Abstract
Interactions between Cauliflower mosaic virus (CaMV) and its aphid vector are regulated by the viral protein P2, which binds to the aphid stylets, and protein P3, which bridges P2 and virions. By using baculovirus expression of P2 and P3, electron microscopy, surface plasmon resonance, affinity chromatography, and transmission assays, we demonstrate that P3 must be previously bound to virions in order that attachment to P2 will allow aphid transmission of CaMV. We also show that a P2:P3 complex exists in the absence of virions but is nonfunctional in transmission. Hence, unlike P2, P3 and virions cannot be sequentially acquired by the vector. Immunogold labeling revealed the predominance of spatially separated P2:P3 and P3:virion complexes in infected plant cells. This specific distribution indicates that the transmissible complex, P2:P3:virion, does not form primarily in infected plants but in aphids. A model, describing the regulating role of P3 in the formation of the transmissible CaMV complex in planta and during acquisition by aphids, is presented, and its consequences are discussed.
Cauliflower mosaic virus (CaMV) belongs to the genus Caulimovirus. It possesses a DNA-based genome but uses RNA templates for replication via reverse transcription. The genome is a double-stranded circular DNA encoding six genes with specific functions (reviewed in ref. 1). Like many other plant viruses, CaMV is transmitted by aphids from one host plant to another (reviewed in refs. 2 and 3). Its mode of transmission is noncirculative, meaning that transmissible virus particles acquired from infected plants are retained only on the cuticle lining the food canal in the aphid stylets. From there, CaMV is released when the vector probes on a new host plant. Transmission of CaMV follows the helper strategy: a helper component (HC), a viral nonstructural protein, is mandatory for mediating vector transmission (4, 5). The HC presumably promotes virus retention in the vector by serving as a reversible molecular bridge between the virus particles and a hypothetical binding site(s) in the aphid stylets. In the case of CaMV, the HC is encoded by the viral gene II (6–8). This gene codes for the 18-kDa nonstructural protein P2 that apparently is involved only in aphid transmission, because P2-deficient CaMV strains are otherwise perfectly infectious although not aphid transmissible (9). It was recently shown, however, that P2 alone cannot mediate aphid transmission of CaMV. In addition, the action of the viral gene III product P3 is needed (10). This 15-kDa protein had earlier been reported to copurify with virus particles and be essential for virus infectivity, although no particular function was attributed to it (11–13). Leh et al. (10, 14) showed a direct interaction between P2 and P3 and between P3 and the virus capsid. They also provided evidence that P2 is unable to associate directly with the virus coat and demonstrated that P3 mediates binding of P2 to the virus. Thus, the components of the CaMV transmissible complex are now identified, but its formation and regulation are not understood.
It has been established that, to allow transmission, P2 can be acquired by aphids either before or together with the other components involved in transmission (4, 5). However, other possible feeding sequences of the three components P2, P3, and virions have not been tested. Therefore, the precise sequence of events resulting in formation of transmissible complexes remains unknown.
In infected plant cells, CaMV proteins are synthesized in the cytoplasm and accumulate in electron-dense (edIBs) and electron-lucent (elIBs) inclusion bodies. The edIBs contain mainly virus particles in a matrix of viral protein P6 (product of gene VI) and are believed to be the site of viral protein synthesis and virion assembly (15). On the other hand, elIBs were described as an electron-lucent matrix made of P2 in which some scattered virions are embedded, and have been suggested to be involved in aphid transmission (16). It is now evident that the possible occurrence of CaMV transmissible complexes in elIBs depends on the presence of P3 in these inclusions. Unfortunately, the intracellular distribution of P3 is not known. Hence, as is the case for any other plant virus transmitted by the helper strategy, it remains unclear whether the complete transmissible viral complexes exist preformed in plant cells or assemble in the aphid vector, or both.
We set out in this report to define precisely the roles of the various CaMV components involved in the transmission process. We provide evidence that the intracellular spatial separation and biochemical properties of these CaMV proteins limit assembly of transmissible complexes in infected plant cells and favor their formation in aphid stylets. The consequences of such an acquisition mode on the way a CaMV population is sampled by the aphid vector during each round of transmission are discussed.
Material and Methods
Propagation and Isolation of CaMV.
CaMV strain CabbS (17) was propagated in turnip plants (Brassica rapa, cv: “Just Right”) as described (7). Virus particles were purified as described (10).
Transmission Assays.
Transmission assays were carried out as described in ref. 7. Briefly, groups of starved aphids of the species Myzus persicae (Sulz.) were allowed to probe consecutively different CaMV protein-containing solutions for 15 min each through stretched Parafilm (American National Can, Menasha, W) membranes. Groups of 10 aphids were then placed on turnip test plants for inoculation and allowed to feed overnight before insecticide treatment. Symptoms were recorded 3 weeks later by visual inspection. In all experiments, P2, P3, and virions were used at concentrations of 0.5, 0.2, and 0.1 mg/ml, respectively.
Plasmids and Recombinant Baculoviruses.
The baculovirus transfer plasmid used in this study is a modification of P10-119pst (18). A DNA fragment containing an ATG codon, a (his)6 tag-encoding sequence, and an additional NotI restriction site was cloned at the NcoI and PstI sites of P10-119pst to yield p119His. The coding sequence, including start and stop codons, of gene III of CaMV CabbS strain was PCR-amplified and cloned into p119His at BglII and PstI sites, resulting in pP3 plasmid. The plasmid construct was verified by sequencing. Cloning of CaMV gene II in p119His and purification of his-tagged P2 (HP2) is reported in ref. 19.
