Abstract
The three PPAR (peroxisome proliferator-activated receptor) isoforms are critical regulators of lipid homeostasis by controlling the balance between the burning and storage of long chain fatty acids. Whereas PPARα and PPARγ have been studied extensively, the function of PPARδ remains the most elusive. Intriguingly, in cotransfection experiments, PPARδ is a potent inhibitor of ligand-induced transcriptional activity of PPARα and PPARγ. This inhibition is achieved, in part, by binding of PPARδ to a peroxisome proliferator response element and the association of nonliganded PPARδ with corepressors SMRT (silencing mediator for retinoid and thyroid hormone receptors), SHARP (SMRT and histone deacetylase-associated repressor protein), and class I histone deacetylases. Stable expression of PPARδ inhibits the expression of endogenous PPARα target gene expression in 3T3-PPARα cells, whereas a PPARδ mutant that does not interact with the corepressor SMRT loses its ability to repress the induction of PPARα target gene. Similarly, stable expression of PPARδ in 3T3-PPARγ cells leads to inhibition of PPARγ target gene expression and PPARγ-mediated adipogenesis. Given the widespread expression of PPARδ and the restricted pattern for PPARα and PPARγ, these results suggest a role for PPARδ as a gateway receptor whose relative levels of expression can be used to modulate PPARα and PPARγ activity.
The PPAR subfamily includes three isoforms (α, γ, and δ) that all bind to the peroxisome proliferator response elements (PPRE) as a heterodimer with retinoid X receptor (RXR), yet exhibit distinct tissue distribution and physiological function. PPARα is highly expressed in the liver and was originally identified as a molecule that mediates the transcriptional effects of drugs that induce peroxisome proliferation in rodents (1). In addition to its activation in response to peroxisome proliferators such as Wy14,643, PPARα is also activated by a variety of medium- and long-chain fatty acids (2) and has been shown to stimulate lipid metabolism by the induction of peroxisomal β-oxidation and fatty acid ω-hydroxylation (3). Mice lacking functional PPARα are incapable of responding to peroxisome proliferators and fail to induce expression of a variety of genes required for the metabolism of fatty acids, including acyl-CoA oxidase (AOX) (4).
PPARγ is highly enriched in adipose tissue and has been shown to play a central role in activating adipogenesis both in vitro and in vivo (5, 6). It is also expressed at high levels in lipid-accumulating macrophages and plays a role in the development of the atherogenic lesion (7). PPARγ has been shown to be activated by 15-deoxy-(12,14)-prostaglandin J2 (15d-PGJ2) or its synthetic analog thiazolidinedione, a novel class of antidiabetic drugs (8, 9). The PPARγ-null mice are embryonic lethal due in part to disrupted placental function (6). Rescue of the placental defect results in lipid dystrophy and neonatal death (10, 11).
PPARδ (also known as PPARβ) is widely expressed with relatively higher levels in brain, colon, and skin (12–14). Although there have been extensive studies on PPARα and PPARγ, much less is known about the function of PPARδ. Recent studies have suggested that PPARδ might play a role in cholesterol efflux (15), colon cancer (16), embryo implantation (17), preadipocyte proliferation (18), and epidermal maturation (19). Whereas the role of PPARγ in adipocyte differentiation is well documented, a role for PPARδ remains controversial (20, 21). Homozygous PPARδ-null term fetuses were smaller than controls, and this phenotype persisted postnatally. Gonadal adipose stores were smaller, yet CD36 mRNA levels were higher in PPARδ-null mice than in controls (22). However, the reduction of adiposity in PPARδ null mice cannot be reconstructed by adipocyte-specific PPARδ deficiency, suggesting that PPARδ controls the process in a systemic fashion, probably through effects on global lipid homeostasis (37).
In contrast to TR and RAR, unliganded PPARγ does not repress PPRE-mediated transcription, presumably because of the inability of PPARγ to bind nuclear receptor corepressors SMRT and NCoR on the PPRE DNA element (23). Nuclear receptor corepressors, such as SMRT and NCoR, have been shown to mediate the transcriptional repression of unliganded nuclear receptors through the recruitment of a histone deacetylase (HDAC) complex (24–27). We recently demonstrated that a novel corepressor SHARP represses both liganded and nonliganded nuclear receptor transcription by association with the corepressors SMRT and HDACs and sequestering the coactivator SRA (28). Whether PPARδ is associated with any of the nuclear receptor cofactors and how PPARδ regulates target gene transcription remains unknown.
