Abstract
Specificity in the interaction between rough lemon (Citrus jambhiri Lush.) and the fungal pathogen Alternaria alternata rough lemon pathotype is determined by a host-selective toxin, ACR-toxin. Mitochondria from rough lemon are sensitive to ACR-toxin whereas mitochondria from resistant plants, including other citrus species, are resistant. We have identified a C. jambhiri mitochondrial DNA sequence, designated ACRS (ACR-toxin sensitivity gene), that confers toxin sensitivity to Escherichia coli. ACRS is located in the group II intron of the mitochondrial tRNA-Ala and is translated into a SDS-resistant oligomeric protein in C. jambhiri mitochondria but is not translated in the toxin-insensitive mitochondria. ACRS is present in the mitochondrial genome of both toxin-sensitive and -insensitive citrus. However, in mitochondria of toxin-insensitive plants, the transcripts from ACRS are shorter than those in mitochondria of sensitive plants. These results demonstrate that sensitivity to ACR-toxin and hence specificity of the interaction between A. alternata rough lemon pathotype and C. jambhiri is due to differential posttranscriptional processing of a mitochondrial gene.
Alternaria alternata (Fr.) Keissl. is commonly known as a cosmopolitan saprophyte, but some species of A. alternata produce host-selective toxins that are selectively toxic to certain plants or certain genotypes of a plant species (1). A. alternata strains producing host-selective toxins are designated as pathotypes of A. alternata (1, 2). Despite the morphological similarity of these host-specific pathotypes, one can be easily distinguished from another on the basis of their host range. Chemical structures of host-selective toxins from six pathotypes of A. alternata have been elucidated (3), including ACR-toxin produced by the rough lemon pathotype (RLP) known to cause citrus brown spot disease.
Citrus brown spot caused by A. alternata RLP is a serious disease of rough lemon (Citrus jambhiri Lush.) in nurseries and seed stocks in many parts of the world. Virulence of A. alternata RLP is due to production of ACR-toxin, which has the same host range as the pathogen (4, 5). Citrus varieties and species that are resistant to A. alternata RLP are insensitive to ACR-toxin, and isolates of A. alternata that do not produce ACR-toxin are not pathogenic on C. jambhiri. The structure of ACR-toxin contains a dihydropyrone ring with a polyalcohol side chain (6, 7). ACR-toxin causes metabolite leakage and uncoupling of oxidative phosphorylation in isolated mitochondria from leaves of sensitive but not resistant citrus, suggesting that the site of action of ACR-toxin is the mitochondrion (8). Electron microscopic examination of toxin-treated cells also showed that mitochondria are disrupted within 1 h after toxin treatment (9). The toxin-treated mitochondria are swollen and have a reduced number of cristae and a decreased matrix density (9). The genetics of sensitivity of Citrus species to ACR-toxin has not been elucidated, but it seemed possible that sensitivity is due to a mitochondrion-encoded gene. In this paper, we identify a mitochondrial gene that confers ACR-toxin sensitivity to Escherichia coli. The mechanism of specificity in plants is an altered transcript processing of the gene conferring ACR-toxin sensitivity.
Materials and Methods
Materials.
A. alternata RLP strain AC325 (4, 5, 8) was provided by the Laboratory of Plant Pathology, Tottori University, Japan. ACR-toxin was purified as described previously (6–8).
Seeds or young plants of rough lemon (C. jambhiri Lush.) were provided by T. Miyoshi, Ehime Prefecture Fruit Research Station, Ehime, and M. Sadano, Tokushima Prefecture Fruit Research Station, Tokushima, Japan. Young plants of Etrog citron (C. limonimedica), grapefruit (C. paradisi), lemon (C. limon), mexican lime (C. aurantifolia), navel orange (C. sinensis), trovita orange (C. sinensis), and yuzu (C. junos) were provided by H. Shiotani, National Institute of Fruit Tree Science, Nagasaki, Japan. Lime (C. latifolia), rangpur lime (C. limonia), and volkamer lemon (C. volkameriana) were kindly provided from the Thai Royal Project Panda Experimental Station, Chiang Mai, and P. Samitamane, Chiang Mai University, Thailand. Young plants of iyokan (C. iyo), satsuma mandarin (C. unshiu), and trifoliate orange (Poncirus trifoliate) were obtained commercially.
Expression of Rough Lemon Mitochondrial Genes in E. coli.