Recombinant baculoviruses were obtained by cotransfecting Spodoptera frugiperda (Sf9) cells with our plasmids and the AcSLP10 DNA, as described (20). Plasmid pP3 resulted in a baculovirus expressing native P3. The baculovirus expressing native untagged P2 was described previously (7).
Production of Recombinant Proteins.
Sf9 cells were infected with recombinant baculoviruses at a multiplicity of infection of 10 and harvested after 48 h incubation at 28°C. For transmission assays, the cells were resuspended in SES buffer (7), ultrasonicated, and stored up to several weeks at −20°C without loss of activity.
For in vitro assays, cells were resuspended in tpP3 buffer [100 mM Tris, pH 8.0/200 mM NaCl/1 mM EDTA/1× Antiprotease Complete (Roche Diagnostics, Meylan, France)] or in DB5 buffer (19) for P3 and HP2, respectively, ultrasonicated, and stored at −20°C until use.
For surface plasmon resonance, heat stable P3 from crude Sf9 extracts was partially purified by a 10-min treatment at 65°C, followed by a 10-min centrifugation at 20,000 × g to remove denatured proteins. The C-terminal 60 aa of P2, containing the P3 binding domain, were produced as described (19) and designated HP2-Cter.
Preparation of P2 Antiserum.
Purified HP2 was subjected to SDS/PAGE, and the major band was excised from the gel and sent to Eurogentec (Seraing, Belgium) where mice were immunized and P2-reactive serum was obtained. The antiserum did not crossreact with any plant or Sf9 proteins. P3 and P4 antisera were kindly provided by T. Hohn (FMI, Basel).
In Vitro Interaction Assays.
To produce HP2-resin, whole cell lysates of HP2-producing Sf9 cells in DB5 buffer were centrifuged for 30 min at 20,000 × g. Ni-nitrilotriacetic acid (NTA) agarose (Qiagen, Chatsworth, CA) preequilibrated with DB5 buffer was added to the supernatant and shaken for 30 min at 4°C. Unbound proteins were removed by three washes with DB5 buffer; then the HP2-charged resin was equilibrated with tpP3 buffer. P3-containing supernatant was obtained by centrifuging lysates, in tpP3 buffer, of Sf9 cells infected with P3-expressing baculovirus. After including or omitting an incubation for 30 min with purified CaMV virions, the solution was mixed with HP2-charged resin and stirred for another 30 min at 4°C. Then the resin was washed three times with tpP3 buffer and resuspended in SES buffer containing 400 mM EGTA to elute Ni-NTA-bound proteins.
Biacore.
This technique, based on surface plasmon resonance, allows real-time visualization of the molecular interactions of a flowing analyte with a ligand immobilized on a gold-coated sensor chip. Mass changes because of binding or dissociation of compounds to the chip are optically measured without the need to label the interacting partners. Experiments were carried out at 25°C by using a BIACORE 2000 instrument (Biacore, Uppsala). HP2, HP2-Cter, or virus particles were immobilized on a CM5 sensor chip via primary amino groups according to the manufacturer's instructions. The running buffer was HBS (10 mM Hepes, pH 7.4/150 mM NaCl/3 mM EDTA/0.005% P20 detergent). Different concentrations of P3, in the presence or absence of virus, were injected simultaneously into the measuring and control (no protein immobilized) flow cells at a flow rate of 30 μl/min. Sensorgrams represent the kinetics of association and dissociation of interacting partners. They were analyzed, by using biaevaluation 3.0 software, to calculate affinities.
Electron Microscopy.
For infected Sf9 cells, preparation of samples for electron microscopy was carried out essentially as previously described (8). Infected turnip leaves were treated similarly except that they were fixed for 4 h at room temperature after vacuum infiltration with 50 mM sodium cacodylate buffer (pH 7.4) containing 0.5% glutaraldehyde and 2% paraformaldehyde. For immunoelectron microscopy, samples were then embedded in Unicryl resin, which was polymerized under UV light at 4°C for 48 h.
Ultrathin sections on grids were quenched with 50 mM (NH4)Cl in PBS and blocked for 30 min in TBS1 (TBS plus 0.1% Tween 20 and 5% skim milk powder). Grids were incubated with primary antisera in TBS1 (1:25 for mouse P2- and 1:50 for rabbit P3-antiserum) for 1 h before three rinses with TBS2 (TBS1 with 0.5% skim milk powder). The grids were finally incubated with gold-conjugated secondary antibodies (30 nm particle rabbit anti-mouse and 10 nm particle goat anti-rabbit) diluted 1:50 in TBS1 for 1 h. After three washes with TBS3 (TBS1 without skim milk powder) followed by several rinses with H2O, the sections were contrasted with 2% uranyl acetate and 1% lead citrate and observed in a Zeiss EM 10C/RC electron microscope operated at 60–80 kV.
Protein Determination.
Protein concentration was measured by using the Bradford assay (21). For determination of P3 in crude extracts, a Coomassie Blue-stained gel was scanned, and the amount of P3 estimated by densitometry by using scionimage (Scion, Frederick, MD), a NIH image software spin-off.
Western Blotting.
Samples were separated by 12% SDS/PAGE (22), and the proteins were transferred to nitrocellulose membranes by using a semidry blotting apparatus (CBS) according to the manufacturer's instructions. After blocking the membranes with TBS1, they were incubated with primary antibody (dilution 1:2,000 for rabbit P3 and P4 antisera, 1:1,000 for rabbit anti-P2) in TBS1 for 1 h. After three rinses with TBS3 and 1 h incubation with alkaline phosphatase-conjugated anti-rabbit IgG in TBS1, the membranes were rinsed three times with TBS3, and bound antibodies were detected by using the NBT/BCIP (nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate) color reaction.
