Among the most highly conserved biochemical pathways in free living organisms are those involved in DNA repair (1). The ubiquitous pathway of nucleotide excision repair (NER) is responsible for the removal of environmentally induced DNA damage, such as the DNA lesions resulting from sunlight exposure or chemical carcinogens. Mutations or deficiencies in specific NER genes can lead to premature aging and cancer in humans (2, 3). The study of DNA repair in the bacterium Escherichia coli has helped us to understand the corresponding repair pathways in humans (4). NER can be viewed in four basic steps: (i) damage recognition and lesion verification; (ii) incision; (iii) excision; (iv) repair synthesis and ligation, as proposed nearly four decades ago. Damage recognition and verification are achieved by a protein machine that utilizes several components to sense a distortion in the double-helical duplex DNA. In E. coli the UvrA and UvrB proteins carry out these functions. If a putative lesion is identified by UvrA, the repair complex enlists the strand-opening activity of UvrB that helps to verify that the distortion is, in fact, due to a damaged nucleotide. It is believed that the beta-hairpin domain of UvrB is inserted into the DNA helix both to verify the damaged nucleotide and to establish which strand has been damaged (5–9). In both bacterial and eukaryotic species, strand opening and processing of the damage serves to further change the conformation of the DNA to help recruit nucleases to the lesion site to produce two endonucleolytic incisions in the phosphodiester backbone of the damaged strand, one on each side of the altered nucleotide(s). In E. coli, Bacillus caldotenax (9), and presumably in all other free living bacterial species, UvrB recruits the UvrC protein, which contains two functional endonuclease domains. The N-terminal part of this protein is responsible for cutting the damaged strand four or five nucleotides 3′ to the altered nucleotide, while the C-terminal part is necessary and sufficient to produce the second incision, some eight nucleotides 5′ from the damaged nucleotide; about one turn of the helix from the first cut. (In apparent contrast, human cells require two separate protein activities to produce dual incisions roughly three turns of the DNA helix apart.) Once incision has occurred, the damaged oligonucleotide is excised by the UvrD helicase, just before or during the repair synthesis step, which is normally carried out by DNA polymerase I. DNA ligase then seals the newly created repair patch at its 3′ end to complete the process.
How important is Cho? From where did this UvrC homolog arise and what function does it serve?
Even though the complete genome of E. coli has been known since 1997 (10), this species continues to yield surprises about the complexity and inner workings of bacteria. Nora Goosen and her coworkers in a recent issue of PNAS report their discovery of a new NER incision enzyme that they have designated Cho because it is a UvrC homolog (11). The gene for Cho was first identified as an ORF, ydjQ, a new DNA damage-inducible gene in E. coli (12–14). Nucleotide sequence analysis revealed that this gene encodes a protein of 295 aa and that it is homologous to the N-terminal portion of UvrC. Cho is surprising for a number of reasons. First, purified Cho only produces incisions on the 3′ side of the lesion, at the ninth phosphodiester bond 3′ to the altered nucleotide, four nucleotides beyond the site of the normal UvrC incision. Second, some types of DNA lesions lead to more efficient incision by Cho than by UvrC. Third, like UvrC, Cho requires the formation of the UvrB-DNA damage verification complex (6) but does not require the UvrC binding domain of UvrB. Finally, UvrC can make the 5′ incision on a substrate already incised 3′ by Cho.