In this study, we demonstrate that PPARδ functions as a potent transcriptional repressor with two interrelated properties. First, on its own it is able to repress basal transcription. Second, coexpression of PPARδ with PPARα or PPARγ leads to dramatic inhibition of PPAR target gene expression. Consequently, PPARδ is able to block the expression of acyl-CoA oxidase and PPARγ-mediated adipogenesis in cultured cells. This anti-lipid oxidation and anti-adipogenic feature of PPARδ might represent a useful strategy to gate the activities of PPARα and PPARγ and could be exploited as a potential therapeutic approach in the control of obesity and related type II diabetes.
Materials and Methods
Plasmids.
CMX-GAL4 DBD, GAL4-SMRT RID, ID1, ID2, ID1+2, and ID1 mutants, CMX-VP, MH100-tk-luc, Flag-HDAC1, HDAC2, HDAC3, HDAC5, and HDAC7, Flag-SMRT, GST-SMRT RID, CMX-PPARα, PPARγ, and PPARδ, PBabe-PPARα and PPARγ have been described (5, 28). GAL4-PPARα, γ, and δ were generated by cloning the corresponding cDNA into CMX-GAL4 DBD vector. Hemagglutinin (HA)-PPARδ was constructed by cloning PPARδ cDNA into CMX-HA vector. VP-PPARδ was constructed by cloning PPARδ cDNA into CMX-VP vector. The point mutations of PPARδ were created by site-directed mutagenesis by using Quickchange site-directed mutagenesis kit (Stratagene). pLHCX-PPARδ was made by subcloning the PPARδ cDNA into pLHCX vector (CLONTECH).
Transient Transfections.
CV-1 cells were transiently transfected as described before (28). Luciferase activity of each sample was normalized by the level of β-galactosidase activity. Each transfection was carried out in triplicate for at least three times.
Northern Blot Analysis.
Total RNA from tissue-cultured cells was isolated by using Trizol reagent (Life Technologies, Grand Island, NY). Northern blot and hybridization were carried out as described (28). The probe for aP2 was a 600-bp EST fragment. The probe for CD36 was a fragment of its cDNA. The probes for PPARγ and PPARδ are their full-length cDNA.
Glutatione S-Transferase (GST) Pull-Down Assay.
GST fusion proteins were expressed in Escherichia coli BL21 strain and affinity purified by glutathione-Sepharose 4B beads (Amersham Pharmacia). GST pull-down assays were carried out by incubating GST fusion proteins with 35S-labeled in vitro translated proteins in TEN100 buffer (29) at 4°C for 1 h.
Coimmunoprecipitation Assay.
Human 293 cells in a 10-cm plate were transfected with 10 μg of appropriate plasmids with Targefect F1 (Targeting Systems). Cells were harvested 48 h posttransfection and lysed in RIPA buffer (29). Cell lysates were incubated with either HA-agarose (Santa Cruz Biotechnology) or M2 (Flag)-agarose (Sigma) at 4°C for 2 h. The agarose-bound proteins were analyzed by Western blot analysis by using the ECL detection system (Amersham Pharmacia).
Histone Deacetylase Assay.
Histone deacetylase assays were performed as described (28) with 60,000 cpm of 3H-labeled histones. Enzyme activity was measured in a scintillation counter by the release of 3H-labeled acetate.
Gel Shift Analysis.
In vitro translated nuclear receptors were mixed with 1 μg of poly (dI:dC) in a 20-μl reaction containing 10 mM Hepes, pH 7.9/50 mM KCl/10% glycerol/200 μg/ml BSA/1 mM DTT. 32P-labeled double-stranded AOX probes were then added and incubated at room temperature for 20 min. The protein–DNA complex was resolved on a 6% native polyacrylamide gel and visualized by autoradiography.
Cell Culture and Induction of Differentiation.
The NIH 3T3-PPARα or NIH 3T3-PPARγ2 cells were infected with pLHCX-PPARδ containing recombinant virus. Cells were selected with 2 μg/ml puromycin and 200 μg/ml hygromycin. Following drug selection, NIH-PPARα/γ-PPARδ stable cell lines were cultured in DMEM containing 10% calf serum, 2 μg/ml puromycin, and 200 μg/ml hygromycin. The differentiation condition for NIH-PPARγ-PPARδ cells and Oil Red O staining were as described (5).
