targeted disruption of BNC1 gene alters pH-evoked current in DRG neurons in a manner consistent with expression of the remaining subunits. (A) Representative currents evoked by changing the solution from pH 7.4 to pH 6 in wild-type and BNC1-null DRG neurons demonstrating similar desensitization rates. Twenty-six percent of BNC1-null DRG (n = 82) responded to pH 5 with greater than 150 pA transient current, compared with 59% of wild type (n = 112). Vertical scale bar: 1 nA wild type, 0.38 nA BNC1 null. (B) pH dose-response and (C) recovery from desensitization data for BNC1-null DRG neurons (n = at least eight cells for all data points) compared with wild type. (D) Superimposed currents evoked by a solution change from pH 7.4 to pH 6 in a BNC1-null DRG neuron, and COS-7 cells expressing indicated subunits demonstrating that only coexpression of subunits reproduced the fast kinetics of the BNC1-null neuron. Vertical scale bar: 1.12 nA BNC1-null neuron, 0.283 nA DRASIC, 0.787 nA ASICα, 1 nA DRASIC + ASICα. (E) pH dose-response data and (F) recovery from desensitization data for DRASIC and ASICα coexpressed in COS-7 cells (n = at least six cells for all data points). Also shown are the BNC1-null DRG data from B and C and the fits of the data for subunits expressed individually for comparison. Data were collected and fit as per Fig. 1.
We therefore tested the hypothesis that two remaining subunits, ASIC and DRASIC, generate the acid-evoked currents in the BNC1-null mice. Coexpression of ASICα and DRASIC generated acid-evoked currents that desensitized faster than currents from either subunit expressed alone, reproducing the fast kinetics of currents of BNC1-null mice (Fig. 3D). Coexpression also reproduced the pH sensitivity and recovery kinetics of the BNC1-null mice (Fig. 3 E and F). These data suggest that ASIC and DRASIC form heteromultimeric channels in BNC1-null neurons.
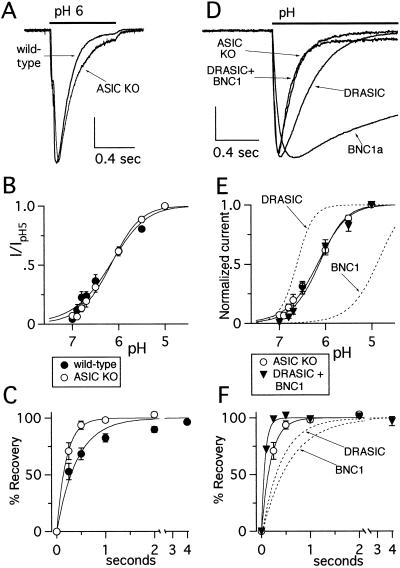
pH-Evoked Current in DRG Neurons from Neurons ASIC-Null Mice.
To test whether ASIC contributes to DRG acid-evoked currents, we studied neurons from mice containing a targeted deletion of ASIC. Similar to BNC1-null neurons, deletion of ASIC did not eliminate acid-evoked current (Fig. 4A), but altered its properties. First, current amplitude was reduced (ASIC null, 1.44 ± 0.18 nA; wild type, 4.07 ± 0.45 nA, P < 0.01). Second, currents in the ASIC-null neurons desensitized more slowly than wild-type neurons (ASIC null, 0.20 ± 0.02 sec; wild type, 0.15 ± 0.01 sec, P = 0.014), although still faster than individual DEG/ENaC subunits (Fig. 4A). Third, the ASIC-null cells recovered faster from desensitization (ASIC null, τ = 0.20 sec; wild type, τ = 0.42 sec) (Fig. 4C). These altered properties indicate that ASIC contributes to the acid-evoked current in wild-type DRG neurons. In contrast to BNC1-null neurons, deletion of ASIC did not alter pH sensitivity (pH0.5 = 6.2) (Fig. 4B).
Figure 4.
