Abstract
Cryptic invasions are a largely unrecognized type of biological invasion that lead to underestimation of the total numbers and impacts of invaders because of the difficulty in detecting them. The distribution and abundance of Phragmites australis in North America has increased dramatically over the past 150 years. This research tests the hypothesis that a non-native strain of Phragmites is responsible for the observed spread. Two noncoding chloroplast DNA regions were sequenced for samples collected worldwide, throughout the range of Phragmites. Modern North American populations were compared with historical ones from herbarium collections. Results indicate that an introduction has occurred, and the introduced type has displaced native types as well as expanded to regions previously not known to have Phragmites. Native types apparently have disappeared from New England and, while still present, may be threatened in other parts of North America.
Biological invasions threaten species and ecosystems worldwide (1). An estimated 50,000 exotic species have been introduced to the United States, of which 5,000 are plant species that have escaped and now exist in natural environments (2). Both the actual number of invaders and the impacts of these species may be underestimated because of the presence of cryptic invaders, or species that cannot be easily classified as native or introduced (3). Over the past decade, use of PCR-based molecular techniques have revealed repeated occurrences of such invasions in marine ecosystems (4–6), and studies have demonstrated both genetic and physiological differences between invading and native populations (6). Given that cryptic invaders typically are unrecognized or are mistaken for native species, knowledge of historical trends in geographic distribution and population genetic structure in cases of suspected introductions are of particular interest when trying to reconstruct the invasion history of a species. In such cases, museum or herbarium specimens are an invaluable resource for reconstructing population history.
Common reed, Phragmites australis (Cav.) Trin. ex Steudel (hereafter referred to as Phragmites), has a cosmopolitan distribution and is abundant in marsh communities and along the borders of lakes, ponds, and rivers. It is a perennial grass that reproduces primarily through vegetative growth, although dispersal by seeds may occur at low frequencies. In North America, the fossil record indicates that it has been present in the southwestern United States for at least 40,000 years (7). Paleoecological investigations have shown it to have been present along both the Atlantic and Pacific coasts for several thousand years (8–10). However, over the last 150 years its distribution and relative abundance has increased dramatically, particularly along the Atlantic coast. Botanical records from the 1800s typically describe Phragmites as being rare or not common (11–14), and a historical gap in its distribution was found in the southeastern states (15). By the early 1900s the species was considered more common and spreading (16, 17). Today it exists in all of the mainland United States as well as throughout southern Canada and is considered an indicator of wetland disturbance. It is also expanding into undisturbed sites, particularly in inland areas. To explain the spread of Phragmites, it has been suggested that the rapid expansion could be the result of human activities causing habitat disturbances or stresses such as pollution, changes in hydrologic regimes, and increased soil salinity (18). Alternatively, non-native genotypes of the species may have been introduced to North America sometime during the past 200 years (19–21), although to date no studies have adequately supported this hypothesis.
This research asks the question of whether or not non-native strains of P. australis exist in North America by using sequencing of two chloroplast DNA markers. Although the rate of evolution of the chloroplast genome is relatively conservative, variation has been found in chloroplast DNA at the intraspecific level (22). It is maternally inherited in angiosperms and has been shown to be geographically structured in a diverse array of plant species (23, 24), and therefore is an effective marker for use in the study of intraspecific phylogeography. In this study, modern samples of Phragmites, collected across the continent, were compared with historical specimens, collected before 1910, to examine changes in the genetic structure of the North American population over the past 150 years. Modern samples were also obtained worldwide for comparison and to determine the source of the introduction.
Materials and Methods
Leaf tissues were collected from green Phragmites plants during the growing seasons of 1997–2001 by the author and collaborators worldwide, with a particular emphasis on obtaining samples from the present-day range of Phragmites in North America and Europe. When available, herbarium specimens also were obtained to increase the number of samples from locations outside of North America. Fresh specimens were dried by using silica gel and frozen on receipt in the laboratory. Total DNA was extracted from 2 cm2 of fresh or dry leaf tissues using a 2% cetyltrimethylammonium bromide extraction protocol (25). Herbarium specimens were pretreated by scrubbing with 10% bleach to remove mounting glue, placed under an UV light for 5 min to remove surface contaminants, and extracted by using the same protocol. All herbarium samples used in historical comparisons were collected before 1910, which is before the time period when references to expansion of Phragmites populations began to appear in the literature (16). Where possible, modern samples also were collected at sites from which herbarium samples were obtained. Sample collection locations, herbarium accession numbers, and haplotype designations for each sample are available on request.