Results
Localization of P2, P3, and Virions in Infected Turnip Cells.
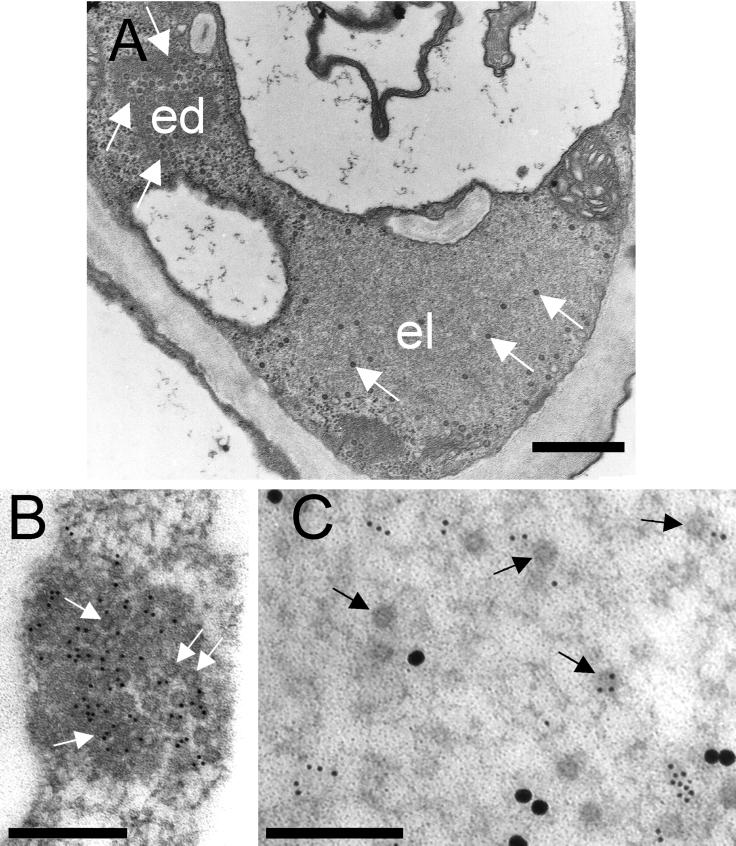
We determined by immunoelectron microscopy of numerous cells, from four independently infected plants, the intracellular location of CaMV components involved in transmission to find out what aphids might take up during a single cell puncture. Fig. 1A displays a cell with typical edIBs and elIBs containing highly concentrated virus particles in a dark matrix and some scattered virions in a light matrix, respectively. By Immunogold double-labeling, we detected P2 exclusively in the matrix of elIBs but not of edIBs (compare Fig. 1 B and C), as previously shown by Espinoza et al. (16). High amounts of P3, on the other hand, were found in both types of inclusions. In edIBs (Fig. 1B), P3 colocalized with CaMV particles whereas, in elIBs, most of P3 was, like P2, distributed in the light matrix (Fig. 1C). Beyond the fact (see below) that these results suggest that edIBs and elIBs are likely composed mainly of P3:virions and P2:P3 aggregates, respectively, they also imply a spatial separation of P2 and virus particles within infected plant cells.
Figure 1.
Differential localization of P2 and P3 in infected turnip leaf mesophyll cells. (A) Overview of a mesophyll cell showing electron-lucent (el) and electron-dense (ed) viral inclusion bodies. Immunogold double-labeling of an edIB (B) and an elIB (C) shows that P2 (30 nm gold) is detected exclusively in elIBs, whereas P3 (10 nm gold) is found in both types of inclusions. The arrows point to virus particles. Bar = 0.6 μm (A) and 0.3 μm (B and C).
To substantiate this important finding, we performed Immunogold single-labeling of P2 with high titer rabbit P2 antiserum and 10 nm gold-conjugated secondary antibody to increase sensitivity. Twelve thin sections from four different infected plants were gold-labeled. We consistently observed heavy labeling of elIBs, whereas the label in edIBs was never above background levels (not shown). We conclude that P2 is absent from mature edIBs and discuss this further below. We also quantified the distribution of virions in edIBs and elIBs by counting CaMV particles in 119 inclusion bodies (edIBs and elIBs) from randomly chosen cells of four independently infected plants. Table 1 shows that edIBs are more numerous than elIBs. The average size of edIBs is smaller, but the total surface area observed for each inclusion type is comparable. More importantly, in the tissues examined, 94% of virus particles were in the edIBs. Thus, both the enormous size of elIBs and the fact that they contain only about 6% of the total virions suggest that they mainly consist of large P2:P3 aggregates, although the presence of host plant components cannot be excluded.
Table 1.
Distribution of virions in viral inclusion bodies
| Number (%) | Virions (%) | Total size* (%) | Mean size/IB* | |
|---|---|---|---|---|
| edIB | 94 (79) | 11,735 (94) | 66.7 μm2 (53) | 0.7 μm2 |
| elIB | 25 (21) | 695 (6) | 58.9 μm2 (47) | 2.4 μm2 |
Ultrathin sections from different locations from four independently infected plants were screened for edIBs and elIBs, and their number and the quantity of contained virions were counted.
Total size refers to the combined sum of the surface of all edIBs or elIBs, mean size to total surface divided by the number of the corresponding inclusions (IB).