Results
PPARδ Associates with SMRT, SHARP, and HDACs.
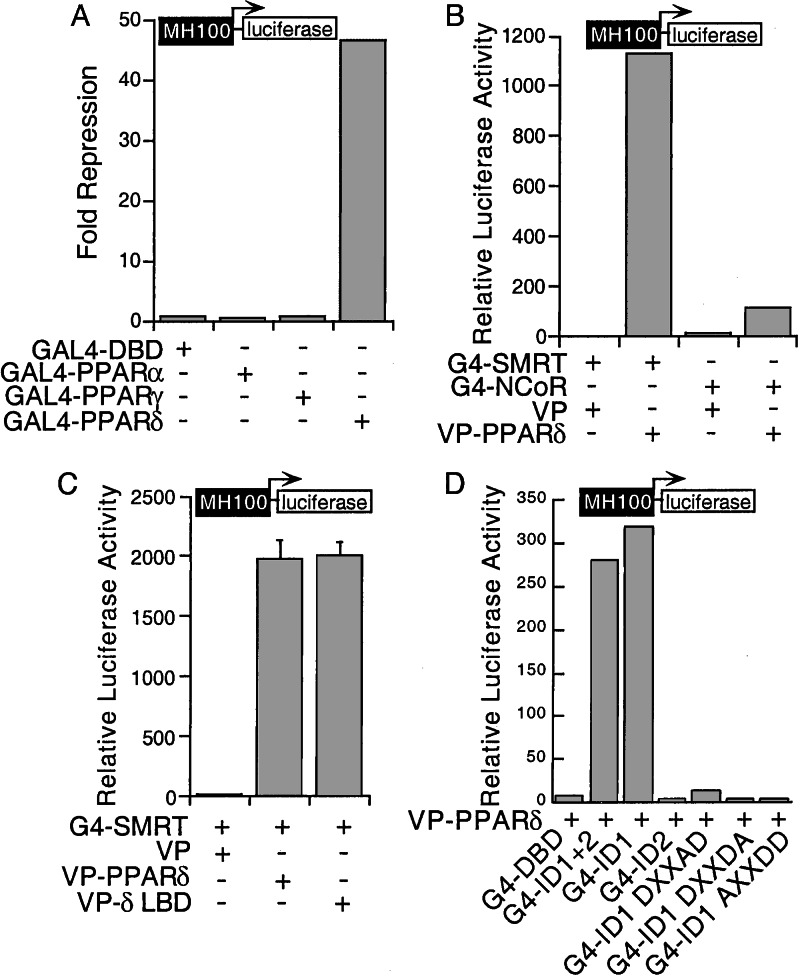
Unliganded nuclear receptors, such as RAR and TR, are able to repress basal transcription through recruitment of corepressors (30, 31). To examine the potential effect of unliganded PPARs on basal transcription, full-length PPARα, γ, and δ were fused to the GAL4 DNA binding domain (DBD) and tested for their effect on GAL4-mediated transcription by using a GAL4-responsive reporter gene MH100-luc. Among the three PPARs, PPARδ distinguished itself by displaying remarkably potent transcriptional repression activity. In contrast, PPARα and PPARγ had no significant effect on the same reporter (Fig. 1A).
Figure 1.
PPARδ represses transcription and interacts with corepressors. (A) PPARδ represses basal transcription. CV-1 cells were transfected with GAL4 DBD or GAl4-PPAR fusions. Normalized luciferase activity was 8.94 for GAL4 DBD, 12.58 for GAL4-PPARα, 9.45 for PPARγ, and 0.19 for PPARδ. Fold repression was determined relative to the GAL4 DBD activity. (B) PPARδ interacts with SMRT and NCoR. CV-1 cells were transfected with GAL4-SMRT RID (G4-SMRT) or NCoR RID (G4-NCoR) together with VP or VP-PPARδ. (C) PPARδ LBD interacts with SMRT. CV-1 cells were transfected with G4-SMRT and VP, VP-PPARδ, or VP-PPARδ LBD (VP-δLBD). (D) The ID1 of SMRT interacts with PPARδ. Transfection was performed with VP-PPARδ and GAL4 DBD or GAL4 fusion of SMRT ID1, ID2, and three ID1 mutants.