Two noncoding chloroplast regions were PCR-amplified by using the primer pairs trnT(UGU) “a”–trnL(UAA)5′ “b” (26) and rbcL–psaI (27) with annealing temperatures of 56°C and 54°C, respectively. Smaller fragments were amplified in the herbarium samples by using the primer pairs trnT(UGU) “a”-trnTaR (5′-TAGATTATTCSTCCGAGCC), trnL(UAA)5′ “b”-trnLbR (5′-GGAGAAGATAGAATCATAGC), rbcL2F (5′-CGCAGCTTGTGAAATATGG)–rbcL2R (5′-CGTATTTGATTCCATTATCGT), and psaI2F (5′-TGTCATAGAATAGGTGTCTC)–psaI2R (5′-GATTAGAAGGATAGAAAGGC), which were designed around the variable regions found in the larger fragments. Double-stranded PCR amplifications were sequenced directly in both directions on an Applied Biosystems 377 sequencer using the amplification primers and two internal primers in the rbcL-psaI region (rpL23F 5′-AGGTAGTAGCTGTGAATAGC and rpL23R 5′-AGTCGATGGCTATTCACAGC). In total about 2,000 bp were sequenced for each modern sample and 1,400 bp for herbarium samples.
Because of the high incidence of large insertion/deletion mutations (indels), sequences were aligned by eye with SEQUENCHER 4.1. Two mononucleotide repeat regions in the trnL region, which showed intrahaplotype length variation, were not used when distinguishing haplotypes. Before analysis all indels were coded as single characters to treat indels as single events rather than multiple independent events. Where indels were composed of several copies of a multiple-site insertion, each copy was treated as a single unit and gaps were inserted in haplotypes with fewer copies of the indel (28). Parsimony networks were obtained with the software tcs (29), using the algorithms of Templeton et al. (30). Haplotype diversity measures (31), analysis of molecular variance (32), and an exact test for population differentiation (33) were calculated by using ARLEQUIN 2.000 (34). All analyses were performed on the combined data set.
Results
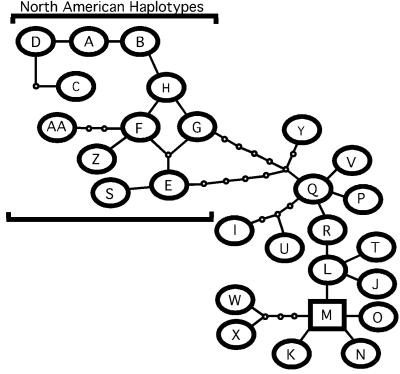
Based on sequencing of 283 modern samples and 62 herbarium samples collected before 1910, a total of 27 haplotypes were identified worldwide (Fig. 1). Where only partial sequences were obtained, samples were assigned to the most likely haplotype class based on the available sequence and geographic origin. There are 26 variable characters, of which 15 are indels (four type Ia, which are mononucleotide repeats, and nine type Ib, which are deletions or duplications of adjacent sequences, and two type II, which are all other types of indels (35) and 11 are base substitutions. Eleven haplotypes are unique to North America (haplotypes A-H, S, Z, and AA) and are considered to be native to the continent. These 11 are distinguished from all other haplotypes by five shared indels (two type Ia, one type Ib, and two type II). Two haplotypes have a widespread distribution on multiple continents (haplotypes I and M), with haplotype M being the most common type in North America, Europe, and Asia today (Table 1). This type is most closely related to other haplotypes found in Europe, Asia, and Africa (Fig. 1). It is also the predicted ancestral haplotype based on coalescent theory (36).
Figure 1.