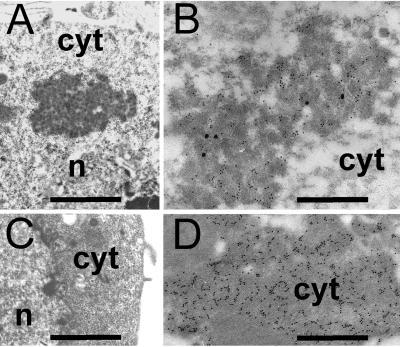
Because a possible P2:P3 complex has not been reported in plant cells before, although in vitro data (10) indicate that it might exist, we sought to confirm its existence in vivo. With this aim, we coinfected Sf9 cells with P2- and P3-encoding baculoviruses. Fig. 2 A and B shows that, in coinfected Sf9 cells, P2 and P3 indeed colocalize in de novo inclusion bodies whereas infection solely with a P2-encoding baculovirus resulted in formation of typical paracrystals as reported (8, 23). In contrast, infection with a P3-encoding baculovirus alone gave rise to uniform distribution of P3 throughout the cell (Fig. 2 C and D). When the P2157m mutant (24), which does not bind P3 in vitro (10), was coexpressed with P3 in Sf9 cells, no such inclusion bodies were observed and P3 remained evenly dispersed throughout the cell (not shown). Thus, our results indicate that a P2:P3 complex exists in Sf9 cells.
Figure 2.
P2 and P3 form aggregates in baculovirus-infected Sf9 cells. Coinfection of Sf9 cells with recombinant baculoviruses encoding P2 and P3 results in formation of inclusions (A) that are (B) labeled by P2 (30 nm gold) and P3 (10 nm gold) antisera whereas P3 in singly infected Sf9 cells does not form inclusions (C) but (D) gives rise to a uniform P3 label (10 nm gold). Bar = 1 μm (A and C) and 0.5 μm (B and D); cyt, cytoplasm; n, nucleus.
P3 Must Be Supplied with Virions To Be Active in Transmission.
The previous section shows a spatial separation in plant cells of P2, probably in a complex with P3 in elIBs, from the P3:virion complexes in edIBs. Consequently, P2:P3 and P3:virions might be independently acquired by aphids. We therefore tested all possible combinations of P2, P3, and virions in different acquisition sequences to determine what components must be taken up in which sequence for CaMV transmission to occur.
First, the experimental system was checked to ensure that P3 expressed in Sf9 cells is active and that the virion preparations were P3 free (not shown). After confirming previous results (4, 5) that P2 must be acquired either before or together with P3 and virions (not shown), we tested other probing sequences. In a first sampling period we offered P2—as this is a prerequisite for transmission—either alone or together with virions or P3 or both. In the following acquisition phase, the aphids were allowed to acquire the remaining components. The transmission results presented in Table 2 (first five rows) indicate that the virions and P3 must not be separated during the acquisition process. We verified this conclusion by a three stage acquisition experiment: aphids were first permitted to sample P2, then P3, and, in a third acquisition period, virions. This combination did not result in transmission, but addition of P3 to the CaMV particles reestablished transmission (Table 2, last three rows).
Table 2.
Determination of the acquisition sequence of P2, P3, and virus particles resulting in aphid transmission of CaMV
| 1st probe* | 2nd probe | 3rd probe | Infected/total plants† | % infection |
|---|---|---|---|---|
| P2 | P3 + VP‡ | — | 60/101 | 63 |
| P2 + VP | P3 | — | 0/120 | 0 |
| P2 + P3 | VP | — | 0/160 | 0 |
| P2 + P3 | P3 + VP | — | 74/120 | 62 |
| P2 + P3 + VP | — | — | 81/120 | 68 |
| P2 | P3 | VP | 0/160 | 0 |
| P2 | P3 | P3 + VP | 46/160 | 29 |
| P2 | Sucrose | P3 + VP | 84/158 | 53 |
Aphids were allowed to feed 15 min on each probing solution containing the indicated CaMV proteins for acquisition before they were transferred (10 aphids per plant) to healthy plants for virus inoculation.
Plants were inoculated in batches of 40 plants per experiment, and each experiment was repeated several times. The numbers represent the combined results of all tests.
VP, purified virus particles.
Taken together, our results show that P3 is lost or otherwise inactivated and does not participate in the transmission process when acquired in the absence of virus particles, regardless of the presence of P2 in the feeding solution or stylets.
Assessment of Transmission Activity of P2:P3 Complexes.
The above findings suggest that, in the absence of virions, a functional P2:P3 complex does not form in aphids. We wondered whether such a complex, preformed in vivo as reported in Fig. 2, would support aphid transmission of CaMV. Accordingly, total extracts of Sf9 cells coexpressing P2 and P3 were used in transmission assays. When offered to aphids together with virus particles in a single probe, transmission was observed, demonstrating that the extracts contained biologically active P2 and P3 (Table 3). However, as was the case for a mixture of independently produced P2 and P3, no transmission was observed when coproduced P2:P3 was offered first, followed by virions alone in a second acquisition phase. Transmission was restored when either P3 or P2:P3 from (co)infected Sf9 cell extracts was added to the virions. The fact that additional P3 was again required in the second acquisition period, together with virions, indicated that P3 of the P2:P3 aggregates was lost or inactivated during the first probing stage. The most likely explanation is that P2:P3 complexes are not stable outside the cellular context. Evidence for this interpretation came from an experiment where whole cell extracts of coinfected Sf9 cells were centrifuged for 30 min at 20,000 × g. This experiment resulted in segregation of P2 and P3, with P2 paracrystals accumulating in the pellet and soluble P3 remaining in the supernatant as revealed by Western blotting and electron microscopy (not shown). The resuspended P2-containing pellet followed by the P3-containing supernatant supplemented with virus were offered to aphids, and, as expected, transmission was observed (Table 3, last row).
Table 3.