Targeted disruption of the ASIC gene alters pH-evoked current in DRG neurons in a manner predicted by expression of the remaining subunits. (A) Representative currents evoked by a solution change from pH 7.4 to pH 6 in wild-type and ASIC-null DRG neurons demonstrating slightly slower desensitization of the ASIC-null neurons. Forty-eight percent of ASIC-null DRG (n = 48) responded to pH 5 with greater than 150 pA transient current. Vertical scale bar: 1 nA wild type, 0.47 nA ASIC null. (B) pH dose-response and (C) recovery from desensitization data for ASIC-null DRG neurons (n = at least 12 cells for all data points) compared with wild type. (D) Currents evoked by application of pH 6 from pH 7.4 in an ASIC-null DRG neuron and COS-7 cells expressing indicated subunits demonstrates that only coexpression of subunits reproduced the fast kinetics of the ASIC-null DRG. Vertical scale bar: 0.33 nA ASIC-null neuron, 0.4 nA DRASIC, 5.5 nA BNC1a, 0.41 nA DRASIC + BNC1a. (E) pH dose-response data and (F) recovery from desensitization data for DRASIC and BNC1a coexpressed in COS-7 cells (n = at least seven cells for all data points). Also shown for comparison are the ASIC-null DRG data from B and C and the fits of the data for subunits expressed individually. Data were collected and fit as per Fig. 1.
Two findings in ASIC-null neurons could not be explained by the summation of currents from individual DEG/ENaC subunits; the currents desensitized (Fig. 4D) and recovered from desensitization (Fig. 4F) faster than either DRASIC or BNC1a currents. Coexpression of DRASIC and BNC1a reproduced both properties (Fig. 4 D and F). Moreover, the pH sensitivity of ASIC-null neurons was reproduced by coexpression of BNC1a and DRASIC, but not by either subunit alone (Fig. 4E). These data suggest that DRASIC and BNC1 coassemble as heteromultimers to generate H+-gated channels in ASIC-null mice.
pH-Evoked Current in DRG from DRASIC-Null Mice.
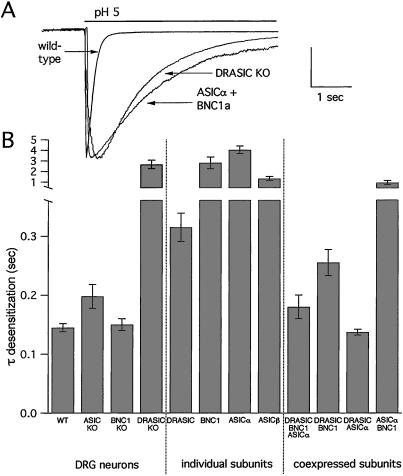
To test the contribution of DRASIC, we studied DRG currents in mice with a targeted deletion in this subunit. We focused on the rate of desensitization, because this rate was a key property in differentiating heteromultimeric channels from individual DEG/ENaC subunits.‖ H+-gated currents in DRASIC-null mice desensitized almost 10-fold slower than neurons from wild-type, BNC1-null, or ASIC-null mice (Fig. 5A). Coexpression of ASICα and BNC1 reproduced these slow kinetic values. Thus, DRASIC also contributes to the formation of H+-gated channels in wild-type DRG neurons. Moreover, DRASIC is a necessary component of the native channel; the fast desensitization kinetic values were only generated when DRASIC was present in the cell (Fig. 5B).
Figure 5.
Deletion of the DRASIC gene slows the desensitization kinetics of pH-evoked current in DRG neurons. (A) Superimposed currents evoked by a solution change from pH 7.4 to pH 5 in a wild-type and DRASIC-null DRG neurons, and a COS-7 cell coexpressing ASICα and BNC1a. Vertical scale bar: 4 nA wild-type DRG, 2.2 nA DRASIC-null DRG, 1.39 nA COS-7 cell. (B) Mean time constants of desensitization of the acid-evoked currents from wild-type and knockout DRG neurons (n = at least 13 cells), and DEG/ENaC subunits expressed individually (n = at least five cells), and coexpressed in COS-7 cells (n = at least six cells) demonstrates that only DRASIC coexpressed with other subunits reproduced the fast desensitization kinetics of the DRG neurons. Currents were evoked by pH 6, except BNC1a currents, which were evoked by pH 4.