Parsimony network of Phragmites chloroplast haplotype diversity obtained from sampling 345 populations worldwide. Each link between haplotypes represents one mutational difference, following coding of indels as single characters. Unlabeled nodes indicate inferred steps not found in the sampled populations. Loops in the network are the result of homoplasies in the number of repeats in some indels. The ancestral haplotype, or root of the network, is indicated by a square. Geographic distribution of haplotypes is as follows: North America = haplotypes A-H, S, Z, AA, I, and M; South America = I and Y; Europe = L-O, and T; Asia/Australia = I, J, L, M, O, P, Q, U, W, and X; Africa = K, M, R, and V.
Table 1.
Frequencies of Phragmites haplotypes worldwide
| Geographic region | Total no. of samples | % Regional haplotypes | % Haplotype I | % Haplotype M |
|---|---|---|---|---|
| North America, after 1960 | 195 | 31.3 | 7.2 | 61.5 |
| North America, before 1910 | 62 | 83.9 | 9.7 | 6.4 |
| South America | 11 | 27.2 | 72.7 | 0 |
| Europe | 41 | 39.0 | 0 | 61.0 |
| Asia/Australia | 27 | 55.6 | 11.1 | 33.3 |
| Africa | 9 | 88.9 | 0 | 11.1 |
Regional haplotypes refer to those found only in the corresponding geographic region.
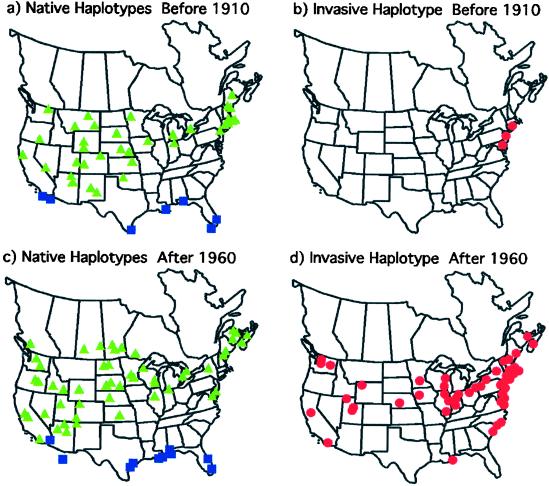
Within North America, the exact test of population differentiation indicates significant changes in haplotype frequencies between the historical and modern samples [P < 0.001 (33); Fig. 2]. The pre-1910 populations showed a widespread distribution of the 11 native haplotypes across North America from New England west to the Pacific coast (Fig. 2a). Haplotype I, which also is found in South America and Asia (Fig. 1), was distributed along the Gulf Coast. Haplotype M was found at four sites of the 62 sampled (New Haven, CT; Madison, CT; Camden, NJ; Chesapeake Beach, MD; Fig. 2b). In comparison, whereas the native haplotypes and haplotype I remain throughout much of their original range, modern populations show a striking pattern of expansion in the range of haplotype M (Fig. 2 c and d). This type has replaced native types in New England and expanded to the southeast where Phragmites historically did not grow. It is presently expanding to the west and becoming prevalent in the Midwestern states.
Figure 2.
Distribution of Phragmites haplotypes in North America. Green triangles represent the 11 native haplotypes, blue squares represent haplotype I, and red circles represent the invasive haplotype M. (a and b) The distribution of haplotypes in the 62 herbarium samples collected before 1910. (c and d) The distribution of haplotypes in 195 samples collected after 1960.
Measures of haplotype diversity show a decline in diversity between the historical population and the present (0.80 ± 0.04 vs. 0.58 ± 0.04). Further, analysis of molecular variance shows that among-population variation accounts for a larger proportion of the total variance in the historical population (35%) than the modern population (−9%) when compared with worldwide populations (P < 0.001), indicating that today the genetic structure of North American Phragmites populations is more like that of Europe and the rest of the world. Within-population variation increased between the historic and modern populations (52% vs. 79% of the total variance), reflecting the spread of haplotype M into new regions.