Many questions arise: How important is Cho? From where did this UvrC homolog arise and what function does it serve? Why is it damage inducible while UvrC is not? With regard to the first question, Goosen and her coworkers show that the cho gene contributes to UV survival, but that its expression cannot completely compensate for the loss of UvrC. This is reminiscent of early studies in several laboratories that indicated that uvrC mutants are less UV-sensitive than are uvrA or uvrB mutants. The possible answers to the second question are a bit perplexing. Analysis of all bacterial genomes sequenced to date reveals that although Cho and UvrC coexist in a few different bacterial species (Listerias, Clostridia, E. coli, and Mycobacterium tuberculosis), many more species only have UvrC, and surprisingly, it appears that the mycoplasmas and Borrelia burgdorferi only have Cho (see Fig. 2, which is published as supporting information on the PNAS web site, www.pnas.org). It is also provocative to note that in the Mycobacterium species the Cho protein is predicted to be larger than the E. coli version of Cho (≈69 kDa), and it has an additional domain with strong homology to the epsilon 3′ exonuclease, which is the proofreading subunit of the DNA polymerase III holoenzyme. This added domain might explain how some bacteria that lack the normal copy of UvrC could still complete NER. Once the 3′ incision is produced by Cho, the putative exonuclease activity of Cho would digest in the 3′ direction through the lesion site and leave a 3′ OH end that could serve as a primer for repair synthesis. Alternatively the excision of the lesion could be performed by another 3′ exonuclease. In this regard it is interesting to note that the gene encoding epsilon, dnaQ, is damage inducible (ref. 15; Fig. 1). Although we are unaware of any demonstration that epsilon can digest lesion-containing polynucleotides, as a predictive model it has been established that the associated 3′–5′ proofreading exonuclease activity of the phage T7 DNA polymerase can hydrolyze DNA containing several different kinds of lesions (16).
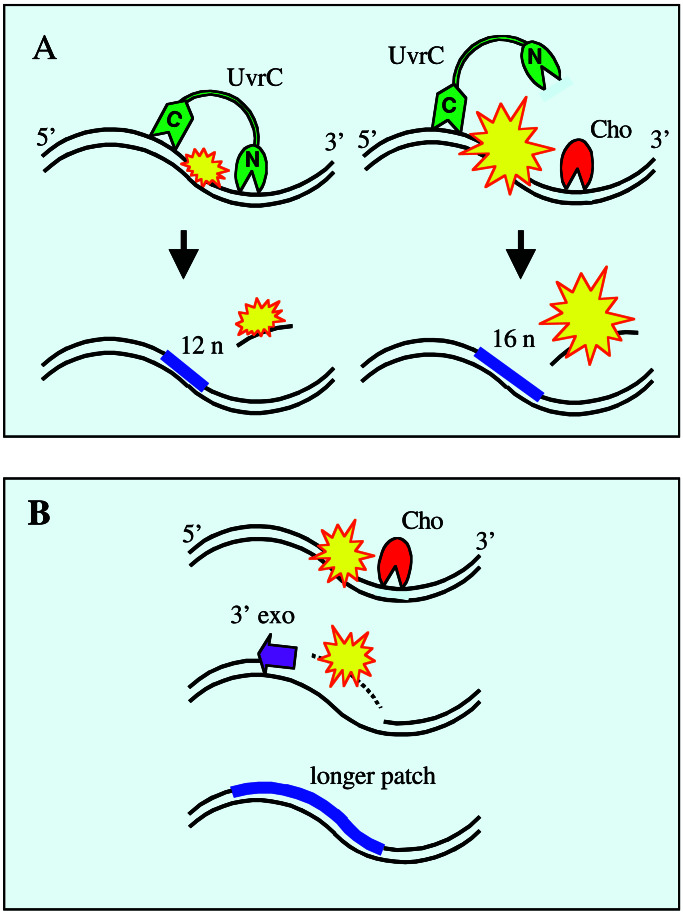
Figure 1.
A schematic depiction of the steps of nucleotide excision repair in E. coli that follow the initial recognition/verification of a lesion. (A) Dual incisions by UvrC (green) at a lesion (yellow) or a 3′ side incision by Cho (red) followed by the 5′ side incision by UvrC (A Right), at a lesion that obscures the normal 3′ incision site for UvrC. Incisions, in both situations, are followed by excision, repair replication, and ligation, but a four-nucleotide-longer patch results from the participation of Cho. (B) Putative steps when UvrC is missing and where Cho makes only a 3′ incision. In this case it is supposed that a 3′ exonuclease removes a stretch of DNA of indeterminate length containing the lesion, and that a much longer patch would be introduced by repair replication.
