Abstract
The identification of two Arabidopsis thaliana genes involved in determining recessive resistance to several strains of the causal agent of bacterial wilt, Ralstonia solanacearum, is reported. Dominant (RRS1-S) and recessive (RRS1-R) alleles from susceptible and resistant accessions encode highly similar predicted proteins differing in length and which present a novel structure combining domains found in plant Toll-IL-1 receptor–nucleotide binding site–leucin-rich repeat resistance proteins and a WRKY motif characteristic of some plant transcriptional factors. Although genetically defined as a recessive allele, RRS1-R behaves as a dominant resistance gene in transgenic plants. Sequence analysis of the RRS1 genes present in two homozygous intragenic recombinant lines indicates that several domains of RRS1-R are essential for its resistance function. Additionally, RRS1-R-mediated resistance is partially salicylic acid- and NDR1-dependent, suggesting the existence of similar signaling pathways to those controlled by resistance genes in specific resistance.
Plants have developed a wide array of defense responses to control pathogen invasion. Among those, the presence or absence of complementary pairs of resistance genes (R) in the host and avirulence genes (avr) in the invading microorganisms determine the outcome of many plant–pathogen interactions. In the elicitor–receptor model proposed to account for this gene-for-gene theory (1), avr genes encode elicitors that serve as ligands for receptors encoded by R genes, which trigger a complex defense response (2). Although several plant R genes from different plant species and corresponding avr genes have been isolated (3, 4), a direct interaction between the products of such genes has been demonstrated in only a few cases (5, 6).
Many R genes conferring race-specific resistance to viral, bacterial, fungal, nematode, and insect pathogens have been characterized (7, 8). Most of these genes contain a leucin-rich repeat (LRR) domain, which functions in mediating protein–protein interactions (9) and plays a direct role in determining specificity in gene-for-gene interactions (8, 10). Indirect evidence indicates that the LRR domains also may be involved in downstream signaling (11). Several cloned R genes belong to the nucleotide binding site (NBS)–LRR class (12) and contain an internal NBS present in several protein families including the RAS group, ATPases, G proteins, and elongation factors (13). NBS motifs also share sequence similarities with the NBS regions of cell death genes such as CED4 from Caenorhabditis elegans and Apaf-1 from humans (14, 15), which suggests that R proteins may bind ATP or act as ATPases. This NBS-LRR class of R genes can be further divided into leucine zipper (LZ)-NBS-LRR and Toll-IL-1 receptor (TIR)-NBS-LRR according to their N-terminal domain. The LZ-NBS-LRR subclass contains in its N-terminal part a LZ sequence, whereas R proteins of the TIR-NBS-LRR class contain a domain similar to the cytoplasmic domains of the Toll, IL-1 receptor, and Myd88 (16, 17). Several studies underline the importance of the conserved TIR, NBS, and LRR domains in pathogen recognition and in triggering transduction pathways leading to induction of defense responses (18–20). Furthermore, the striking degree of similarity in the structural components of R proteins suggests some conservation in the signaling events leading to resistance to various pathogens.
However, the simplified gene-for-gene theory does not provide a clear explanation for all types of disease resistance in plants. For example, many resistances to various plant pathogens including fungi, oomycetes, and viruses are conferred by recessive genes, suggesting a greater mechanistic complexity. Molecular mechanisms involved in this type of resistance remain hypothetical (21), but the recent identification of recessive mutations conferring high levels of resistance to various pathogens provides some insight into the diversity of mechanisms involved. The recessive mlo mutation in barley confers broad spectrum resistance to several isolates of the fungus Erysiphe graminis f sp. hordei, and the Mlo gene encodes a novel class of plant-specific integral membrane proteins anchored in the plasma membrane by seven transmembrane domains (22, 23). Mlo is hypothesized to be a negative regulator of defense responses and/or cell death such that the null mlo alleles mediate resistance by allowing abnormal defense responses to occur both spontaneously and during infection by the pathogen. An Arabidopsis thaliana recessive mutant, edr1, presents a higher level of resistance to some bacterial and fungal pathogens (24). EDR1 encodes a raf-like mitogen-activated protein kinase kinase kinase that may also function as a negative regulator of disease resistance (25). These two examples illustrate recessive mutations that affect the control of defense responses and/or cell death. Other mutants such as some of the A. thaliana pmr mutants, which do not support growth of a fungal pathogen, Erysiphe cichoracearum, are probably altered in plant genes required for growth and reproduction of pathogens. Resistance in this case is not caused by the constitutive activation of known defense pathways (26).