Washed mitochondria were prepared by the method described previously (8). The mitochondria were resuspended in Tris-EDTA (TE) buffer (pH 8.0) containing 10% (wt/vol) N-lauroylsarcosine and 0.5 mg/ml proteinase K, and incubated at 37°C for 3 h. The solution was extracted with phenol once, phenol/chloroform/isoamyl alcohol [25:24:1 (vol/vol/vol)] twice, and chloroform once, and mitochondrial DNA was precipitated with sodium acetate and EtOH. Mitochondrial DNA was then digested with 15 units of BamHI, and the reaction was stopped by heating at 85°C for 10 min. BamHI-digested mitochondrial DNA fragments were subcloned randomly into vector pGEX-3X (Amersham Pharmacia Biotech) and transformed into E. coli (XL1-Blue MRF′; Stratagene) cells. Transformed cells were placed with 1 mM of isopropyl 1-thio-β-d-galactoside (IPTG) on both LB/ampicillin plates with or without ACR-toxin (1 μg/ml). E. coli colonies that grew poorly on LB plates containing ACR-toxin, but those that grew normally in the absence of the toxin were further examined for oxygen uptake measured with a Clark (Clark Electromedical Instruments, Pangbourne, U.K.) oxygen electrode at 25°C. Respiration rates are given as nanomoles of O2 consumed per minute per milligram of E. coli protein. Respiration rates of toxin-sensitive cells, toxin-insensitive cells, or cells transformed by vector alone were measured separately, and each time the respiration rates of cells incubated with ACR-toxin or methanol were compared with those of same cells without the additions to elucidate their effects. Cell viability after toxin treatment was also determined by streaking the cell solutions used for oxygen uptake tests on LB plates. ACR-toxin- or methanol-treated cell suspensions with or without addition of IPTG were streaked on LB plates and incubated at 37°C for 12 h.
Mitochondrial DNA Extraction and Analysis.
Purified mitochondrial DNA was prepared by the method of Lu and Hanson (10) with some modifications. Washed mitochondria prepared by the method described previously (8) were treated with RNase A (10 μg/ml) and DNase I (20 μg/ml) for 1 h on ice, and fractionated on a discontinuous percoll gradient [15%, 22%, 27%, and 60% (vol/vol)] in resuspension buffer (250 mM sorbitol/2.5 mM Hepes-Tris, pH 7.2/1 mM DTT/0.5% BSA; ref. 8) by centrifugation at 33,000 × g for 45 min. Percoll-purified mitochondria were washed three times with resuspension buffer, and the final pellet was resuspended in 0.4 M mannitol, 10 mM Tricine (pH 7.2) and 1 mM EGTA. Mitochondria were lysed by adding one-quarter volume of lysis buffer [25 mM Tris⋅HCl, pH 7.5/20 mM EDTA/10% (wt/vol) SDS] for 10 min at 25°C. The lysed solution was extracted two times with phenol/chloroform (1:1) and one time with chloroform. Mitochondrial DNA was precipitated with EtOH/sodium acetate from the aqueous phase. The DNA was resuspended in TE, and used as purified mitochondrial DNA.
DNA probes were labeled with the non-isotope digoxigenin system (Roche). Restriction enzyme digestion, hybridization, and detection conditions were described previously (11, 12).
Mitochondrial Genomic Cloning and Sequencing.
The fragments of mitochondria genome were subcloned into the Bluescript SK(+) (Stratagene), and the sequences were obtained from both strands by the dideoxy chain termination method (13) with the use of an Applied Biosystems PRISM Dye Termination Cycle Sequencing Ready Reaction Kit and an automated fluorescent DNA sequencer (Model 310; Applied Biosystems). DNA sequences were aligned with clustal w (14), and homology analysis was performed with blast at the DNA Data Bank of Japan.
Analysis of Mitochondrial RNA.
Posttranscriptional modifications of mitochondrial RNA were determined by 3′ rapid amplification of cDNA ends (RACE). For RNA purification, washed mitochondria were treated with RNase A and DNase I, and purified by a discontinuous percoll gradient. The percoll-purified mitochondria were lysed as described in DNA extraction section. The lysed cell solution was extracted twice with phenol/chloroform (1:1) and once with chloroform, and nucleic acids were precipitated with EtOH/sodium acetate. Total RNA was extracted from the precipitates by using the MagExtractor RNA Purification Kit (Toyobo, Tokyo). Purified total RNA (about 1 μg) was mixed with MgCl2 (9 μl) to a final concentration of 5 mM and RNase free-DNase I (5 units/μl; Takara, Shiga, Japan) for 2 h at 37°C, followed by 15 min at 85°C. Ten microliters of the solution was then mixed with poly(A) polymerase (0.2 to 2.0 units/μl; Takara) in 39 μl of poly(A) polymerase buffer [final composition of 50 mM Tris⋅HCl, pH 7.9/10 mM MgCl2/2.5 mM MnCl2/250 mM NaCl/1 mM DTT/0.05% BSA/0.1 mM ATP). The mixture was incubated for 10 min at 37°C followed by 15 min at 85°C. The polyadenylated RNAs were purified by using the Oligotex dt30-Super-mRNA Purification Kit (Takara), and redissolved in RNase-free water. First strand cDNA synthesis used ThermoScript RT (Life Technologies, Rockville, MD) with Oligo(dT)20. Second strand synthesis of the cDNA, adapter ligations, and RACE were performed with the Marathon cDNA Amplification Kit (CLONTECH). For 3′ RACE, ACRS-F2 primer (5′-CCAGGAACGGAGAGCTTTCC-3′) and the nested adaptor primer 2 (5′-ACTCACTATAGGGCTCGAGCGGC-3′) were used. PCR conditions were as follows: denaturation for 1 min at 94°C, followed by 5 cycles of 30 s at 94°C and 4 min at 72°C, another 5 cycles of 30 s at 94°C and 4 min at 70°C, followed by 25 cycles of 20 s at 94°C and 4 min at 68°C. The products were separated on 10% (wt/vol) acrylamide gels transferred to Hybond-N+ membrane, and probed with the ACRS (ACR-toxin sensitivity gene). The RACE products were also subcloned into pT7Blue-2 T-vector (Novagen), and sequenced as described above.