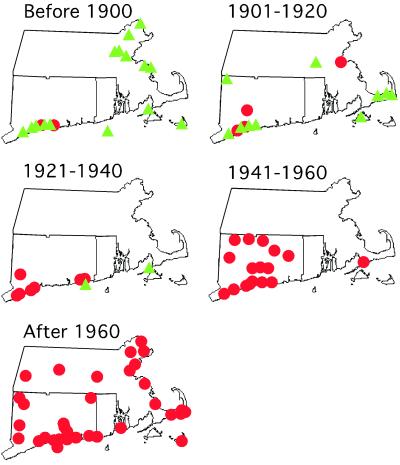
To examine the time frame over which the rapid expansion of haplotype M occurred, a chronology of haplotype frequencies in 20-year increments was examined for populations in Connecticut and Massachusetts (Fig. 3). Although the 19th-century samples were primarily native types (haplotype E = 47%, haplotype F = 29%, haplotype AA = 12%, and haplotype M = 12%), a changeover is seen and by 1940 all samples displayed haplotype M.
Figure 3.
Changes in Phragmites haplotype distribution patterns over 20-year time intervals in Connecticut, Massachusetts, and Rhode Island. Green triangles represent native haplotypes; red circles represent the invasive haplotype M.
Discussion
Geographic structuring in Phragmites was found worldwide at continental scales. Closely related unique haplotypes were found in each geographic region with haplotype M being the most common worldwide (Table 1, Fig. 1). Haplotype M was found across Europe and continental Asia in high frequencies and is closely related to all other haplotypes found in these areas. It was not found in the islands of the Pacific or Indian oceans but it was present at one site in New Zealand. However, Phragmites may be a recent addition to the New Zealand flora (Paul Champion, personal communication). The estimation of haplotype M being the ancestral one, based on coalescent theory, corresponds well with the finding that higher levels of isozyme diversity are found in Phragmites populations from western and central Asia (37).
Today haplotype M is the most common and has the most widespread distribution of any haplotype in North America (Fig. 2d). However, network analysis suggests that it is not closely related to other North American haplotypes, which cluster together and are quite distinct from other Phragmites haplotypes (Fig. 1). Its extremely limited distribution in the historic sample (Fig. 2b) indicates that the present-day distribution of this haplotype does not reflect historical trends in North American Phragmites populations.
Haplotype I was found along the Gulf Coast of North America, as well as in South America, where it is the dominant type, and on several islands in the southern Pacific. The distribution of this type in North America appears to be the same as the Gulf Coast phenotype identified by Pellegrin and Hauber (20) based on isozymes. These data support their suggestion that the presence of this type in a wide variety of habitats across southern North America may be the result of the establishment of a single genetic lineage with broad ecological tolerances that has spread throughout the region. However, because of its prevalence in other parts of the world, it is not possible to assign haplotype I to a category of native or introduced to North America although its distribution remained the same between the historic and modern samples (Fig. 2 a and c). Given that its mostly closely related haplotype is found only in Asia (Fig. 1), it is possible that haplotype I originated there but it is not known when it arrived in North America.
The 11 native North American types show little change in their distribution between the historic and modern samples from the Midwest to the Pacific Coast (Fig. 2 a and c). However, the three native haplotypes that were found in the historical sample from southern and central New England were not detected in the modern sample, despite resampling of all of the sites from which 19th-century herbarium specimens were available (Fig. 3). Further, haplotype AA was restricted to this region in the historical samples and is not found in any of the modern samples. Thus, in addition to local changes in haplotype frequencies, extinction of Phragmites lineages may have occurred over the past century. Native haplotypes were found in only two sites along the Atlantic coast (Allen, MD and Chance, VA) in the modern sample in contrast to their widespread distribution throughout eastern North America in the historic sample (Fig. 2 a and c). The lack of persistence of native types is surprising given the clonal nature of this species and suggests that haplotype M is highly competitive and aggressive. This is further evidenced by the rapid replacement of native lineages by the invasive one seen in marshes of Connecticut and Massachusetts by 1940 (Fig. 3).