Ralstonia solanacearum (previously named Pseudomonas solanacearum) (27) is the causal agent of bacterial wilt, one of the most important bacterial diseases worldwide. Hundreds of different plant species, including many important agricultural crops such as potato, tomato, banana, pepper, and even trees such as eucalyptus are affected by this vascular pathogen (28). Because of the soil-borne nature of the pathogen, the favored strategy to control bacterial wilt is the breeding of resistant cultivars. In tomato, resistance to R. solanacearum is polygenic, and several loci governing resistance to bacterial wilt have been identified (29, 30). An important step in understanding the molecular basis of host responses to R. solanacearum was the demonstration that different strains of the pathogen induce wilt symptoms on various A. thaliana ecotypes (31). Upon root inoculation, virulent bacteria were found predominantly in the xylem vessels and spread systemically throughout the plant, leading to the complete wilting of susceptible ecotypes within 5–10 days. A. thaliana resistance to R. solanacearum differed markedly from that to most other bacterial pathogens of this Crucifer because no hypersensitive response was observed. In this study, the cloning and characterization of RRS1-R, a gene conferring broad spectrum resistance to R. solanacearum and encoding a novel R protein are reported.
Materials and Methods
Plant Materials and Bacterial Strains.
A. thaliana accessions used in this study were Col-5 (a glabrous derivative of Col-0 used for the generation of recombinant inbred lines) and Nd-1. Disease-resistance phenotypes were determined by root inoculation of 4-week-old plants with R. solanacearum strains as reported (31). Bacterial internal growth curves were performed as described (31).
Recombinant DNA Methods.
Molecular biology techniques were performed by using standard protocols unless otherwise noted (32, 33).
Constructs Used for Plant Transformation.
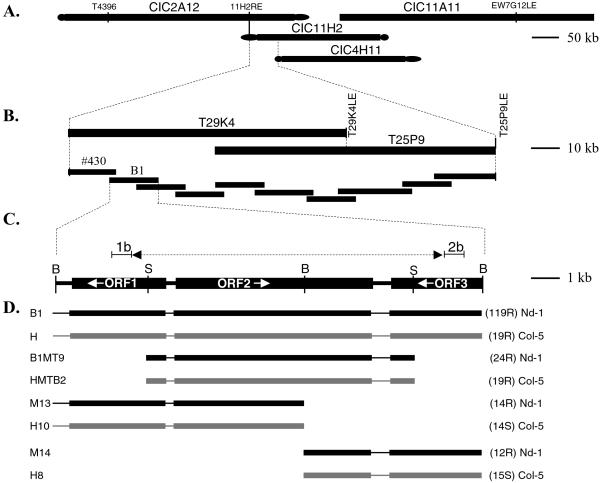
Two DNA fragments of 10 and 8 kb resulting from a BamHI digestion of cosmids B1 (Col-5) and H (Nd-1) were subcloned in the pDHB321.1 binary vector. Subclones M13 and H10 contained the 10-kb fragment from cosmids B1 and H, respectively, whereas subclones M14 and H8 contained the 8-kb fragment from cosmids B1 and H, respectively (Fig. 1D). Two 9.3-kb SphI fragments from cosmids B1 and H were subcloned into pUC19 vector. One SphI site is located 1.1 kb upstream of the predicted ATG translation start codon of the RRS1 genes, and the second SphI site is located 1.8 kb after their polyadenylation signal. The SphI fragments were then cloned into a pDHB321.1 binary vector digested with BamHI and ligated with a BamHI/SphI adapter (primer 1: 5′-GATCGCGGCCGCCATG-3′; primer 2: 5′-GCGGCCGC-3′). The resulting constructs, B1MT9 and HMTB2, corresponding to the genomic clones obtained from the B1 and H cosmids, respectively, were introduced into strain c58pMP90 of Agrobacterium tumefaciens (Fig. 1D). The parental accessions were then transformed by whole-plant infiltration (34). Plants transformed with constructs obtained by using the pSLJ75515 and pDHB321.1 vectors were selected by immersing young seedlings in a 15 mg/l solution of gluphosinate. The copy number of the transgenes was determined on the T2 plants by genetic segregation of BASTA resistance and Southern experiments.
Figure 1.