P2:P3 aggregates do not support aphid transmission though their components are functional
| 1st probe* | 2nd probe | Infected/ total plants† | % infection |
|---|---|---|---|
| P2P3coinf. + VP | — | 53/80 | 66 |
| P2P3coinf.‡ | VP | 0/120 | 0 |
| P2P3coinf. | P3 + VP | 18/80 | 23 |
| P2P3coinf. | P2P3coinf. + VP | 29/77 | 38 |
| Pellet§ P2P3coinf. | Supernatant§ P2P3coinf. + VP | 98/120 | 82 |
† See Table 2.
The probing solution contained a total extract of Sf9 cells coinfected with P2- and P3-encoding baculoviruses.
A total extract of P2- and P3-expressing coinfected Sf9 cells was centrifuged for 30 min at 20,000 × g. The pellet (resuspended in an equal volume of SES buffer) and supernatant were offered for acquisition.
Affinities Between P2, P3, and Virions.
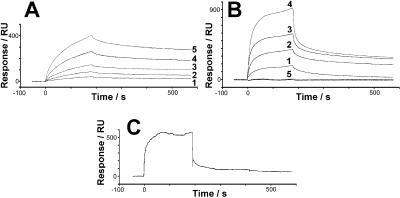
The electron microscopic data and aphid transmission assays indicated that P2 and P3 likely interact in vivo but not outside the cellular context, unless virus particles are present. To investigate this possibility, we measured the affinities between the viral components involved in transmission by Biacore (see Materials and Methods). Whereas a kD of ≈20 nM was determined for the affinity between P3 and virions (Fig. 3A), the interaction between P2 and P3 was found to differ, depending on whether or not P3 was complexed with virions. Fig. 3B shows that P3 together with virus interacted strongly (kD ≈20 nM) with HP2, the biologically active his-tagged form of P2 (19), whereas P3 alone did not measurably bind to HP2 (track 5 in Fig. 3B). Thus, a changed affinity of P3 when complexed to virions, as compared with free P3, may account for the differences observed in its functional interaction with P2 in our transmission assays (Tables 2 and 3). To determine whether the absence of in vitro interaction of native P2 and P3 is due to steric reasons, we made additional experiments with the P2 deletion mutant HP2-Cter (19). P3 interacted with the mutant P2 in the absence of virions (Fig. 3C).
Figure 3.
Affinities between P2, P3, and virions. Solutions containing test proteins were passed over virions, HP2, or HP2-Cter immobilized on Biacore sensor chips. The change in mass of the sensor chip because of binding/dissociation of protein was recorded in real-time as change in surface plasmon resonance in response units (RU). (A) Increasing concentrations of P3 (5, 10, 20, 40, and 80 μg/ml; tracks 1–5) were injected on immobilized virions. (B) Increasing concentrations of P3 (4, 8, 16, and 32 μg/ml) together with an excess amount of virions (150 μg/ml; tracks 1–4) were injected on immobilized HP2. In track 5, 32 μg/ml P3 was injected on HP2 in the absence of virus; (C) 32 μg/ml P3 was injected on immobilized HP2-Cter. All injections were started at t = 0 s and stopped at t = 180 s. The control sensorgrams (no protein immobilized) were subtracted from the measuring sensorgrams illustrated.
Significance of a Preformed P2:P3:Virion Complex in Aphid Transmission.
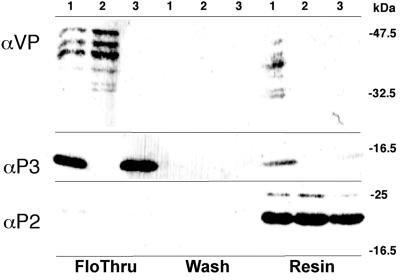
Our data indicate that virus transmission probably results from sequential acquisition of P2:P3 and P3:virion complexes from spatially separated elIBs and edIBs, respectively. However, an alternative acquisition mode exists: about 6% of all virus particles are present in elIBs, and it is possible that some rare preformed P2:P3:virion complexes may also be directly acquired by aphids. To assess the transmissibility of this putative complex, we reconstituted it in vitro. HP2 was reversibly immobilized on Ni-NTA resin and incubated with P3 or virus particles or both, and proteins specifically retained by the resin were analyzed by Western blotting. Fig. 4 shows that only the CaMV particles that were preincubated with P3 bound to immobilized HP2. Consistent with results presented above, virus particles alone did not bind to the HP2-charged resin, and P3 alone was barely retained if at all, as judged by immunoreaction. When eluted P2:P3:virion complexes were offered to aphids, CaMV was transmitted (36 plants infected of 80 tested), demonstrating that preformed HP2:P3:virion complexes are transmissible.
Figure 4.
Reconstitution of the transmissible complex. HP2-charged resin was incubated with virus particles and P3 (lanes 1), virus particles alone (lanes 2), or with P3 alone (lanes 3). The figure shows immunoblots of the flow-through fractions (first three lanes), the last wash fractions (next three lanes), and of the resin (last three lanes) that were analyzed with antisera directed against virus particles (Top), P3 (Middle), and P2 (Bottom). Positions of marker proteins, in kDa, are indicated at the right.
Discussion
The Role of P3 in Transmission.
Classical in vitro transmission assays were used to study the role of the CaMV proteins involved in the transmission process. While confirming that P2 can be acquired alone before the other viral components involved in aphid transmission, we show here that no transmission occurs when P3 and virus particles are acquired separately by aphids. This result was interpreted as P3 having to be previously attached to virions for transmission to occur. That P3 binds indeed to CaMV particles was recently shown by Leh et al. (14) in membrane overlay assays and quantified here by affinity measurements. What we show here is that P3 must first bind to virions to be able to attach to native P2 and allow transmission.