The rapid proliferation of haplotype M throughout the Atlantic Coast could result from either an introduction of this type from elsewhere or a range expansion of an existing native type. Because this haplotype was present in historical samples it is possible that human-induced changes in the landscape or other unidentified causes gave it an advantage that allowed it to expand rapidly. It is more likely that an introduction of Phragmites has occurred because (i) haplotype M shares none of the mutations that link the 11 native North American haplotypes; (ii) it is most closely related to EurAsian types (Fig. 1); and (iii) population structuring has declined significantly between the pre-1910 and modern samples from North America. This introduction probably occurred sometime during the early part of the 19th century, most likely at one or more coastal ports along the Atlantic coast. In the 1800s, Phragmites was documented growing in places where ships ballast was dumped or used to fill marsh lands being converted to railroad and shipping hubs (38). Because Phragmites already grew in coastal marshes as a native component of the plant community and the introduced variety showed little or no morphological differences with native types, the establishment of non-native populations was not recognized. After several decades of persisting in low densities, rapid expansion of the type began and was probably facilitated by human dispersal by means of the widespread construction of railroads and major roadways across North America in the late 19th and early 20th centuries. Given the aggressive patterns of spread seen over the past century, it is likely that this expansion will continue to occur into western and northern parts of the continent. The presence of native Phragmites lineages throughout these areas will only complicate efforts to control this spread.
It has been difficult for scientists to predict whether or not a species will become invasive upon entering a new habitat (1). Detection of cryptic invasions is critical for quantifying both the numbers of invaders and their impacts. For species with widespread native distributions, genetic diversity may play an important role in their behavior when establishing at new sites. Differences in physiological tolerance and behavior may give non-native genotypes unforeseen advantages allowing them to proliferate and changing the genetic structure of the species. This study presents compelling evidence of a cryptic invasion in a terrestrial plant species. This invasion is on a scale comparable to (if not greater than) other known wetland invaders, such as purple loosestrife (Lythrum salicaria) and salt cedar (Tamarix sp.), but appears to still be in a phase of expansion into new areas. It is important to recognize that the structure and function of native terrestrial communities may be influenced by both cryptic and easily recognized invaders.
Acknowledgments
Thanks to J. R. Powell, G. Caccone, D. Skelly, M. Donoghue, J. Gleason, J. S. Hall, and two anonymous reviewers for comments and discussions. J. R. Powell kindly provided the laboratory space and facilities for this work. I thank the U.S. National Herbarium, Harvard University Herbaria, New England Botanical Society, George Safford Torrey Herbarium at the University of Connecticut, Connecticut Botanical Society, and Yale University Herbarium for providing herbarium samples. This research was funded by an Environmental Protection Agency Science to Achieve Results graduate fellowship, The Nature Conservancy Connecticut chapter, The Long Island Sound Fund administered by the Connecticut Department of Environmental Protection through the sale of Long Island Sound license plates and contributions, the New Jersey Public Service, Electric and Gas Company, and the U.S. Fish and Wildlife Service through the Biological Control of Nonindigenous Plant Species Program at Cornell University, and the National Oceanic and Atmospheric Administration Office of Sea Grant and Extramural Programs, U.S. Department of Commerce, under Grant MERP/SG 99–21.
Footnotes
References