Map-based cloning of the RRS1-S locus from A. thaliana Col-5 accession. (A) Contig of the four A. thaliana yeast artificial chromosome (YAC) clones CIC2A12, CIC11H2, CIC4H11, and CIC11A11 spanning the RRS1 locus. (B) Contig of cosmid clones generated from BAC clones T29K4 and T25P9 that span the RRS1 locus from the YAC contig. (C) Structural organization of the B1 cosmid. The direction of transcription of the three genes present on B1 is indicated by arrows. (D) Resistant Nd-1 plants were transformed with cosmid B1 or with different constructs derived from it (M13, M14, and B1MT9, black lines) and tested for their response to strain GMI1000. Similar constructs were also generated from Nd-1 cosmid H and used to transform susceptible Col-5 plants (H8, H10, and HMTB2, gray lines). For each construct, the number of independent transgenic lines tested, their genetic background, and their responses to strain GMI1000 (R, resistant; S, susceptible) are indicated on the right.
Results and Discussion
Positional Cloning of the RRS-1 Locus.
Resistance to R. solanacearum strain GMI1000, normally virulent on tomato, segregated as a simply inherited recessive trait in a genetic cross between A. thaliana accessions Col-5 (susceptible) and Nd-1 (resistant). The Col-5 RRS1 locus was localized previously between restriction fragment length polymorphism (RFLP) markers mi61 and mi83 (31) (Fig. 1A). Other RFLP markers tested in this interval, T43968 and EW7G12LE, positioned RRS1 on a yeast artificial chromosome (YAC) contig including CIC2A12, CIC11H2, CIC4H11, and CIC11A11 clones (35) (Fig. 1A). 11H2RE, a marker generated by inverse PCR from the right end of the YAC clone CIC11H2, was used as an RFLP marker on the recombinant inbred population (31). At this stage, no recombination event was detectable between this marker and the RRS1 locus. A total of 650 F2 plants were then screened to identify lines presenting recombination events between the T43968 and EW7G12LE markers and the RRS1 locus. This strategy led to the isolation of 14 F2 plants containing informative recombinants. The Texas A&M University bacterial artificial chromosome (BAC) library was screened by using 11H2RE and a BAC clone, T29K4, was identified (Fig. 1B). A DNA marker, T29K4LE, corresponding to right end of this BAC clone, was generated and used as a probe to identify an overlapping BAC clone, T25P9 (Fig. 1B).
An RFLP cosmid marker, 430, and T25P9LE, derived from the left end of BAC clone T25P9, further narrowed the physical genomic region containing RRS1 to a 150-kb region (Fig. 1B). A contig of overlapping cosmid clones was then generated by subcloning these two BAC clones by a partial BamHI digestion into the vector pSLJ75515 (Fig. 1B). Several of these cosmid clones, when used as RFLP markers, allowed us to position the RRS1 locus on an 18-kb cosmid called B1 whose nucleotide sequence had been determined and contained two R gene-like ORFs and a third unrelated gene (Fig. 1C). ORF1 is RPS4 (GenBank accession number AJ243468), a R gene of the TIR-NBS-LRR family (36), whereas ORF2 and ORF3 are two previously uncharacterized genes. ORF2 is full length and contains features of previously described R genes, whereas ORF3 is truncated. The development of two additional CAPS markers, 1b and 2b, corresponding to ORF1 and ORF3, respectively, defined a mapping interval that contained only ORF2 (Fig. 1C). Therefore, ORF2, which is 6.3 kb long and contains six introns (Fig. 2), unambiguously represents the RRS1-S gene.
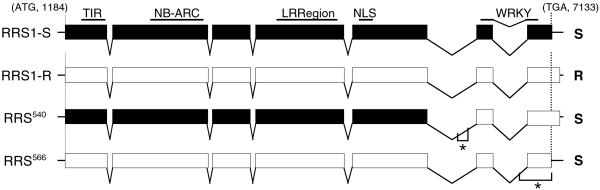
Figure 2.
Schematic representation of RRS1 allelic structures in accessions Col-5 and Nd-1 and in two intragenic recombinant lines. Filled and open boxes indicate exons corresponding to a Col-5 and a Nd-1 genetic background, respectively. Introns are indicated as gaps. Localization of TIR, NB-ARC [nucleotide binding adapter shared by APAF-1 R proteins and CED-4 (14, 15)], LRR, nuclear localization signal (NLS), and WRKY (consensus domain of WRKY protein family) domains on the genes is indicated. In the RRS540 gene, the recombination event occurred between nucleotides 13018 (Col-5) and 13408 (Nd-1) and between nucleotides 13403 and 13686 in RRS566 gene (as indicated by *). The response to strain GMI1000 of homozygous plants at the RRS1 locus is indicated on the right (R, resistant; S, susceptible).