Because the distribution of P3 in infected plant cells was previously unreported, we looked for its location in infected turnip cells. Fig. 1 shows that P3 colocalizes with the virus capsid in situ in edIBs and, to a much lesser extent, in elIBs. This finding extends older data that P3 tends to copurify with virus particles (12) and makes it probable that this P3:virion complex is the viral form binding to P2 in P2-loaded aphids and in elIBs, as was also the case in the transmission assays, in our in vitro reconstitution experiment of the HP2:P3:virion complex, and in the affinity measurements.
Our failure to detect P2 in edIBs confirms previous findings (16). This finding reopens the apparent contradiction with other reports indicating that absence (25) or mutation (26) of P2 decreases edIB stability, thus suggesting P2 may be present in mature edIBs. This question has been discussed previously (2, 3), and a hypothesis reconciling these data has been proposed: like the other viral gene products, P2 is indeed produced in edIBs, but is then rapidly exported and accumulates in elIBs. In this hypothesis, a possible role of P3 in the export and storage of P2 remains to be investigated.
Interaction Between P2 and P3 in the Absence of Virions.
Immunoelectron microscopy revealed that P2 and P3 coexpressed in Sf9 cells colocalize in inclusion bodies presumably as they do in elIBs of CaMV-infected plant cells. We believe that this interaction is specific and points to the existence of a virion-free P2:P3 complex in inclusion bodies, presumably because the P2157m mutant, which does not bind P3, failed to form such inclusions when coexpressed with P3 in Sf9 cells.
In sharp contrast to this finding, we were unable to detect a measurable in vitro interaction between native P2 and P3 in the absence of virus particles, either by Biacore or by affinity chromatography. However, HP2-Cter, a P2 deletion mutant retaining just the P3-binding C-terminal domain, did interact with P3 in Biacore assays. Apparently, outside the cell, this motif in native P2 is accessible to P3 only when P3 is bound to virions. A conformational change of P3, when attaching to virions, could account for this differential behavior. Indeed, a best fit alignment of the association-dissociation curves (Fig. 3) points to this possibility (M.P., unpublished results). Such an assumption would also explain the apparent discrepancy between our in vitro results (no P2-P3 binding) and those of Leh et al. (P2-P3 binding, ref. 10). Whereas we used immobilized native proteins for Biacore assays and affinity chromatography, Leh and coworkers used membrane-immobilized proteins. We hypothesize that these were only partially renatured before the assays, leaving the usually inaccessible binding sites of P2 and P3 exposed.
We have thus far no experimental data explaining formation of virion-free P2:P3 complexes in vivo, either in Sf9 cells or in elIBs of CaMV-infected plant cells. We therefore postulate that either an unknown cellular factor present in both plant and Sf9 cells catalyzes their formation or, alternatively, that P3—being synthesized in plant and Sf9 cells before or simultaneously with P2 (27, 28)—scavenges nascent, unfolded P2 polypeptides in a chaperone-like manner.
The Role of P3 in elIBs.
Transmission assays with P2:P3-containing inclusions from Sf9 cells showed that these structures do not support transmission per se. In agreement with affinity data discussed above, a simple segregation experiment indicated that P2:P3 aggregates are unstable outside the cell. We believe this instability of virus-free P2:P3 complexes can explain their inactivity in transmission and opens an elegant but hypothetical way for P3 to regulate formation of transmissible complexes in plant cells. In the intracellular environment, the large amount of P3 in elIBs perhaps forms a loose mesh with P2 and impedes binding of P3:virions to P2 until this is needed, i.e., in aphids. Thus, the large amount of P3 in elIBs could keep P2 and virions apart and work against large scale formation of transmissible complexes in plant cells. In this manner, the majority of virus particles could be kept in a P2-free state, able to fulfill other viral functions such as cell-to-cell movement during the infection. In aphid stylets, the P2:P3 complexes probably dissociate because of the action of the aphid saliva or simply by a dilution effect, as observed when disrupting P2:P3-coproducing Sf9 cells, and so enabling transmissible complexes to form. Thus, a new role for P3 emerges where it functions as a regulator to control transmission versus other steps in the CaMV infection cycle.
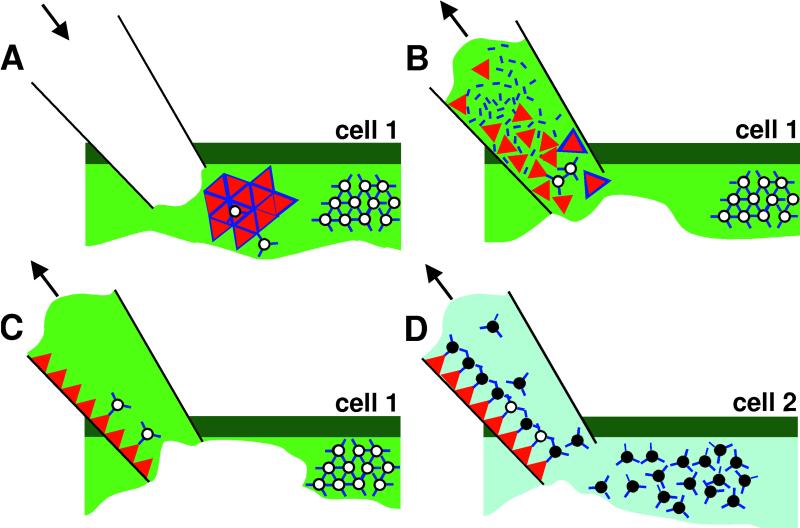
A Model for CaMV Acquisition.