Western Blotting.
Polyclonal antibodies were raised in mouse by injections of inclusion bodies of protein produced by overexpression in E. coli. The inclusion bodies from E. coli were partially purified by four rounds of centrifugation and then fractionated by SDS/PAGE (10% acrylamide). Bands corresponding to the protein product were excised from the gel, homogenized in PBS, and injected s.c. into mice with four boosts at 1-week intervals. For protein analysis, mitochondria were prepared from rough lemon, grapefruit, lemon, and navel orange. The mitochondria were mixed with SDS/PAGE sample buffer containing a final concentration of 4% (wt/vol) SDS, 12% (vol/vol) glycerol, 50 mM Tris (pH 6.8), 2% (vol/vol) β-mercaptoethanol, and 0.01% bromophenol blue. The mixture was heated for 30 min at 40°C and fractionated by tricine-SDS/PAGE with a separating gel of 10% (wt/vol) total monomer (acrylamide + bis), 3% bis (wt/wt), and a stacking gel of 4% total monomer, 3% bis, as described by Schagger and von Jagow (15). The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane with an electrotransfer unit (LKB2117 multiphor II) by using a transfer buffer composed of 99 mM Tris, 192 mM glycine, and 20% (vol/vol) methanol. The PVDF membrane was blocked with 3% (wt/vol) BSA in Tris-buffered saline (TBS) overnight at room temperature. The membrane was washed several times with TBS plus 0.05% Tween 20 (TBS-Tween) and incubated with anti-ACRS antiserum at 1:1000. After additional washings with TBS-Tween, the membrane was treated with anti-mouse IgG-alkaline phosphatase conjugate (Sigma) at 1:10000 dilution. Alkaline phosphatase was detected with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium in buffer containing 100 mM Tris (pH 9.5), 100 mM NaCl, and 10 mM MgCl2.
Preparation of ACRS Deletion Clones.
pACRS-AflII, a plasmid with deleted ACRS in vector pGEX-3X, was created from the plasmid of toxin-sensitive strain J104 by digestion of ACRS at the internal AflII site and EcoRI site in multiple cloning site of the vector, and re-ligation after filling overhangs of the restriction sites. Deleted ACRS in pACRS-DL10 was amplified from ACRS in the plasmid from J104 by PCR by using GEX primer (5′-ATCGGATCTGATCGAAGG-3′) designed from −23 to −6 upstream of BamHI-cloning site of pGEX-3X vector and DL10 primer containing EcoRI site (5′-CGGAATTCTTACTCATTCTTA-3′) at the end, under denaturing conditions at 94°C for 2 min, followed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C. The PCR product was digested with BamHI and EcoRI and ligated to pGEX-3X. A stop codon (TGA) was created at the asparagine codon (AAT) that is located at the 3′-end by U.S.E. Mutagenesis Kit (Amersham Pharmacia Biotech). Deleted ACRS in pACRS-DL9 was also made by PCR by using the GEX primer and DL9 primer containing BamHI site (5′-CGGGATCCAGAACCCTGC-3′) under conditions described above. pACRS-NcoI was created from the plasmid of J104 by digestion at NcoI site in ACRS and EcoRI in multiple cloning site of pGEX-3X vector, and re-ligation after filling overhangs of the restriction sites. Every mutation was confirmed by sequencing. All deleted ACRS constructs were transformed into E. coli (XL1-Blue) cells, and the transformants were designated deletion clones ACRS-AflII, ACRS-DL10, ACRS-DL9, and ACRS-NcoI, respectively. ACR-toxin sensitivity of each deletion clone was measured by O2 consumption as described above.
Results and Discussion
Expression of a Rough Lemon Mitochondrial Gene Confers Sensitivity to ACR-toxin in E. coli.
DNA was isolated from mitochondria of rough lemon, and random BamHI fragments were expressed in E. coli, which is normally resistant to ACR-toxin. The E. coli transformants (2406 total) were plated on ACR-toxin, and one strain, designated J104, was found that was sensitive to ACR-toxin. The toxin-sensitive strain J104 grew normally on plates without ACR-toxin or in the absence of the inducer IPTG, and E. coli was insensitive when transformed with the expression plasmid alone (Fig. 1).