Deduced Primary Structures of RRS1-R and RRS1-S, Genes Corresponding to the Alleles Present in Susceptible Col-5 and Resistant Nd-1 Plants, Respectively.
The deduced protein corresponding to ORF2 presents a modular organization. In its NH2 terminus, RRS1-S contains motifs common to the TIR-NBS-LRR class of R gene products (8). However, its C-terminal region contains a potential nuclear localization signal and a WRKY domain. The WRKY domain is a 60-aa conserved motif characteristic of transcription factors identified only in plants and involved in many biological processes (Figs. 2 and 3) (37, 38).
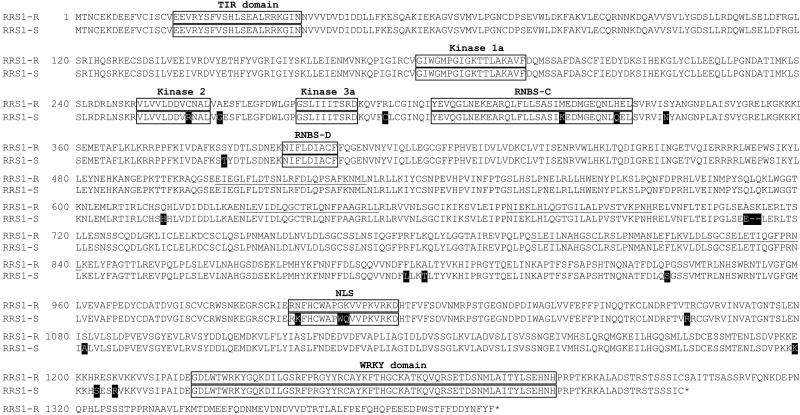
Figure 3.
Amino acid comparison of RRS1-R and RRS1-S. The open boxes indicate the different conserved domains. The six imperfect LRR domains are underlined and the amino acids differing between RRS1-R and RRS1-S are shown as black boxes.
A cosmid, H, carrying the allele associated to resistance, RRS1-R, was isolated by screening a Nd-1 cosmid library using the RRS1-S gene as a probe. Its nucleotide sequence indicated a high level of identity (98%) between the sequences of the two alleles. This level of conservation (98%) extends through 1.1 kb of the potential promoter region 5′ to the gene. Despite this overall conserved organization, the two genes differ in the position of a stop codon that leads in RRS1-S, the Col-5 allele, to the formation of a protein truncated by 90 aa (Figs. 2 and 3). Both proteins had 65% identity with an A. thaliana putative protein of similar structure (The Institute for Genomic Research accession number AT5 g45050).
Analysis of the derived amino acid sequences of the RRS1 proteins revealed the presence of several domains found in many resistance proteins (Fig. 3). Based on structural and functional similarities with the signaling domains of the Drosophila Toll protein and mammalian IL-1 receptors, the TIR domain present in the NH2 terminus of the RRS1 proteins (Fig. 2) may play a role in R gene specificity and in disease resistance signal-transduction pathways (20, 39, 40). The putative NBS domain (Fig. 2) contains the conserved motifs found in a large number of proteins binding ATP or GTP (41). The consensus LRRs of RRS1 proteins consists of six imperfect 23- to 25-aa motifs. All of the domains mentioned above are extremely well conserved among the two RRS1 proteins (Fig. 3), and structure-function analysis of RRS1-R and RRS1-S should bring some insight into the importance of the various minor amino acid differences detected between the two proteins on RRS1-R function.
RRS1 represents therefore the first R protein containing a group III conserved WRKY domain (42), suggesting a regulatory role in the expression of the signaling pathways leading to resistance/susceptibility. This potential role in transcriptional activation is enhanced by the presence of a putative nuclear localization signal (Figs. 2 and 3) suggesting that this protein is targeted to the nucleus. Interestingly, some avr gene products such as AvrBs3 have been shown to be nuclear localized (43, 44), indicating that a direct or indirect interaction between products of some avr genes and those of the corresponding R genes may occur in the plant nucleus.
RRS1-R Confers Resistance to Strain GMI1000 of R. solanacearum.