Previous results (10, 14) clearly indicated that P2 bound via P3 to virions constitutes the transmissible complex. We believe that the data presented here enlarge the so far static picture of this complex by giving important insights into dynamics and regulation of its formation with far-reaching consequences for the mode of CaMV transmission by aphids. Our results lead us to propose a model for CaMV acquisition by aphids where every step is experimentally supported.
The situation in infected plant cells (Fig. 1) is a compartmentalized location of viral components in elIBs and edIBs and thus a spatial separation of P2 and the vast majority (94%) of P3:virions as depicted in Fig. 5A. As a consequence and because only small samples of a cell's contents are taken up by probing aphids (29, 30), it is likely that aphids acquire only one or very few viral inclusions during a single cell puncture. In the next step (Fig. 5B), when the ingested inclusion body is an elIB, it will disintegrate because of the weak extracellular interaction between P2 and virus-free P3 (Figs. 3 and 4) and expose free P2 that then is ready to bind to the aphid stylet cuticle. P3 devoid of virions will be ingested and lost or otherwise be inactivated as was the case in our experiments presented in Table 3. In the third step (Fig. 5C) we considered the possibility that some scattered virus particles from elIBs are retained in the stylets and will subsequently be transmitted. Because we showed that HP2:P3:virions reconstituted in vitro (Fig. 4) were aphid-transmitted, it is likely that a few P2:P3:virion complexes exist in elIBs and directly attach to the stylets. However, the extent of this single phase acquisition of P2:P3:virion complexes seems to be limited by the relatively small number of virions in elIBs, so favoring instead the loading of aphids with liberated P2. The final step (Fig. 5D) shows acquisition of further P3:virion complexes (probably from edIBs) by P2-loaded aphids from the same or other cells during subsequent brief intracellular punctures. This secondary acquisition of P3:virions is undoubtedly very efficient because it has been reported many times both from plant and membrane probing assays (e.g., Table 2 and refs. 4 and 5).
Figure 5.
Model of sequential acquisition of CaMV by aphids from infected cells. (A) In infected plant cells, the viral components involved in transmission are spatially separated in elIBs (Left structure) and edIBs (Right structure). Whereas most virus particles (open circles) complexed with P3 (blue bars) are stored in edIBs, P2 (red triangles) in association with P3 and a few virus particles are located in elIBs. When an aphid stylet (at the left) pierces the plasma membrane, saliva is injected into the plant cell. (B) After salivation, the aphid ingests some of the plant cell's contents through its stylet and together with viral inclusion bodies. If an elIB is taken up, it disintegrates and sets free its components P2, P3, and some P2:P3:virion complexes. (C) Although the liberated P3 is lost, the released P2, together with a few P2:P3:virion complexes from the elIB, attaches to the aphid stylet cuticle. The aphid is now P2-loaded, thus, transmission-competent, and ready to acquire more P3:virions (filled circles) from either the same cell or (D) in subsequent punctures, from another cell(s). This model has also been published in a compendium (36).
Our new data therefore indicate that sequential acquisition of helper (P2) and virus by the vector might be an important natural mode of acquisition. Indeed, whereas our model supports coexistence of two acquisition modes—en bloc and sequential—the spatial separation in edIBs and elIBs of the viral players involved in transmission might result in sequential acquisition being predominant. Previous reports of an “irregular biphasic” transmission of CaMV by aphids (31–33) are compatible with this model. The first peak of transmission efficiency observed after short acquisition probes by aphids may correspond to the en bloc acquisition from elIBs (Fig. 5 A–C), whereas the second and larger peak (32) observed after longer aphid feeding periods might represent subsequent acquisition by P2-carrying aphids of P3:virion complexes from edIBs in other plant cells, or even from phloem sieve tubes (Fig. 5D).
The fact that P2 is mainly found virion free in elIBs and may subsequently assist the transmission of P3:virion complexes collected at various locations in the host may have consequences far beyond the molecular and cellular level. Indeed, in the acquisition mechanism described here, the virions of a given population are not competing for the binding sites in the vector but rather cooperating through the action of the HC (here P2). This finding obviously will influence the sampling of the virus population transmitted by the vector. In particular, this finding will influence the genetic diversity within a population that is sampled by the vector, and hence the population genetics of subsequent generations and the evolution of fitness of the virus (for reviews see refs. 34 and 35).
We suggest that there is a need for research to study how the mode of vector transmission influences the population genetics of viruses that do not replicate in their vectors.
Acknowledgments
This work is dedicated to the memory of Jean-Claude Mani. We are very grateful to Yannis Michalakis for critical reading of the manuscript, to Thomas Hohn and Denis Leclerc for kindly providing P3 and P4 antiserum, and to Takii Seed Co. (Japan) for generously providing turnip seeds. Also, thanks to Stefan Drucker for a stimulating idea, and to T. E. Quila for moral support.