To explore the mechanisms underlying PPARδ-mediated repression, a mammalian two-hybrid assay was used to examine its potential association with the nuclear receptor corepressors SMRT and NCoR. As shown in Fig. 1B, PPARδ associated strongly with SMRT but only weakly with NCoR. The ligand binding domain (LBD) of PPARδ appears sufficient to mediate the association with SMRT (Fig. 1C).
Deletion mapping confirmed that the association with PPARδ was mediated by the SMRT RID, which is composed of ID1 and ID2 (32). Remarkably, ID1 alone is sufficient to mediate the full interaction with PPARδ, whereas ID2 is not involved (Fig. 1D). The L/I-X-X-I/V-I motifs found in the IDs of SMRT have been shown to be critical for the receptor interaction (32–34). Accordingly, mutations within the conserved hydrophobic residues of ID1 from IXXVI to DXXAD, DXXDA, or AXXDD abolish the interaction with PPARδ, supporting the importance of this motif in mediating SMRT-dependent repression by PPARδ.
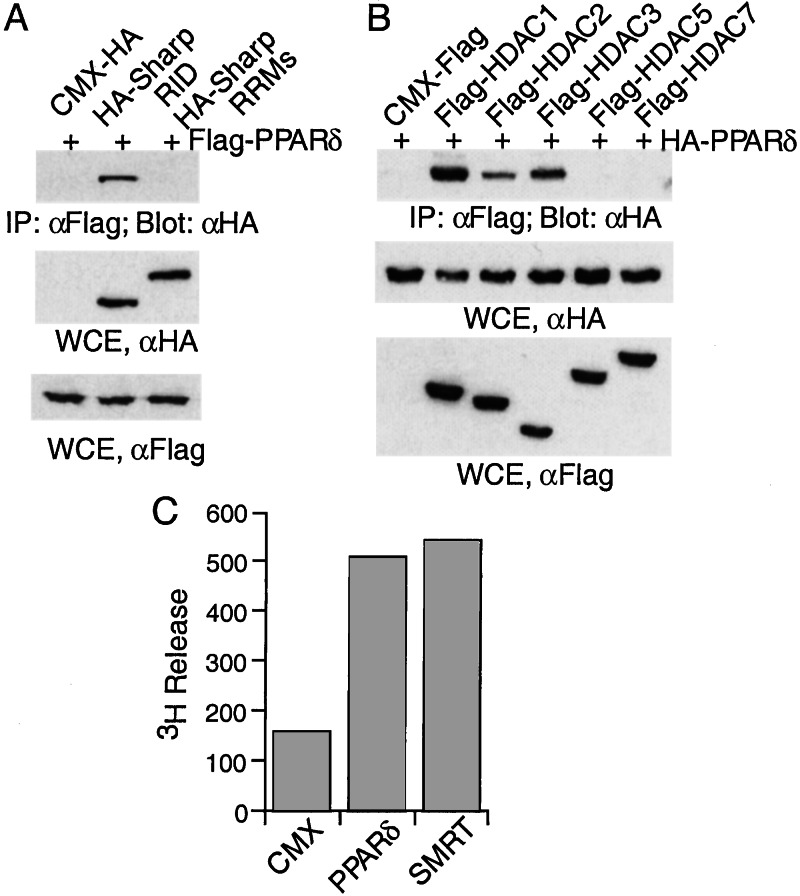
Recently, we have cloned a novel SMRT-associated protein SHARP, which functions as a potent transcriptional corepressor for nuclear receptors. SHARP is composed of a receptor interaction domain (RID) and three RNA recognition motifs. To examine the interaction of SHARP with PPARδ, HA-tagged SHARP RID was cotransfected into 293 cells with Flag-tagged PPARδ. The cell lysates were immunoprecipitated with a Flag-specific antibody, and the immunoprecipitates were analyzed by Western blot analysis with an HA-specific antibody. PPARδ is indeed associated with the SHARP RID, whereas no association was detected between PPARδ and the SHARP RNA recognition motifs as a control (Fig. 2A).
Figure 2.