To confirm the function of RRS1-R and RRS1-S, constructs were introduced into both parental accessions (Fig. 1D). Nd-1 plants containing either a cosmid (B1) containing the RRS1-S gene and the two ORFs previously mentioned (119 independent transformants) or a 9.3-kb genomic fragment (B1MT9) containing only the RRS1-S gene and its flanking regions (24 independent transformants) did not develop wilt disease. This finding indicated that RRS1-S is not in itself a susceptibility gene capable of suppressing the resistance phenotype of Nd-1. These results contradict previous genetic data that heterozygous Col-5/Nd-1 plants are fully susceptible to the pathogen (31). Because position effects or gene silencing in the transgenic plant analysis may explain this discrepancy, reverse transcriptase–PCR experiments were performed to estimate the transcript levels corresponding to both alleles in transgenic and parental lines. Results indicated that transcripts corresponding to RRS1-R and RRS1-S were detected in resistant transgenic plants and that no correlation could be established between the RRS1-R and RRS1-S transcript levels and the plant response to strain GMI1000 (data not shown). Thus, position effects or gene silencing are not responsible for the lack of suppression of the resistance phenotype. Furthermore, we found no genetic evidence for semidominance of RRS1-R in 12 independent F1 progenies obtained by crossing the two parental ecotypes and inoculated with 105, 106, 107, and 108 colony-forming units/ml of strain GMI1000, this latter concentration being the one used routinely in our pathogenicity tests. Wilt disease developed at all inoculum concentrations in the F1 lines, demonstrating genetic dominance of the RRS1-S allele from Col-5 plants.
Susceptible Col-5 plants were transformed with either the H cosmid containing RRS1-R, or HMTB2, a 9.3-kb genomic clone corresponding to the RRS1-R gene and its flanking regions (Fig. 1D). All of the transgenic lines (19 independent for each construct) were resistant and failed to develop wilt symptoms upon inoculation with strain GMI1000 (Fig. 1D). Col-5 plants transformed with constructs H8 and H10, which contain a truncated RRS1-R gene remained fully susceptible to the pathogen. These data demonstrated that RRS1-R is an R gene. Reverse transcriptase–PCR experiments indicated that, as in the case of Nd-1 plants transformed with RRS1-S, both alleles were transcribed in transgenic Col-5 lines (data not shown).
Several Domains of RRS1-R Are Essential for Resistance.
Structure-function analysis of two susceptible homozygous intragenic recombinant lines identified during RRS1-S positional cloning provided further evidence for the role of RRS1-R in resistance to R. solanacearum. Line 566 contains a chimaeric RRS1 gene whose nucleotide sequence corresponds to RRS1-R up to the stop codon of RRS1-S (Fig. 2). The 90 aa at the C-terminal end of the resistant allele appear therefore to be essential for the function of the protein in disease resistance. Line 540 contains the C-terminal WRKY domain of RRS1-R and the TIR-NBS-LRR domains of RRS1-S and yet does not act as a resistance protein (Fig. 2). This finding suggests that other domains of the resistance protein are also indispensable for RRS1-R function.
RRS1-R Restricts Pathogen Growth in Infected Plants and Confers Broad Spectrum Resistance to R. solanacearum.
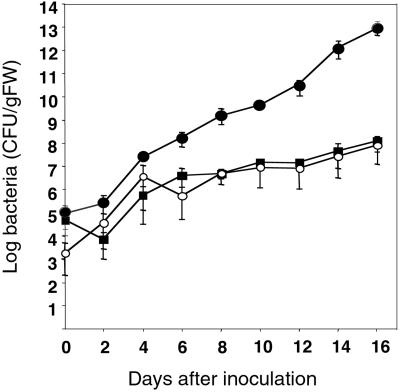
The growth of the pathogen within control plants and a selected transgenic line, CH1.2, homozygous for both RRS1-R and RRS1-S, was monitored (Fig. 4). Bacterial multiplication in Col-5 plants containing the RRS1-R transgene was comparable with that obtained in Nd-1 resistant plants, and lower by more than 5 orders of magnitude than that in susceptible Col-5 plants. Therefore, the RRS1-R gene behaves as a dominant resistance gene capable of limiting bacterial growth in a Col-5 genetic background.
Figure 4.
Internal bacterial growth curves of R. solanacerum strain GMI1000 in Col-5 plants (●), CH1.2, a selected transgenic Col-5 line containing the cloned RRS1-R gene (○), and Nd-1 plants (■). Root inoculations with strain GMI1000 were performed as described (31).
To assess the ability of RRS1-R to confer resistance to a variety of R. solanacearum strains, Col-5 transgenic plants carrying the resistance allele RRS1-R were inoculated with four strains of the pathogen that induce differential responses on accessions Nd-1 and Col-5 (31). Transgenic Col-5 plants were shown to respond in a similar way to Nd-1 plants and were fully resistant to strains AW1, GA4, GT4, and 0170 isolated from different host plants in various geographic areas. This finding implies that all of these pathogen isolates directly interact with the RRS1-R pathway and, in fact, may contain the same avirulence protein. Alternatively, these strains may have developed similar strategies to colonize various plants that involve interactions with regulatory pathways controlled by RRS1-R. RRS1-R, therefore, provides resistance to several strains of R. solanacearum.