Discussion
Our data indicate that three DEG/ENaC subunits, BNC1, ASIC, and DRASIC, combine to produce H+-activated channels in mouse DRG neurons. Coexpression of all three subunits reproduced the biophysical properties of the DRG currents. Moreover, targeted disruption of BNC1, ASIC, or DRASIC did not abolish the acid-evoked currents, but altered the properties in a manner predicted by residual expression of the other two subunits.
Do DEG/ENaC Channels Assemble as Homomultimers or Heteromultimers in DRG Neurons?
There are two general ways the DEG/ENaC subunits might generate H+-gated channels. BNC1, ASIC, and DRASIC might form homomultimeric channels. Conversely, two or more subunits might assemble to form heteromultimeric channels.
In distinguishing between these two alternatives, the rate of current desensitization was the most informative property. Acid-evoked currents in DRG neurons desensitized faster than any individual DEG/ENaC homomultimeric channel. This property is not explained by the sum of multiple homomultimeric channels; the desensitization rate in this case would be intermediate between the rates for the individual subunits. On the other hand, coexpression of two or more DEG/ENaC subunits reproduced the fast desensitization kinetics. Desensitization was also fast in mice lacking either BNC1 or ASIC, suggesting the remaining subunits formed heteromultimers in these mice.
The rate of current recovery from desensitization was also informative in differentiating homo- from heteromultimers. Currents in ASIC-null mice recovered from desensitization faster than any individual DEG/ENaC homomultimeric channel. Coexpression of DRASIC and BNC1a reproduced this property, suggesting that they form heteromultimeric channels. Our data, showing that DRASIC (11) and BNC1 (9) are expressed in most medium to large DRG neurons, support these findings.
Heteromultimerization seems to be a common arrangement for the DEG/ENaC family of channels: the mammalian epithelial Na+ channel (ENaC) is formed by three subunits (α, β, and γ) (19, 20), and the degenerins in Caenorhabditis elegans are composed of multiple related subunits (21).
What Is the Subunit Composition of the Heteromultimeric DEG/ENaC Channels in DRG Neurons?
Coexpression of DRASIC, ASICα, and BNC1a reproduced relatively well the properties of the transient acid-evoked currents in mouse DRG neurons. Thus, it is possible that these three subunits form a heteromultimeric channel. However, other subunit combinations could also form H+-gated channels in the DRG.
First, channels might be composed of two subunits. For example, the cell might express a mixture of BNC1/DRASIC and ASIC/DRASIC channels. We cannot exclude this possibility because coexpression of these combinations produced currents with properties similar to coexpression of the three subunits. Moreover, our knockout animals do not allow us to differentiate the relative contribution of the BNC1 and ASIC splice variants. Nevertheless, our data suggest that DRASIC is necessary to reproduce the fast desensitization kinetics of DRG neurons; targeted disruption of DRASIC markedly slowed desensitization. However, DRASIC alone is not sufficient to explain fast desensitization; currents produced by DRASIC expressed alone desensitized more slowly than those from DRG neurons, whereas coexpression of DRASIC with either ASIC or BNC1 reproduced the fast desensitization of native currents.
Second, additional DEG/ENaC subunits might contribute to the channel complex. For example, ENaC subunits are located in sensory nerve endings (22, 23), and transcripts of BLINaC (24) and SPASIC (25, 26) have been detected in the nervous system. Although protons do not gate these subunits by themselves, they could coassemble with BNC1, ASIC, and DRASIC to form H+-gated channels. Other as-yet-unidentified DEG/ENaC family members might also contribute to the channel complex. Although we cannot exclude these possibilities, currents in BNC1, ASIC, and DRASIC knockout mice had biophysical properties that matched remarkably well with coexpression of the other two known H+-gated subunits. Thus, BNC1, ASIC, and DRASIC apparently are the predominant DEG/ENaC subunits responsible for transient H+-gated currents in DRG neurons.