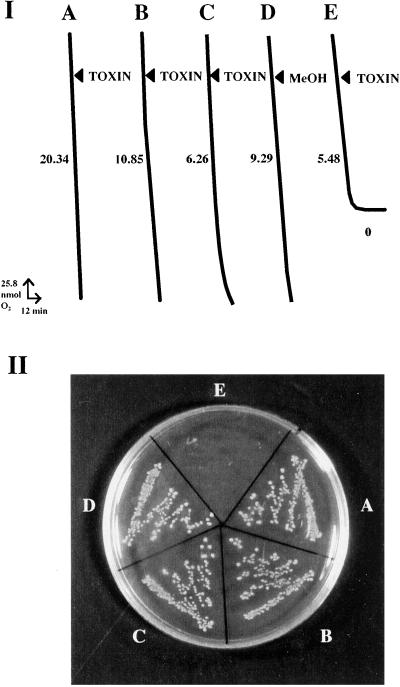
Figure 1.
ACR-toxin sensitivity of E. coli strain J104 expressing the 355-bp rough lemon mitochondrial DNA sequence, called ACRS. The E. coli cells (XL1-blue MRF′) were transformed with (I, A) pGEX-3X vector alone, (I, B) pGEX-3X with insertion of a random DNA sequence from rough lemon mitochondrial DNA, and (I, C–E) pGEX-3X containing ACRS. Cell cultures of A, B, D, and E had been induced for 2 h by 1 mM IPTG, whereas C was not induced by IPTG. ACR-toxin (TOXIN; final concentration 1 μg/ml in 0.01% methanol; A–C and E) or methanol (MeOH; final concentration 0.01%; D) was added to E. coli cells growing in LB medium containing 10 to 50 μg of E. coli protein. Oxygen uptake (I) was measured by a Clark (Clark Electromedical Instruments, Pangbourne, U.K.) oxygen electrode, and respiration rates were indicated as nanomoles of O2 consumed per minute per milligram of E. coli protein. Respiration rate of cells in each treatment (A–E) was measured separately, and the rates with addition of ACR-toxin or methanol were compared each time to those of the same cells without the additions to confirm the effects of toxin or methanol in respective treatments (A–E). The measurements of each treatment were repeated at least five times, and one of these results was shown in this figure. Cell viability (II) after toxin treatment was also determined by streaking the cell solutions described above (A–E) on an LB plate.
Addition of the toxin completely abolished oxygen uptake by toxin-sensitive E. coli strain J104 (Fig. 1I, E). Oxygen uptake of J104 was slightly inhibited in the absence of the inducer IPTG; the residual inhibition in the absence of IPTG could be explained by a basal level of expression (Fig. 1I, C). No inhibition occurred when the toxin was added to E. coli cells transformed with vector with an insert of other regions of mitochondrial DNA or with the vector alone (Fig. 1I, A and B). The minimum concentration of ACR-toxin required to inhibit oxygen uptake of toxin-sensitive strain J104 was 50 nM, which is a similar concentration causing necrosis on rough lemon leaves (4, 6–8).
ACRS Is Located in the Group II Intron Region of tRNA-Ala in Rough Lemon Mitochondrial Genome.
The plasmid in toxin-sensitive E. coli strain J104 that conferred sensitivity to ACR-toxin contained a 355-bp insert. This insert was named ACRS (ACR-toxin sensitivity gene). Southern blot analysis with purified rough lemon mitochondrial DNA identified a 4.3-kb EcoRI-XhoI fragment that contained ACRS. The subcloned 4.3-kb fragment was digested with KpnI to make a 2.6-kb KpnI-KpnI fragment (called ACRS22Kpn32) containing ACRS. A 2,303-bp internal fragment of ACRS22Kpn32 (called 2303K) was sequenced (DNA databank accession number AB061306; Fig. 2). A search of the nonredundant databases with the sequence of 2303K indicated that it is highly homologous to chloroplast and mitochondrial genes for tRNA-Ala and tRNA-Ile. Sequences showing more than 94% identity to ACRS and its flanking regions in 2303K were Helianthus annuus mitochondrion DNA for tRNA-Ile and tRNA-Ala (DNA databank accession number, X95260), Oenothera lamarckiana chloroplast tRNA-Ile and tRNA-Ala (X97295), tobacco chloroplast tRNA-Ile and tRNA-Ala (V00166), and Arabidopsis thaliana chloroplast genomic DNA (AP000423). ACRS was located within the intron of the tRNA-Ala gene. This intron, called a self-splicing group II intron, catalyzes its own splicing (16–21). This type of intron has previously been found in tRNA genes of plant mitochondrial and chloroplast DNAs (e.g., refs. 16–21), and many of these introns have been reported to contain ORFs for polypeptides (e.g., refs. 22–26).
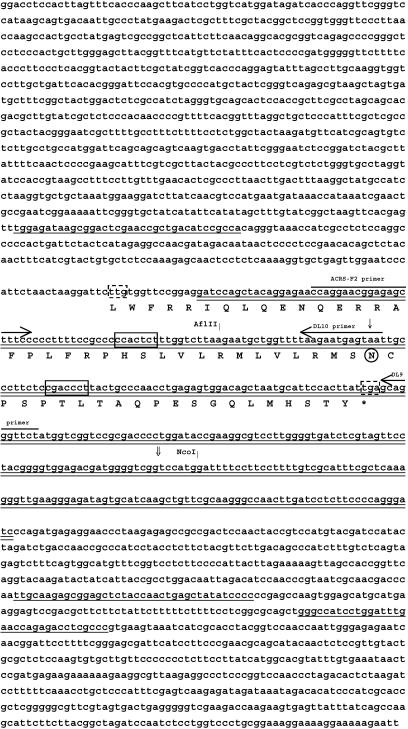
Figure 2.