RRS1-R-Mediated Resistance Depends on Salicylic Acid and NDR1.
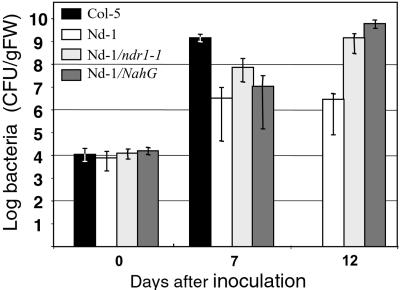
Most A. thaliana R genes characterized so far require salicylic acid and the signal pathway genes NDR1 or EDS1 (45). Their role in RRS1-R-mediated resistance that does not involve apparent hypersensitive response upon root inoculation, was tested. Salicylic acid appeared to play a role in RRS1-R-mediated resistance because Nd-1 plants containing salicylate hydroxylase, which converts salicylic acid into inactive catechol, developed wilt symptoms. Resistance was also abolished when Nd-1 plants were crossed into the ndr1/ndr1 mutant background and RRS1-R/RRS1-R ndr1/ndr1 offspring were selected (Fig. 5). In both cases, however, complete wilting of the plant occurred 4–7 days after the death of susceptible Col-5 plants, implying that other signaling components play a role in RRS1-mediated resistance.
Figure 5.
Effect of salicylic acid and ndr1–1 on RRS1-R-mediated resistance to R. solanacearum. Plants were inoculated with strain GMI1000. Bacterial growth was estimated as described (16) at the time of inoculation, 7 days after inoculation at which time 75–100% of Col-5 leaves were wilted and no symptoms were visible on Nd-1, Nd-1/NahG, and RRS1-R/ndr1–1 plants (T7), and 12 days after inoculation at which time 75–100% of Nd-1/NahG and RRS1-R/ndr1–1 leaves were wilted whereas no symptoms were detectable on Nd-1 plants (T12).
Conclusions
In summary, this study led to the identification of a class of proteins conferring resistance to several strains of R. solanacearum and combining structural motifs found in the TIR-NBS-LRR subclass of R gene products and a putative transcriptional domain. The structure of the RRS1 alleles is consistent with RRS1-R being a recessive gene. RRS1-S is probably not a host susceptibility protein required by the pathogen for disease development because transgenic Nd-1 plants carrying the RRS1-S gene failed to develop wilt disease. In addition, structural similarities between the RRS1 genes and other cloned R genes, as well as results obtained from Col-5 complementation experiments with the RRS1-R gene, make this hypothesis rather unlikely. Additionally, establishment of RRS1-R-mediated resistance requires NDR1 and salicylic acid, both essential for many A. thaliana R gene-mediated resistances. We favor a model in which the truncated RRS1-S protein would be a dominant negative regulator of the function of the full-length RRS1-R protein in F1 and F2 heterozygous plants. Additionally, the high similarity between the RRS1 proteins strengthens the possibility that they may compete for some essential component(s) involved in the pathogen perception or in signal transduction pathways. For instance, despite a deletion of 90 aa, which does not eliminate the WRKY motif, RRS1-S may still be capable of binding its DNA target(s) but would be inefficient in transcription of these genes. In this case, RRS1-R function may be affected/inhibited as the DNA binding sites are occupied by RRS1-S. In addition, minor differences in the NH2-terminal region of RRS1 proteins may also modify their respective affinities, either for an Avr- or a pathogen-derived factor, or for a plant protein. However, reasons for which RRS1-S cannot exert its direct or indirect repressor activity on the function of resistance of RRS1-R in Col-5 resistant transgenic plants remain unexplained. Several studies highlight the complexity of plant R gene expression: for example, the tobacco N gene, another member of the TIR-NBS-LRR subclass of R genes, encodes two transcripts by means of alternative splicing of an alternative exon present in intron III (46). The relative ratio of both transcripts is regulated by tobacco mosaic virus infection and is critical for complete resistance to the virus. The possible involvement of posttranscriptional/translational controls has also been proposed for Xa21 (47), whose transcript levels during plant development are not correlated with the expression of disease resistance (48). Future studies evaluating the importance of such mechanisms on RRS1 gene expression should bring some insights into the underlying mechanisms responsible for the observed contradiction between genetic and complementation data. Identification of null mutants and their complementation by the RRS1-R or RRS1-S genes also should provide some indications about the molecular mechanisms involved.