PPARδ is associated with SHARP and HDACs. (A) PPARδ interacts with SHARP. Two hundred ninety-three cells were transfected with CMX-HA, HA-SHARP RID, or HA-SHARP RNA recognition motifs, along with Flag-PPARδ. The cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA antibody. (B) PPARδ interacts with class I HDACs. Two hundred ninety-three cells were transfected with CMX-Flag, or Flag tagged-HDAC1, HDAC2, HDAC3, HDAC5, and HDAC7, along with HA-PPARδ. The cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA antibody. Aliquots of the cell lysates were assayed in parallel by Western blot with HA-specific antibody or Flag-specific antibody. (C) PPARδ is associated with histone deacetylase activity. Lysates from cells expressing CMX, Flag-PPARδ, or Flag-SMRT were immunoprecipitated with anti-Flag antibody and assayed for HDAC activity.
As both SHARP and SMRT have been shown to mediate transcriptional repression through association with HDACs, we therefore examined whether PPARδ interacts with HDACs by using a similar coimmunoprecipitation assay. PPARδ was found to specifically interact with class I HDACs such as HDAC1–3 (Fig. 2B), whereas association with class II HDACs, HDAC5 and HDAC7, was not detected. This interaction with HDACs prompted us to examine whether PPARδ might be associated with histone deacetylase activity. Indeed, the immunocomplex of PPARδ displayed histone deacetylase activity as measured by the release of 3H-labeled acetate (Fig. 2C), further strengthening the link between HDAC interaction and the repression activity of PPARδ. Together, these results demonstrate that PPARδ exhibits potent transcriptional repression activity via its association with corepressors SMRT, SHARP, and class I HDACs.
PPARδ Represses PPARα- and PPARγ-Mediated Transcription.
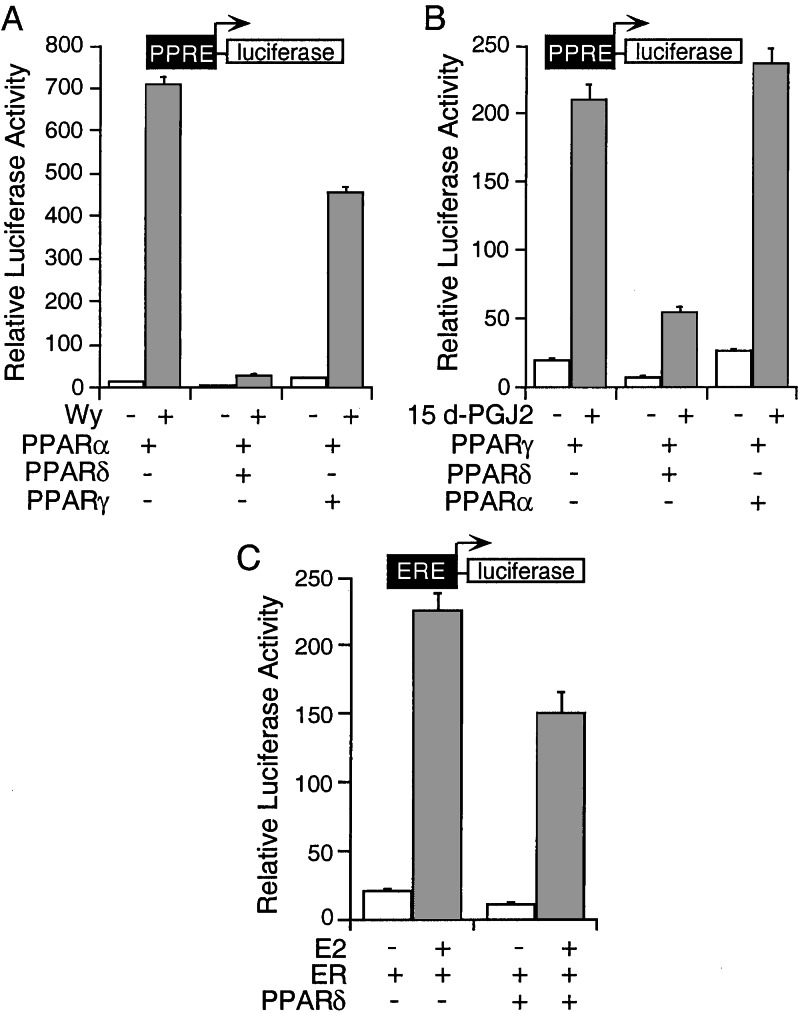
Having established the ability of PPARδ to repress basal transcription, we next examined whether it modulates transcriptional activation by PPARα or PPARγ. Interestingly, coexpression of PPARδ with PPARα leads to a dramatic repression of both basal and ligand (Wy)-induced PPARα transcriptional activity on a tk-PPRE-luc reporter gene (>20×), whereas coexpression of PPARγ has only a modest effect on PPARα activity (Fig. 3A). Likewise, PPARδ represses both basal and 15dPGJ2-induced PPARγ transcriptional activity (≈4×), whereas PPARα shows no effect on PPARγ activity (Fig. 3B), suggesting that the isoform repression depends on the intrinsic repression activity of PPARδ. In contrast to the dramatic repression of PPARα and PPARγ activity, PPARδ only slightly reduces ERE- and TRE-mediated transcription (Fig. 3C and data not shown), indicating that the repression is PPRE-dependent.