Nucleotide and deduced amino acid sequences of rough lemon mitochondrial genomic region containing ACRS. The 355-bp region (ACRS) that conferred sensitivity to ACR-toxin in E. coli is double-underlined. The deduced amino acid sequence of the predicted 171-bp ORF (ACR-ORF6.7) is indicated underneath the nucleotide sequence. The putative start (TTG; 27) and stop codons are boxed with dotted-lines. Exons of tRNA-Ala and -Ile are single-underlined. Processing motifs of 5′-CNACNNU-3′ (35) are boxed with solid lines. AflII and NcoI restriction sites in ACRS are shown by vertical bars. Regions used for design of PCR primers are indicated by horizontal arrows. Transcript ends identified by 3′ RACE are indicated by vertical arrows; ↓ indicates transcript end identified in ACR-toxin insensitive mitochondria, and ⇓ indicates transcript end identified in ACR-toxin sensitive mitochondria. A codon AAT at the circled asparagine is converted to a stop codon (TGA) to create the deletion clone ACRS-DL10 in the experiment described in Fig. 4.
The 355-bp DNA sequence that confers ACR-toxin sensitivity contains an ORF of 171 bp, based on a predicted plant mitochondrial initiation codon of TTG (27) and stop codon of TGA (Fig. 2). The deduced amino acid sequence of this ORF, called ACR-ORF6.7, contains 26 hydrophobic residues out of 56, which suggests that, if it were translated, it would likely be a mitochondrial membrane protein.
To investigate further the origin of ACRS and its relation to selective toxicity of ACR-toxin and hence susceptibility to A. alternata RLP, we investigated this DNA region in other, resistant cultivars and species of citrus. All tested species of citrus, including citron, grapefruit, iyokan, lemon, lime, mexican lime, navel orange, satsuma mandarin, trifoliate orange, trovita orange, rangpur lime, yuzu, and volkamer lemon, contained DNA that hybridized to ACRS on a mitochondrial DNA restriction fragment of the same size as in rough lemon (4.3 kb; Fig. 3A). The sequences of the 355-bp region of iyokan (DNA databank accession number AB061307), grapefruit (AB061308), lemon (AB061309), and navel orange (AB061310), all of which are resistant to A. alternata RLP and ACR-toxin, were identical to that of rough lemon.
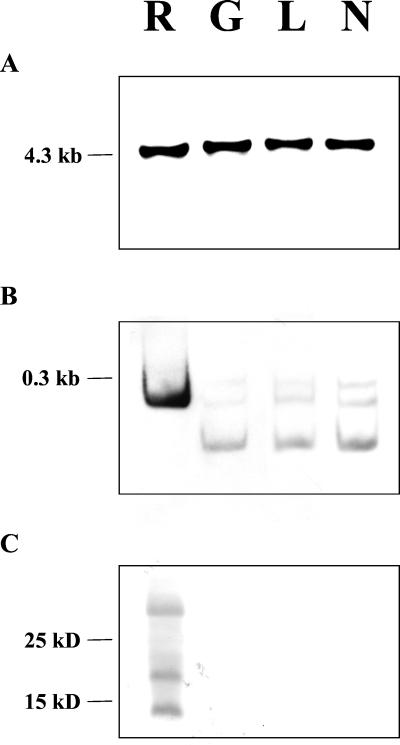
Figure 3.
Analysis of mitochondrial genomes, transcript modifications, and translations of ACRS in mitochondria from ACR-toxin-sensitive and -insensitive citrus. (A) DNA blot analysis of ACRS in citrus mitochondria. Purified mitochondrial DNA from rough lemon (R), grapefruit (G), lemon (L), or navel orange (N) was digested with EcoRI and XhoI, separated by agarose electrophoresis, blotted, and probed with ACRS. (B) Posttranscriptional modifications of ACRS determined by 3′ RACE. The 3′ RACE products from total RNA extracted from mitochondria from rough lemon (R), grapefruit (G), lemon (L), or navel orange (N) were fractionated by acrylamide gel electrophoresis, blotted, and probed with ACRS. (C) Detection of the product of ACRS in citrus mitochondria by immunoblot analysis. Washed mitochondria from rough lemon (R), grapefruit (G), lemon (L), or navel orange (N) were heated for 30 min at 40°C before fractionation by tricine-SDS/PAGE. The gel was blotted and probed with polyclonal antibodies raised against the ACRS products expressed in toxin-sensitive E. coli strain J104.