RRS1-R is the first R gene of the TIR-NBS-LRR subclass for which a functional NDR1 gene is required. This observation may, however, be explained by the presence in the C-terminal part of this R protein of the additional WRKY domain, which may alter its mode of action. Alternatively, the NDR1 requirement may be caused by the atypical nature of RRS1-R-mediated resistance, which is not associated with hypersensitive response development. Attempts to test the effect of EDS1, a gene required by most R genes of the TIR-NBS-LRR subclass, were unsuccessful because of the presence of the eds1 mutation in two Arabidopsis ecotypes, WS and La-er, which tolerate very poorly the conditions used to perform pathogenicity tests.
RRS1 is the first TIR-NBS-LRR resistance protein that contains a putative transcriptional activation domain. Potentially, their modular structure suggests that RRS1-R and RRS1-S have a dual function: the NH2 terminus may bind a pathogen-derived signal by means of the LRR motifs known to mediate protein–protein interactions. This recognition event may lead to the activation of the C-terminus WRKY transcriptional factor domain. This, in turn, would activate a signaling cascade, or directly activate defense-related genes, leading to the plant resistance response. The importance of some WRKY DNA-binding proteins in pathogen-induced signaling pathways has recently been highlighted by several studies. Thus, certain WRKY factors act upstream of some defense gene products such as pathogenesis-related proteins (44) or NPR1, a positive regulator of inducible plant disease resistance, and regulate their expression (49). Additionally, transcriptome analysis of A. thaliana during systemic acquired resistance (50) led to the identification of WRKY transcription factors as potential common regulators of genes in the PR-1 regulon, which contains known pathogenesis-related proteins as well as novel genes likely to function during systemic acquired resistance and disease resistance (51).
The identification of RRS1-S and RRS1-R exemplifies a type of resistance in which both recessive and dominant alleles are probably functional and may compete for a given DNA target or a limiting factor necessary either for the growth/propagation of a pathogen or for the establishment of resistance/susceptibility in the infected plants. Although purely speculative at this stage, it is tempting to hypothesize that such a mechanism may constitute the molecular basis of some recessive resistance. Finally, RRS1-R is the first characterized R gene conferring resistance to R. solanacearum, and its characterization, the elucidation of its mode of action, as well as its intergeneric transfer in various crops are important steps in facilitating the elaboration of new approaches to disease control.
Acknowledgments
We thank D. Bouchez for the pDHB321.1 vector, B. J. Staskawicz and W. Gassmann for several cosmid clones used as RFLP markers and ndr1–1 plants, C. R. Buell and E. Drenkard for RFLP markers, Ohio State University's Arabidopsis Stock Center for various RFLP markers and BAC filters containing Texas A&M University libraries, J. Jones for the pSLJ75515 vector, J. Dénarié, R. Ranjeva, and M. Whalen for critical reading of the manuscript, and Génoplante for financial support.
Abbreviations
- R
resistance gene
- avr
avirulence gene
- TIR
Toll–IL-1 receptor
- NBS
nucleotide binding site
- LRR
leucin-rich repeat
- BAC
bacterial artificial chromosome
- RFLP
restriction fragment length polymorphism
Footnotes
References
- 1.Flor H. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 2.Gabriel D, Rolfe B. Annu Rev Phytopathol. 1990;28:365–391. [Google Scholar]
- 3.Baker B, Zambrisky P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 4.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 5.Tang X Y, Frederick R D, Zhou J M, Halterman D A, Jia Y L, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 6.Jia Y, MacAdams S A, Bryan G T, Hershey H P, Valent B. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis J, Jones D. Curr Opin Plant Biol. 1998;1:288–293. doi: 10.1016/1369-5266(88)80048-7. [DOI] [PubMed] [Google Scholar]
- 8.Bent A F. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobe B, Deisenhofer J. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 10.Jones D A, Jones J D G. Adv Bot Res Incorp Adv Plant Pathol. 1997;24:90–167. [Google Scholar]