Although most transient H+-gated currents from DRG have properties similar to those we observed, previous studies described small subsets of cells with different properties. For example, a minority of rat DRG cells have markedly slower kinetics of desensitization (2, 16, 27). Although in this study of mouse neurons we did not observe such currents, they are likely generated by ASIC subunits; the slow kinetic values are consistent with ASICα and ASICβ, and these currents are blocked by psalmotoxin 1, a tarantula toxin specific for ASICα homomultimers (27). Other subunit combinations may be responsible for H+-gated currents in select DRG subpopulations. For example, currents in most cardiac afferent neurons were reproduced by expression of DRASIC alone or in combination with BNC1b (14, 16). Thus, the relative expression levels of BNC1, ASIC, and DRASIC in DRG neurons might determine the functional properties of their H+-gated currents. In turn, these properties might tune specific populations of neurons to respond in a graded fashion to a variety of stimuli.
In addition to the transient currents studied here, protons also produce sustained currents in some DRG neurons (28). Although DEG/ENaC channels can generate sustained currents (5, 6), the vanilloid receptors (including VR1) are important contributors to these currents, and to acid-mediated responses in cutaneous nociceptive neurons (17, 18). Sorting out the relative contribution of DEG/ENaC channels and vanilloid receptors to H+-gated currents, and the larger question of their relative functional roles as pH sensors in specific afferent neuron populations, are key issues for the future.
Physiological Implications.
Our data are consistent with the initial phenotypic descriptions of BNC1- and DRASIC-null mice. Animals of both genotypes showed specific sensory deficits. BNC1 knockout mice displayed reduced sensitivity to touch in large myelinated mechanoreceptors (9). In DRASIC-null mice, rapidly adapting Aβ mechanoreceptors exhibited enhanced sensitivity, Aδ fibers showed reduced mechanosensitivity, and polymodal C-fiber nociceptors displayed impaired acid sensitivity (11). Thus, disruption of these genes failed to abolish any one sensory modality completely; instead, the loss of these subunits modified sensory transduction. Similarly, we found that deletion of any single subunit did not abolish pH-evoked currents, but altered their biophysical properties. These data suggest that DEG/ENaC subunits may have both overlapping functions and some functional redundancy. Future studies of double and triple knockout animals may be of value in addressing these issues.
Understanding the molecular composition of the DEG/ENaC channels in mammalian sensory neurons is critical to understanding the physiological role they play in sensory transduction. It also has important therapeutic implications. As sensory transducers, DEG/ENaC channels provide potential targets for the pharmacological modulation of sensory stimuli, including pain. Their function can be altered by several agents, including FMRFamide (Phe–Met–Arg–Phe–NH2) and related neuropeptides (15), psalmotoxin 1 (27), cold temperature (29), Zn2+ (30), lactate (31), and nonsteroidal anti-inflammatory drugs (32). In some cases, modulation is specific to the subunit composition of the channel. For example, FMRFamide potentiates pH-evoked currents generated by ASICα and DRASIC, but not BNC1. Similarly, psalmotoxin 1 inhibits ASICα homomeric channels, but not channels containing ASICβ, BNC1a, or DRASIC. Thus, understanding how DEG/ENaC subunits assemble to form H+-gated channels is critical to understanding their function and regulation, and to the development of molecules to modulate their activity.