Specific Sensitivity of Citrus Mitochondria to ACR-toxin Is Regulated by a Posttranscriptional Event.
tRNAs in plant mitochondria, like those in animal and yeast mitochondria, are excised from longer transcripts (28). It is not known whether the tRNA-Ala of citrus mitochondria is active or not. However, it is possible that ACRS located in the tRNA-Ala intron is transcribed as part of the longer tRNA-Ala, spliced out, and then translated. We examined the RNA that resulted from ACRS by 3′ RACE with a primer (ACRS-F2; see Fig. 2) from both toxin-sensitive and toxin-insensitive species of citrus. The results showed that a RACE product of 232 bp that contained the predicted ACR-ORF6.7 was present only in rough lemon mitochondria, whereas in mitochondria from grapefruit, lemon, and navel orange, all of which are toxin insensitive, the major transcript was only 75 bp (Fig. 3B). Smaller amounts of RNA species of approximately 300 and 200 bp were also present only in toxin-insensitive citrus (Fig. 3B), and these bands, like the 75-bp band, were consistently less abundant than the 232-bp band in rough lemon mitochondria. RACE products from toxin-insensitive citrus were detectable only when a minimum of 40 ng of double stranded cDNA was used as template in the PCR reaction, whereas 8 ng of RACE product from rough lemon was sufficient to detect a product.
Posttranscriptional RNA maturation events, such as editing and processing, are known to be common in plant mitochondria (16, 17). Processing of mitochondrial transcripts has been shown to be involved in cytoplasmic male sterility in rice, sorghum, and maize (29–31). Mitochondrial RNA editing can introduce initiation or stop codon into transcripts and induce tRNA excision or intron splicing (16, 17, 32–34). However, sequencing of RACE products from both rough lemon (toxin-sensitive) and iyokan (insensitive) citrus indicated that neither RNA was edited (data not shown).
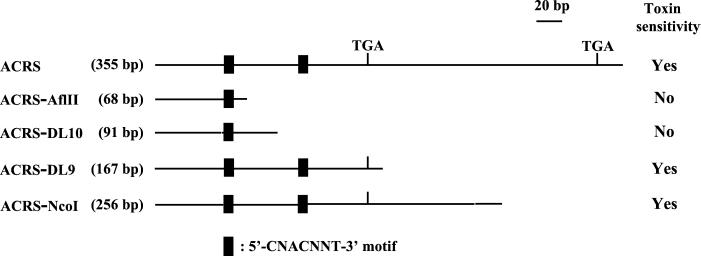
ACRS contains two copies of a mitochondrial processing motif, 5′-CNACNNU-3′ (Figs. 2 and 4; ref. 35). The transcript end of the 75-bp RACE product from toxin-insensitive citrus mitochondria indicated that the processing occurred 35-bp downstream of the first processing motif (Fig. 2). Therefore, the effect of processing on the 355-bp sequence was examined in E. coli by using various subclones. One, called ACRS-AflII (68 bp), started 10 bp downstream of the first processing site (5′-CCACTCT-3′) and another, called ACRS-DL10 (91 bp) started 35 bp downstream of the same site, which is equal to the transcript-end of the 75-bp RACE product from resistant citrus. Deletion clones expressing a region ending 6 bp downstream of the RACE product from rough lemon transcripts (ACRS-NcoI: 256 bp), a region ending 10 bp from the putative stop codon (Fig. 2; ACRS-DL9: 167 bp), were as sensitive as toxin-sensitive strain J104 expressing the full 355-bp ACRS, whereas deletion clones expressing fragments ACRS-AflII and ACRS-DL10 were insensitive to ACR-toxin (Fig. 4). These results suggest that the differential processing of the ACRS is the cause of ACR-toxin sensitivity in the mitochondrion.
Figure 4.
Effects of deletions of ACRS on ACR-toxin sensitivity in E. coli. Deleted ACRSs in plasmid vector pGEX-3X. pACRS-AflII [ACRS was deleted to 68 bp at AflII site locating 10 bp downstream of the first processing site (5′-CCACTCT-3′; Fig. 2], pACRS-DL10 [deleted ACRS to 91 bp at 35 bp downstream from the site, which is equal to the transcript-end of the 75-bp RACE product from resistant citrus (Fig. 3B)], pACRS-DL9 [deleted ACRS to 167 bp, expressing a region ending at 10 bp from the putative ACRS stop codon (Fig. 2)], and pACRS-NcoI [deleted ACRS to 256 bp, expressing a region ending at 6 bp downstream of the RACE product from rough lemon transcripts (Fig. 3B)] were transformed into E. coli cells to create deletion clones ACRS-AflII, ACRS-DL10, ACRS-DL9, and ACRS-NcoI. ACR-toxin sensitivity of each deletion clone was measured by an O2 consumption of the E. coli cells. For each mutation, toxin-sensitive mutants are indicated by Yes, whereas insensitive mutants are indicated by No. The lengths in nucleotides (bp) are listed for each deleted ACRS.