- 11.Warren B C, Henk A, Mowery P, Holub E, Innes R W. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyers B C, Dickerman A W, Michelmore R W, Sivaramakrishnan S, Sobral B W, Young N D. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313x.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- 13.Saraste M, Sibbald P R, Wittinghoher A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 14.van der Biezen E A, Jones J D. Curr Biol. 1998;8:R226–R227. doi: 10.1016/s0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- 15.Aravind L, Dixit V M, Koonin E V. Trends Biochem Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 16.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slack J L, Schooley K, Bonnert T P, Mitcham J L, Qwarstrom E E, Sims J E, Dower S K. J Biol Chem. 2000;275:4670–4678. doi: 10.1074/jbc.275.7.4670. [DOI] [PubMed] [Google Scholar]
- 18.Dinesh-Kumar S P, Wai-Hong T, Baker B. Proc Natl Acad Sci USA. 2000;97:14789–14794. doi: 10.1073/pnas.97.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botella M A, Parker J E, Frost L N, Bittner-Eddy P D, Beynon J L, Daniels M J, Holub E B, Jones J D. Plant Cell. 1998;10:1847–1860. doi: 10.1105/tpc.10.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis J G, Lawrence G J, Luck J E, Dodds P N. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser R S S. Annu Rev Phytopathol. 1990;28:179–200. [Google Scholar]
- 22.Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 23.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 24.Frye C A, Innes R W. Plant Cell. 1998;10:947–956. doi: 10.1105/tpc.10.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frye C A, Tang D, Innes R W. Proc Natl Acad Sci USA. 2001;98:373–378. doi: 10.1073/pnas.98.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel J, Somerville S. Proc Natl Acad Sci USA. 2000;97:1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuki Y. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 28.Hayward H C. Annu Rev Phytopathol. 1991;29:65–87. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 29.Thoquet P, Olivier J, Sperisen C, Rogowsky P, Laterrot H, Grimsley N. Mol Plant-Microbe Interact. 1996;9:826–836. [Google Scholar]
- 30.Thoquet P, Olivier J, Sperisen C, Rogowsky P, Prior P, Anaïs G, Mangin G, Bazin B, Nazer R, Grimsley N. Mol Plant-Microbe Interact. 1996;9:837–842. [Google Scholar]
- 31.Deslandes L, Pileur F, Liaubet L, Camut S, Can C, Williams K, Holub E, Beynon J, Arlat M, Marco Y. Mol Plant-Microbe Interact. 1998;11:659–667. doi: 10.1094/MPMI.1998.11.7.659. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 33.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J G, Struhm K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 34.Bechtold N, Pelletier G. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 35.Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, Le Paslier D, Cohen D, Chaboue M E, Durr A. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- 36.Gassmann W, Hinsch M E, Staskawicz B J. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 37.Rushton P J, Macdonald H, Huttly A K, Lazarus C M, Hooley R. Plant Mol Biol. 1995;29:691–702. doi: 10.1007/BF00041160. [DOI] [PubMed] [Google Scholar]
- 38.Rushton P J, Torres J T, Parniske M, Wernert P, Hahlbrock K, Somssich I. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 39.Dinesh-Kumar S P, Whitham S, Choi D, Hehl R, Corr C, Baker B. Proc Natl Acad Sci USA. 1995;92:4175–4180. doi: 10.1073/pnas.92.10.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker J E, Coleman M Y, Szabo V, Frost L N, Schmidt R, van der Biezen E A, Moores T, Dean C, Daniels M J, Jones J D G. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traut T W. Eur J Biochem. 1994;222:9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- 42.Eulgem T, Rushton P J, Robatzek S, Somssich I. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 43.Bonas U, Van Der Ackerveken G. Plant J. 1997;12:1–7. doi: 10.1046/j.1365-313x.1997.12010001.x. [DOI] [PubMed] [Google Scholar]
- 44.Yang P, Wang Z, Fan B, Chen C, Chen Z. Plant J. 1999;18:141–149. [Google Scholar]
- 45.Glazebrook J. Curr Opin Plant Biol. 1999;2:280–286. doi: 10.1016/S1369-5266(99)80050-8. [DOI] [PubMed] [Google Scholar]
- 46.Dinesh-Kumar S P, Baker B J. Proc Natl Acad Sci USA. 2000;15:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song W Y, Wang G L, Chen L L, Kim H S, Pi L Y, Holsten T, Gardner J, Wang B, Zhai W X, Zhu L H, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 48.Century K S, Lagman R A, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald P, Whalen M. Plant J. 1999;20:231–236. doi: 10.1046/j.1365-313x.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- 49.Yu D, Chen C, Chen Z. Plant Cell. 2001;13:1527–1540. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryals J L. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maleck K, Levine A, Eulgem T, Morgan A, Schmid R, Lawton K A, Dangl J L, Dietrich R A. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]