Acknowledgments
We thank Diane R. Olson, Patrick G. Yoder, and the University of Iowa DNA Core Facility (National Institutes of Health Grant DK25295) for technical assistance and Edwin W. McCleskey and Klaus Bielefeldt for comments. This work was supported by National Heart, Lung, and Blood Institute of the National Institutes of Health Grants HL04349 (to C.J.B.), and HL-58812 and HL-55006 (to P.M.S.), by a Veteran's Administration Research Career Development Award (to J.A.W.), and by the Howard Hughes Medical Institute (M.J.W.). M.J.W. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- DRG
dorsal root ganglion
- DEG/ENaC
degenerins/epithelial sodium channel
Footnotes
BNC1a has also been named MDEG (33), BNaC1 (34), and ASIC2a (8), and its splice variant BNC1b has been called MDEG2 (6) and ASIC2b (8). ASICα has also been named BNaC2 (34) and ASIC1a (8), and its splice variant ASICβ has been termed ASIC1b. DRASIC has also been called ASIC3 (8).
Additional properties are more fully characterized by J.X., M.P.P., A. L. Berger, and M.J.W., unpublished data.
References
- 1.Krishtal O A, Pidoplichko V I. Neuroscience. 1980;5:2325–2327. doi: 10.1016/0306-4522(80)90149-9. [DOI] [PubMed] [Google Scholar]
- 2.Krishtal O A, Pidoplichko V I. Neuroscience. 1981;6:2599–2601. doi: 10.1016/0306-4522(81)90105-6. [DOI] [PubMed] [Google Scholar]
- 3.Price M P, Snyder P M, Welsh M J. J Biol Chem. 1996;271:7879–7882. doi: 10.1074/jbc.271.14.7879. [DOI] [PubMed] [Google Scholar]
- 4.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. Nature (London) 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 5.Waldmann R, Bassilana F, de Weille J, Champigny G, Heurteaux C, Lazdunski M. J Biol Chem. 1997;272:20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 6.Lingueglia E, de Weille J R, Bassilana F, Heurteaux C, Sakai H, Waldmann R, Lazdunski M. J Biol Chem. 1997;272:29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- 7.Chen C C, England S, Akopian A N, Wood J N. Proc Natl Acad Sci USA. 1998;95:10240–10245. doi: 10.1073/pnas.95.17.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldmann R, Lazdunski M. Curr Opin Neurobiol. 1998;8:418–424. doi: 10.1016/s0959-4388(98)80070-6. [DOI] [PubMed] [Google Scholar]
- 9.Price M P, Lewin G R, McIlwrath S L, Cheng C, Xie J, Heppenstall P A, Stucky C L, Mannsfeldt A G, Brennan T J, Drummond H A, et al. Nature (London) 2000;407:1007–1011. doi: 10.1038/35039512. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Anoveros J, Samad T A, Woolf C J, Corey D P. J Neurosci. 2001;21:2678–2686. doi: 10.1523/JNEUROSCI.21-08-02678.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price M P, McIlwrath S L, Xie J, Cheng C, Qiao J, Tarr D E, Sluka K A, Brennan T J, Lewin G R, Welsh M J. Neuron. 2001;32:1071–1083. doi: 10.1016/s0896-6273(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 12.Bassilana F, Champigny G, Waldmann R, de Weille J R, Heurteaux C, Lazdunski M. J Biol Chem. 1997;272:28819–28822. doi: 10.1074/jbc.272.46.28819. [DOI] [PubMed] [Google Scholar]
- 13.Babinski K, Catarsi S, Biagini G, Seguela P. J Biol Chem. 2000;275:28519–28525. doi: 10.1074/jbc.M004114200. [DOI] [PubMed] [Google Scholar]