To determine whether ACRS is translated, proteins from isolated mitochondria of toxin-sensitive rough lemon and toxin-insensitive grapefruit, lemon, and navel orange were analyzed by immunoblotting by using antibodies raised against the 33-kDa protein product of the 355-bp sequence expressed in toxin-sensitive E. coli strain J104 (Fig. 3C). The antiserum detected three proteins with molecular masses of 14, 21, and 28 kDa in extracts from rough lemon mitochondria, but nothing in extracts from the toxin-insensitive citrus mitochondria (Fig. 3C). The calculated molecular weight of the product from the predicted 171-bp ORF, ACR-ORF6.7, is 6683, and therefore the proteins detected by immunoblotting could be the dimer, trimer, and tetramer that are not fully dissociated during SDS/PAGE. SDS-resistant protein oligomers have been reported for many pore-forming transmembrane proteins, e.g., G-protein-coupled receptors, apolipoprotein E, and URF13 of maize (36–39). Pore-forming transmembrane proteins are also known to mediate the biological activity of many fungal and bacterial toxins (40, 41). The known physiological effects of ACR-toxin are consistent with its forming of pores in membranes, because ACR-toxin-treated mitochondria show not only increased permeability to protons but also to NAD+ (8).
Maize containing Texas male sterile cytoplasm (T-cms) is susceptible to Cochliobolus heterostrophus race T (Helminthosporium maydis race T) because it is sensitive to a host-selective toxin made by the fungus called T-toxin. Mitochondria of T-cms maize are sensitive to T-toxin and hence susceptible to the pathogen because of an internal genomic rearrangement resulting in a novel chimeric gene, called T-urf13. Translation of T-urf13 results in the synthesis of a novel 13-kDa protein that inserts into the inner mitochondrial membrane, causing uncoupling of oxidative phosphorylation and metabolite leakage by interactions with T-toxin (42, 43). A host-selective toxin is also the basis of specificity in the interaction between A. alternata RLP and citrus. Here, we have shown that the molecular basis of susceptibility to A. alternata can also be found in the mitochondria of sensitive vs. insensitive citrus species. In both cases, a novel protein is synthesized that confers sensitivity to the toxin. However, the basis of susceptibility to A. alternata RLP is not due to a genomic rearrangement as in the interaction between C. heterostrophus and maize but rather to a difference in RNA processing. Nuclear effects are well established; however, it is not known whether the observed difference in processing between sensitive and insensitive species of Citrus is due to a nuclear or an organellar genetic event.
Acknowledgments
We thank Dr. J. D. Walton, Michigan State University, for valuable discussions and proofreading of the manuscript. We also thank Dr. D. A. Day, University of Western Australia, for critical reading of the manuscript; Ms. N. Itoh, Kagawa University, for technical assistance for ACRS-antibody production; Dr. T. Tsuge, Nagoya University, and Dr. N. T. Keen, University of California, Riverside, for valuable comments and discussions; Dr. H. Kobayashi, University of Shizuoka, and Dr. K. Kohmoto, Tottori University, for valuable comments at initial stages of the project; and Dr. M. Yamamoto, Okayama University, and Dr. H. Otani, M. Kodama, Tottori University, for discussions. This study was supported in part by a grant on priority area (A) to K.A. from the Ministry of Education, Culure, Sports, Science and Technology of Japan.
Abbreviations
- ACRS, ACR-toxin sensitivity gene
Alternaria alternata RLP, Alternaria alternata rough lemon pathotype
- IPTG
isopropyl 1-thio-β-d-galactoside
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Kohmoto K, Otani H, Tsuge T. In: Pathogenesis and Host Specificity in Plant Diseases: Histopathological, Biochemical, Genetic and Molecular Bases. Kohmoto K, Singh M S, Singh R P, editors. II. Oxford: Pergamon; 1995. pp. 51–63. [Google Scholar]
- 2.Nishimura S, Kohmoto K. Annu Rev Phytopathol. 1983;21:87–116. doi: 10.1146/annurev.py.21.090183.000511. [DOI] [PubMed] [Google Scholar]
- 3.Kohmoto K, Otani H. Experientia. 1991;47:755–764. doi: 10.1007/BF01922454. [DOI] [PubMed] [Google Scholar]
- 4.Kohmoto K, Akimitsu K, Otani H. Phytopathology. 1991;81:719–722. [Google Scholar]
- 5.Kohmoto K, Scheffer R P, Whiteside J O. Phytopathology. 1979;69:667–671. [Google Scholar]
- 6.Gardner J M, Kono Y, Tatum J H, Suzuki Y, Takeuchi S. Agric Biol Chem. 1985;49:1235–1238. [Google Scholar]