- 1.Mack R N, Simberloff D, Lonsdale W M, Evans H, Clout M, Bazzaz F A. Ecol Appl. 2000;10:689–710. [Google Scholar]
- 2.Pimentel D, Lach L, Zuniga R, Morrison D. BioScience. 2000;50:53–65. [Google Scholar]
- 3.Carlton J T. Ecology. 1996;77:1653–1655. [Google Scholar]
- 4.Geller J B, Walton E D, Gorsholz E D, Ruiz G M. Mol Ecol. 1997;6:901–906. doi: 10.1046/j.1365-294x.1997.00256.x. [DOI] [PubMed] [Google Scholar]
- 5.Bastrop R, Jurss K, Strumbauer C. Mol Biol Evol. 1998;15:97–103. doi: 10.1093/oxfordjournals.molbev.a025919. [DOI] [PubMed] [Google Scholar]
- 6.McIvor L, Maggs C A, Provan J, Stanhope M J. Mol Ecol. 2001;10:911–919. doi: 10.1046/j.1365-294x.2001.01240.x. [DOI] [PubMed] [Google Scholar]
- 7.Hansen R M. Paleobiology. 1978;4:302–319. [Google Scholar]
- 8.Niering W A, Warren R S, Weymouth C G. Connecticut Arboretum Bull. 1977;22:2–12. [Google Scholar]
- 9.Orson R. Biol Inv. 1999;1:149–158. [Google Scholar]
- 10.Goman M, Wells L. Quat Res. 2000;54:206–217. [Google Scholar]
- 11.Torrey J. Flora of the State of New York. Albany: Carroll and Cook; 1843. [Google Scholar]
- 12.Willis O R. Catalogue of Plants Growing Without Cultivation in the State of New Jersey. New York: J. W. Schermerhorn; 1874. [Google Scholar]
- 13.MaCain J. Catalogue of Canadian Plants: Part 1, Polypetale. Montreal: Dawson Brothers; 1883. [Google Scholar]
- 14.Dame L L, Collins F S. Flora of Middlesex County, MA. Malden, MA: Middlesex Institute; 1888. [Google Scholar]
- 15.Hitchcock A S. Manual of the Grasses of the United States. Washington, DC: U.S. Goverment Printing Office; 1935. , Misc. publication no. 200. [Google Scholar]
- 16.Graves C B, Eames E H, Bissell C H, Andrews L, Harger E B, Weatherby C A. Bulletin of the Connecticut Geological and Natural History Survey No. 14. Brainard, Hartford, CT: Case, Lockwood; 1910. [Google Scholar]
- 17.Stone W. 1910 Annual Report of the New Jersey State Museum. Trenton: New Jersey State Museum; 1911. [Google Scholar]
- 18.Marks M, Lapin B, Randall J. Natural Areas J. 1994;14:285–294. [Google Scholar]
- 19.Metzler K, Rosza R. Newsl Connecticut Bot Soc. 1987;15:1–6. [Google Scholar]
- 20.Pellegrin D, Hauber D P. Aquatic Bot. 1999;63:241–259. [Google Scholar]
- 21.Chambers R M, Meyerson L A, Saltonstall K. Aquatic Bot. 1999;64:261–273. [Google Scholar]
- 22.Soltis D E, Soltis P S, Milligan B G. In: Molecular Systematics of Plants. Soltis D E, Soltis P S, Doyle J J, editors. New York: Chapman & Hall; 1992. pp. 117–150. [Google Scholar]
- 23.Soltis D E, Gitzendanner M A, Strenge D D, Soltis P S. Plant Syst Evol. 1997;206:353–373. [Google Scholar]
- 24.Sahuquillo E, Lumaret R. Mol Ecol. 1999;8:1797–1803. doi: 10.1046/j.1365-294x.1999.00755.x. [DOI] [PubMed] [Google Scholar]
- 25.Doyle J J, Dickson E E. Taxon. 1987;36:715–722. [Google Scholar]
- 26.Taberlet P, Gielly L, Pautou G, Bouvet J. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 27.Saltonstall K. Mol Ecol Notes. 2001;1:76–78. [Google Scholar]
- 28.McGuire G, Denham M C, Balding D J. Mol Biol Evol. 2001;18:481–490. doi: 10.1093/oxfordjournals.molbev.a003827. [DOI] [PubMed] [Google Scholar]
- 29.Clement M, Posada D, Crandall K A. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 30.Templeton A R, Crandall K A, Sing C F. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 32.Excoffier L, Smouse P, Quattro J. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raymond M, Rousset F. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 34.Schneider S, Roessli D, Excoffier L. arlequin: A Software for Population Genetics Data Analysis. Univ. of Geneva, Geneva: Genetics and Biometry Laboratory; 2000. , version 2.0. [Google Scholar]
- 35.Golenberg E M, Clegg M T, Durbin M L, Doebley J, Ma D P. Mol Phylogenet Evol. 1993;2:52–64. doi: 10.1006/mpev.1993.1006. [DOI] [PubMed] [Google Scholar]
- 36.Castelloe J, Templeton A R. Mol Phylogenet Evol. 1994;3:102–113. doi: 10.1006/mpev.1994.1013. [DOI] [PubMed] [Google Scholar]
- 37.Bahrman N, Gorenflot R. Rev Géneral Bot. 1983;90:177–184. [Google Scholar]
- 38.Burk I. Proc Acad Natl Sci Philadelphia. 1877;29:105–109. [Google Scholar]