What purpose could Cho serve in bacterial species, like E. coli, that already contain UvrC? Goosen and her coworkers offer a very plausible explanation that is supported by data supplied in their paper. Some types of bulky DNA adducts may interfere with UvrC incision nearby at the fifth or sixth phosphodiester bond 3′ to the adduct, and this has actually been shown for certain conformations of carcinogen adducts (17). However Cho, which cuts four nucleotides farther away from the lesion, would not be inhibited. Goosen and her coworkers showed that substrates containing either of two large synthetic lesions, a cholesterol or a menthol DNA adduct, resulted in more efficient incision by Cho than by UvrC (11). Another interesting possibility is raised by findings from Tang and coworkers (18, 19). They have shown that E. coli UvrC can be isolated in two states: a monomeric form and a tetrameric form, which has different properties. For example, although both forms can bind to single-stranded DNA, only the tetrameric form can bind to double-stranded DNA. Furthermore, the helix-stabilizing CC-1065-N3-adenine adduct is only incised on the 5′ side by tetrameric UvrC, suggesting that an important role for Cho could be to incise on the 3′ side of this type of damage. It is possible that an unusually large lesion, such as that imposed by a protein–DNA crosslink, might also sterically hinder access by UvrC, but not by Cho to produce the 3′ side incision. It has been recently shown that DNA carrying a covalently trapped T4 phage pyrimidine dimer glycosylase (a 16-kD protein) is subject to dual incisions by UvrC at moderate efficiency (20). Perhaps Cho will incise more efficiently than UvrC at such a protein–DNA crosslink. Another possible explanation for the existence of Cho relates to the manner of its regulation, as discussed below.
The arsenal of lesion recognition and DNA damage processing enzymes is not fully expressed until cells have been genomically stressed, as through environmental genotoxic chemicals or radiation. Thus, the so-called SOS-inducible stress response in UV-irradiated E. coli results in the up-regulated expression of over 40 genes, as a consequence of the arrest of DNA replication forks at the sites of pyrimidine photoproducts (14). The up-regulated genes include uvrA and uvrB that are required for efficient lesion recognition and verification. In fact, the global genomic repair of the predominant photoproducts, cyclobutane pyrimidine dimers, is very poor unless these genes are induced (21). Similarly, in UV-irradiated human cells the p53 tumor suppressor becomes activated/stabilized and then induces the p48 gene whose product is part of a DNA damage binding activity that is required for the efficient recognition of pyrimidine dimers (22, 23). However, in neither E. coli nor human cells has there been any indication that the enzymes required for incisions are also up-regulated under genomic stress. The uvrC gene is not SOS regulated—but now we have the example that cho appears to be as responsive to SOS up-regulation as uvrB (14). What could be the significance of this inducible up-regulation? It could be simply to expand on the options for performing NER once the lesion has been recognized. If UvrC is unable to make the 3′ incision because of steric hindrance, then Cho comes to the rescue with “a cut above” as illustrated in Fig. 1.
One may wonder why the cell does not constitutively maintain its full NER capability An obvious answer is energy economy. It is metabolically expensive to support a large “standing army” of repair enzymes to deal with some future genomic invasion that may never happen. The SOS system has evolved to rapidly call up the “reserves” when needed. These reserves in E. coli include not only the enzymes needed for versatile and efficient NER but also three different DNA polymerases with unique capabilities for translesion DNA synthesis, and a number of enzymes used in genetic recombination. A second important rationale for suppressing the expression of NER enzymes during cellular growth in a nonthreatening environment is to avoid the occurrence of gratuitous DNA repair in undamaged DNA. The damage recognition elements have evolved to deal with a very broad spectrum of lesions, of which some, like the pyrimidine dimer, are only minimally disruptive of the DNA structure. Thus, unique DNA structures such as sequence-dependent cruciforms, hairpins, DNA bends, and Z-DNA might be mistaken as lesions, especially if the levels of repair enzymes are high. Sancar and coworkers (24) have reported that the UvrABC system attacks undamaged DNA at a significant rate and they have documented similar gratuitous repair events in a human in vitro DNA repair assay. Therefore, it makes good sense for the cell to prevent unnecessary tampering with its genomic integrity by not maintaining its repair capabilities at full capacity until a life-threatening situation arises.