- 7.Nakatsuka S, Goto T, Kohmoto K, Nishimura S. In: Natural Products and Biological Activities. Imura H, Goto T, Murachi T, Nakajima T, editors. Tokyo: Univ. of Tokyo Press; 1986. pp. 11–18. [Google Scholar]
- 8.Akimitsu K, Kohmoto K, Otani H, Nishimura S. Plant Physiol. 1989;89:925–931. doi: 10.1104/pp.89.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohmoto K, Kondoh Y, Kohguchi T, Otani H, Nishimura S, Sheffer R P. Can J Bot. 1984;62:2485–2492. [Google Scholar]
- 10.Lu B, Hanson M R. Plant Cell. 1994;6:1955–1968. doi: 10.1105/tpc.6.12.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isshiki A, Akimitsu K, Yamamoto M, Yamamoto H. Mol Plant–Microbe Interact. 2001;14:749–757. doi: 10.1094/MPMI.2001.14.6.749. [DOI] [PubMed] [Google Scholar]
- 12.Masunaka A, Tanaka A, Tsuge T, Peever T L, Timmer L W, Yamamoto M, Yamamoto H, Akimitsu K. Phytopathology. 2000;90:762–768. doi: 10.1094/PHYTO.2000.90.7.762. [DOI] [PubMed] [Google Scholar]
- 13.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 16.Leaver C J, Gray M W. Annu Rev Plant Physiol. 1982;33:373–402. [Google Scholar]
- 17.Gray M W, Hanic-Joyce P J, Covello P S. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:145–175. [Google Scholar]
- 18.Leblanc C, Boyen C, Richard O, Bonnard G, Grienenberger J-M, Kloareg B. J Mol Biol. 1995;250:484–495. doi: 10.1006/jmbi.1995.0392. [DOI] [PubMed] [Google Scholar]
- 19.Manhart J R, Palmer J D. Nature (London) 1990;345:268–270. doi: 10.1038/345268a0. [DOI] [PubMed] [Google Scholar]
- 20.Vogel J, Börner T, Hess W R. Nucleic Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff G, Plante I, Lang B F, Kuck U, Burger G. J Mol Biol. 1994;237:75–86. doi: 10.1006/jmbi.1994.1210. [DOI] [PubMed] [Google Scholar]
- 22.Colleaux L, D'Auriol L, Galibert F, Dujon B. Proc Natl Acad Sci USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.du Jardin P, Portetelle D, Harvengt L, Dumont M, Wathelet B. Curr Genet. 1994;25:158–163. doi: 10.1007/BF00309542. [DOI] [PubMed] [Google Scholar]
- 24.Fassbender S, Bruhl K H, Ciriacy M, Kuck U. EMBO J. 1994;13:2075–2083. doi: 10.1002/j.1460-2075.1994.tb06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr G, Perlman M G, Lambowitz A M. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paquin B, Lang B F. J Mol Biol. 1996;255:688–701. doi: 10.1006/jmbi.1996.0056. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T, Kazama S, Hirai A, Akihama T, Kadowaki K. Curr Genet. 1991;20:331–337. doi: 10.1007/BF00318523. [DOI] [PubMed] [Google Scholar]
- 28.Clayton D A. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- 29.Iwabuchi M, Kyozuka J, Shimamoto K. EMBO J. 1993;12:1437–1446. doi: 10.1002/j.1460-2075.1993.tb05787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang H V, Pring D R, Shaw L C, Salazar R A, Muza F R, Yan B, Schertz K F. Plant J. 1996;10:123–133. doi: 10.1046/j.1365-313x.1996.10010123.x. [DOI] [PubMed] [Google Scholar]
- 31.Dill C L, Wise R P, Schnable P S. Genetics. 1997;147:1367–1379. doi: 10.1093/genetics/147.3.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Binder S, Brennicke A. Nucleic Acids Res. 1993;21:5012–5019. doi: 10.1093/nar/21.22.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson M R, Sutton C A, Lu B. Trends Plant Sci. 1996;1:57–64. [Google Scholar]
- 34.Marchfelder A, Brennicke A, Binder S. J Biol Chem. 1996;271:1898–1903. doi: 10.1074/jbc.271.4.1898. [DOI] [PubMed] [Google Scholar]
- 35.Wise R P, Gobelman-Werner K, Pei D, Dill C L, Schnable P S. J Heredity. 1999;90:380–385. doi: 10.1093/jhered/90.3.380. [DOI] [PubMed] [Google Scholar]
- 36.Golabek A A, Kida E, Walus M, Perez C, Wisniewski T, Soto C. Biophys J. 2000;79:1008–1015. doi: 10.1016/S0006-3495(00)76354-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hebert T E, Moffett S, Morello J-P, Loisel T P, Bichet D G, Barret C, Bouvier M. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 38.Overton M C, Blumer K J. Curr Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- 39.Kaspi C I, Siedow J N. J Biol Chem. 1993;268:5828–5833. [PubMed] [Google Scholar]
- 40.Hoch D H, Romero-Mira M, Ehrlich B E, Finkelstein A, DasGupta B R, Simpson L L. Proc Natl Acad Sci USA. 1985;82:1692–1696. doi: 10.1073/pnas.82.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhoads D M, Kaspi C I, Levings C S, III, Siedow J N. Proc Natl Acad Sci USA. 1994;91:8253–8257. doi: 10.1073/pnas.91.17.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dewey R E, Siedow J N, Timothy D H, Levings C S., III Science. 1988;239:293–295. doi: 10.1126/science.3276005. [DOI] [PubMed] [Google Scholar]
- 43.Levings C S., III Science. 1996;272:1279–1280. doi: 10.1126/science.272.5266.1279. [DOI] [PubMed] [Google Scholar]