- 14.Sutherland S P, Benson C J, Adelman J P, McCleskey E W. Proc Natl Acad Sci USA. 2001;98:711–716. doi: 10.1073/pnas.011404498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Askwith C C, Cheng C, Ikuma M, Benson C, Price M P, Welsh M J. Neuron. 2000;26:133–141. doi: 10.1016/s0896-6273(00)81144-7. [DOI] [PubMed] [Google Scholar]
- 16.Benson C J, Eckert S P, McCleskey E W. Circ Res. 1999;84:921–928. doi: 10.1161/01.res.84.8.921. [DOI] [PubMed] [Google Scholar]
- 17.Caterina M J, Leffler A, Malmberg A B, Martin W J, Trafton J, Petersen-Zeitz K R, Koltzenburg M, Basbaum A I, Julius D. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- 18.Davis J B, Gray J, Gunthorpe M J, Hatcher J P, Davey P T, Overend P, Harries M H, Latcham J, Clapham C, Atkinson K, et al. Nature (London) 2000;405:183–187. doi: 10.1038/35012076. [DOI] [PubMed] [Google Scholar]
- 19.Canessa C M, Schild L, Buell G, Thorens B, Gautschi I, Horisberger J D, Rossier B C. Nature (London) 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 20.McDonald F J, Price M P, Snyder P M, Welsh M J. Am J Physiol. 1995;268:C1157–C1163. doi: 10.1152/ajpcell.1995.268.5.C1157. [DOI] [PubMed] [Google Scholar]
- 21.Tavernarakis N, Driscoll M. Annu Rev Physiol. 1997;59:659–689. doi: 10.1146/annurev.physiol.59.1.659. [DOI] [PubMed] [Google Scholar]
- 22.Drummond H A, Abboud F M, Welsh M J. Brain Res. 2000;884:1–12. doi: 10.1016/s0006-8993(00)02831-6. [DOI] [PubMed] [Google Scholar]
- 23.Fricke B, Lints R, Stewart G, Drummond H, Dodt G, Driscoll M, von During M. Cell Tissue Res. 2000;299:327–334. doi: 10.1007/s004419900153. [DOI] [PubMed] [Google Scholar]
- 24.Sakai H, Lingueglia E, Champigny G, Mattei M G, Lazdunski M. J Physiol (London) 1999;519 (Pt. 2):323–333. doi: 10.1111/j.1469-7793.1999.0323m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akopian A N, Chen C C, Ding Y, Cesare P, Wood J N. NeuroReport. 2000;11:2217–2222. doi: 10.1097/00001756-200007140-00031. [DOI] [PubMed] [Google Scholar]
- 26.Grunder S, Geissler H S, Bassler E L, Ruppersberg J P. NeuroReport. 2000;11:1607–1611. doi: 10.1097/00001756-200006050-00003. [DOI] [PubMed] [Google Scholar]
- 27.Escoubas P, De Weille J R, Lecoq A, Diochot S, Waldmann R, Champigny G, Moinier D, Menez A, Lazdunski M. J Biol Chem. 2000;275:25116–25121. doi: 10.1074/jbc.M003643200. [DOI] [PubMed] [Google Scholar]
- 28.Bevan S, Yeats J. J Physiol (London) 1991;433:145–161. doi: 10.1113/jphysiol.1991.sp018419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Askwith C C, Benson C J, Welsh M J, Snyder P M. Proc Natl Acad Sci USA. 2001;98:6459–6463. doi: 10.1073/pnas.111155398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron A, Schaefer L, Lingueglia E, Champigny G, Lazdunski M. J Biol Chem. 2001;276:35361–35367. doi: 10.1074/jbc.M105208200. [DOI] [PubMed] [Google Scholar]
- 31.Immke D C, McCleskey E W. Nat Neurosci. 2001;4:869–870. doi: 10.1038/nn0901-869. [DOI] [PubMed] [Google Scholar]
- 32.Voilley N, de Weille J, Mamet J, Lazdunski M. J Neurosci. 2001;21:8026–8033. doi: 10.1523/JNEUROSCI.21-20-08026.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waldmann R, Champigny G, Voilley N, Lauritzen I, Lazdunski M. J Biol Chem. 1996;271:10433–10436. doi: 10.1074/jbc.271.18.10433. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Anoveros J, Derfler B, Neville-Golden J, Hyman B T, Corey D P. Proc Natl Acad Sci USA. 1997;94:1459–1464. doi: 10.1073/pnas.94.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]