What surprises await us in the bacterial world of DNA repair? Genome analysis indicates that many species have duplications of uvrA, including, for example, the most radiation-resistant species known, Deinococcus radiodurans (25). UvrA duplications are also found in those Gram-positive bacterial species noted above in which uvrC and Cho coexist. In addition, some bacteria encode close homologs of eukaryotic NER genes (e.g., Saccharomyces cerevisiae RAD25) and some archaea encode both a set of UvrABCD genes and multiple RAD homologs (1). The discovery of Cho reminds us that there is still much to be learned about bacterial NER, and that the continued study of E. coli and other species in the exciting bacterial world will certainly reveal more surprises.
Supplementary Material
Footnotes
See companion article on page 1467 in issue 3 of volume 99.
References
- 1.Eisen J A, Hanawalt P C. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg E C. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers J H J. Nature (London) 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 4.Petit C, Sancar A. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 5.Zou Y, Van Houten B. EMBO J. 1999;18:4889–4901. doi: 10.1093/emboj/18.17.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theis K, Chen P J, Skorvaga M, Van Houten B, Kisker C. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theis K, Skorvaga M, Machius M, Nakagawa N, Van Houten B, Kisker C. Mutat Res. 2000;460:277–300. doi: 10.1016/s0921-8777(00)00032-x. [DOI] [PubMed] [Google Scholar]
- 8.Moolenaar G F, Hoglund L, Goosen N. EMBO J. 2001;20:6140–6149. doi: 10.1093/emboj/20.21.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skorvaga M, Theis K, Kisker C, Van Houten B. J Biol Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- 10.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 11.Moolenaar G F, van Rossum-Fikkert S, van Kestern M, Goosen N. Proc Natl Acad Sci USA. 2002;99:1467–1472. doi: 10.1073/pnas.032584099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis L K, Harlow G R, Gregg-Jolly L A, Mount D W. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez de Henestrosa A R, Ogi T, Aoyagi S, Chafin D, Hayes J J, Ohmori H, Woodgate R. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 14.Courcelle J, Khodursky A, Peter B, Brown P O, Hanawalt P C. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinones A, Piechocki R, Messer W. Mol Gen Genet. 1988;211:106–112. doi: 10.1007/BF00338400. [DOI] [PubMed] [Google Scholar]
- 16.Koehler D R, Hanawalt P C. Biochem J. 1993;293:451–453. doi: 10.1042/bj2930451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou Y, Liu T, Geacintov N E, Van Houten B. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 18.Nazimiec M, Lee C S, Tang Y L, Ye X, Case R, Tang M-S. Biochemistry. 2001;40:11073–11081. doi: 10.1021/bi010953p. [DOI] [PubMed] [Google Scholar]
- 19.Tang M-s, Nazimiec M, Ye X, Iyer G H, Eveleigh J, Zheng Y, Zhou W, Tang Y-Y. J Biol Chem. 2001;276:3904–3910. doi: 10.1074/jbc.M008538200. [DOI] [PubMed] [Google Scholar]
- 20.Minko I G, Zou Y, Lloyd R S. Proc Natl Acad Sci USA. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowley D J, Hanawalt P C. J Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ford J M, Hanawalt P C. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 23.Hwang B J, Ford J M, Hanawalt P C, Chu G. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branum M E, Reardon J T, Sancar A. J Biol Chem. 2001;276:25421–25426. doi: 10.1074/jbc.M101032200. [DOI] [PubMed] [Google Scholar]
- 25.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, et al. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.