Abbreviations
- CaMV
Cauliflower mosaic virus
- edIB
electron-dense inclusion body
- elIB
electron-lucent inclusion body
- Sf9
Spodoptera frugiperda insect cell line 9
- HC
helper component
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rothnie H M, Chapdelaine Y, Hohn T. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 2.Pirone T, Blanc S. Annu Rev Phytopathol. 1996;34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Blanc S, Hébrard E, Drucker M, Froissart R. In: Virus-Insect-Plant Interactions. Harris K, Smith O P, Duffus J E, editors. San Diego: Academic; 2001. pp. 143–166. [Google Scholar]
- 4.Lung M C Y, Pirone T P. Phytopathology. 1973;63:910–914. [Google Scholar]
- 5.Lung M C Y, Pirone T P. Virology. 1974;60:260–264. doi: 10.1016/0042-6822(74)90383-3. [DOI] [PubMed] [Google Scholar]
- 6.Woolston C J, Czaplewski L G, Markham P G, Goad A S, Hull R, Davies J W. Virology. 1987;160:246–251. [Google Scholar]
- 7.Blanc S, Cerutti M, Usmany M, Vlak J M, Hull R. Virology. 1993;192:643–650. doi: 10.1006/viro.1993.1080. [DOI] [PubMed] [Google Scholar]
- 8.Blanc S, Schmidt I, Kuhl G, Esperandieu P, Lebeurier G, Hull R, Cerutti M, Louis C. Virology. 1993;197:283–292. doi: 10.1006/viro.1993.1589. [DOI] [PubMed] [Google Scholar]
- 9.Howarth A J, Gardner R C, Messing J, Shepherd R J. Virology. 1981;112:678–685. doi: 10.1016/0042-6822(81)90313-5. [DOI] [PubMed] [Google Scholar]
- 10.Leh V, Jacquot E, Geldreich A, Hermann T, Leclerc D, Cerrutti M, Yot P, Keller M, Blanc S. EMBO J. 1999;18:7077–7085. doi: 10.1093/emboj/18.24.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon L K, Koenig I, Hohn T. Gene. 1983;25:189–199. doi: 10.1016/s0378-1119(83)80001-8. [DOI] [PubMed] [Google Scholar]
- 12.Dautel S, Guidasci T, Pique M, Mougeot J L, Lebeurier G, Yot P, Mesnard J M. Virology. 1994;202:1043–4045. doi: 10.1006/viro.1994.1435. [DOI] [PubMed] [Google Scholar]
- 13.Jacquot E, Geldreich A, Keller M, Yot P. Virology. 1998;242:395–402. doi: 10.1006/viro.1997.8995. [DOI] [PubMed] [Google Scholar]
- 14.Leh V, Jacquot E, Geldreich A, Haas M, Blanc S, Keller M, Yot P. J Virol. 2001;75:100–106. doi: 10.1128/JVI.75.1.100-106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohn T, Fütterer J. Crit Rev Plant Sci. 1997;16:133–167. [Google Scholar]
- 16.Espinoza A M, Medina V, Hull R, Markham P G. Virology. 1991;185:337–344. doi: 10.1016/0042-6822(91)90781-6. [DOI] [PubMed] [Google Scholar]
- 17.Franck A, Guilley H, Jonard J, Richards K, Hirth L. Cell. 1980;21:285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- 18.Héricourt F, Blanc S, Redeker V, Jupin I. Biochem J. 2000;349:417–425. doi: 10.1042/0264-6021:3490417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hébrard E, Drucker M, Leclerc D, Hohn T, Uzest M, Froissart R, Strub J-M, Sanglier S, van Dorsselaer A, Padilla A, et al. J Virol. 2001;75:8538–8546. doi: 10.1128/JVI.75.18.8538-8546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaabihi H, Ogliastro M H, Martin M, Giraud C, Devauchelle G, Cerutti M. J Virol. 1993;67:2664–2671. doi: 10.1128/jvi.67.5.2664-2671.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;277:680–684. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Blanc S, Schmidt I, Vantard M, Scholthof H B, Khul G, Esperandieu P, Cerutti M, Louis C. Proc Natl Acad Sci, USA. 1996;93:15158–15163. doi: 10.1073/pnas.93.26.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt I, Blanc S, Esperandieu P, Kuhl G, Devauchelle G, Louis C, Cerutti M. Proc Natl Acad Sci USA. 1994;91:8885–8889. doi: 10.1073/pnas.91.19.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Givord L, Xiong C, Giband M, Koenig I, Hohn T, Lebeurier G, Hirth L. EMBO J. 1984;3:1423–1427. doi: 10.1002/j.1460-2075.1984.tb01987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu S G, Wintermantel W M, Sha Y, Schoelz J E. Virology. 1997;227:180–188. doi: 10.1006/viro.1996.8314. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi K, Nakayashiki H, Tsuge S, Mise K, Furusawa I. Microbiol Immunol. 1998;42:65–69. doi: 10.1111/j.1348-0421.1998.tb01972.x. [DOI] [PubMed] [Google Scholar]
- 28.Maule A J, Harker C L, Wilson I G. Virology. 1989;169:436–446. doi: 10.1016/0042-6822(89)90169-4. [DOI] [PubMed] [Google Scholar]
- 29.Tjallingii W F, Hogen Esch T. Physiol Entomol. 1993;18:317–328. [Google Scholar]
- 30.Martin B, Collar J L, Tjallingii W F, Fereres A. J Gen Virol. 1997;78:2701–2705. doi: 10.1099/0022-1317-78-10-2701. [DOI] [PubMed] [Google Scholar]
- 31.Hamlyn B M G. Plant Pathol. 1955;4:13–16. [Google Scholar]
- 32.Chalfant R B, Chapman R K. J Econ Entomol. 1962;55:584–590. [Google Scholar]
- 33.Markham P G, Pinner M S, Raccah B, Hull R. Ann Appl Biol. 1987;111:571–587. [Google Scholar]
- 34.Domingo E. Virology. 2000;270:251–253. doi: 10.1006/viro.2000.0320. [DOI] [PubMed] [Google Scholar]
- 35.Moya A, Elena S F, Bracho A, Miralles R, Barrio E. Proc Natl Acad Sci USA. 2000;97:6967–6973. doi: 10.1073/pnas.97.13.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hull R. Matthews' Plant Virology. 4th Ed. San Diego: Academic; 2001. p. 501. [Google Scholar]