Figure 3.
PPARδ represses PPARα and PPARγ transcriptional activity. (A) PPARδ represses PPARα activity. CV-1 cells were transfected as indicated and treated with solvent or Wy for 24 h. (B) PPARδ represses PPARγ activity. CV-1 cells were transfected as indicated and treated with solvent or 15d-PGJ2 for 24 h. (C) PPARδ does not inhibit ERE/ER transcription. CV-1 cells were transfected as indicated and treated with solvent or 17β-estradiol for 24 h.
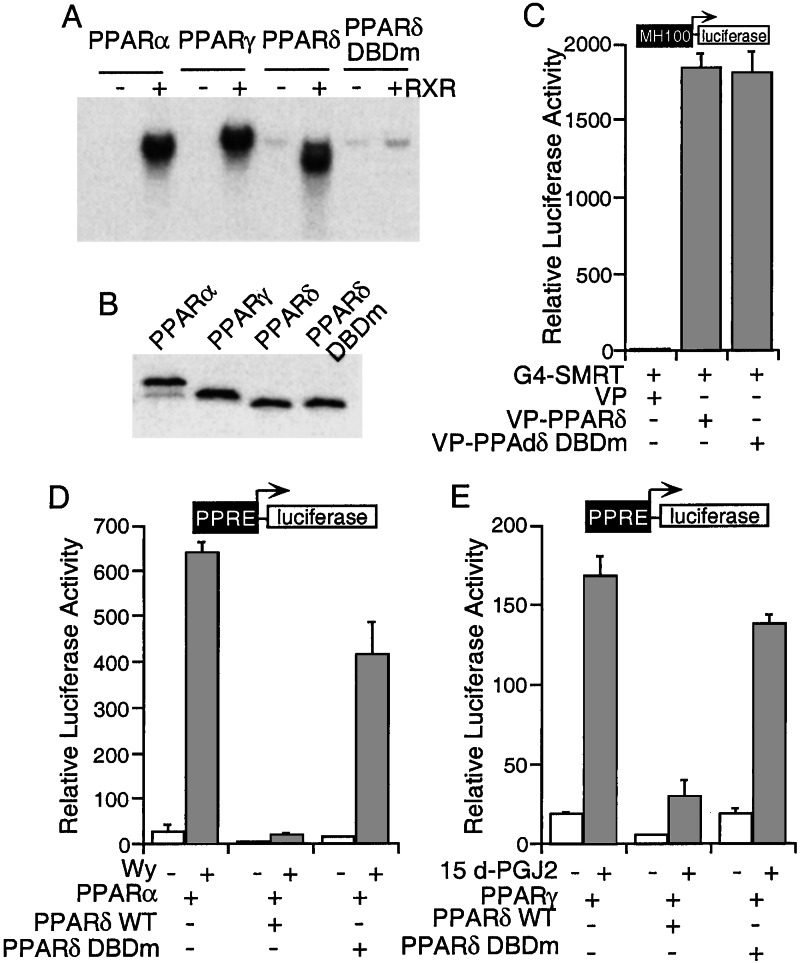
As shown in an EMSA assay, all three PPARs bind to the PPRE element in a similar manner (Fig. 4A), suggesting that competition for DNA binding may represent one of the mechanisms for PPARδ-mediated isoform repression. To further investigate this possibility, we generated a PPARδ mutant in which two conserved cysteine residues (Cys-90 and Cys-93) in the DNA binding domain were mutated to alanines. The PPARδ DBD mutant (PPARδ DBDm) loses its DNA binding activity as expected (Fig. 4 A and B). This mutant is still functional as revealed by its ability to interact with SMRT (Fig. 4C) and repress basal transcription when tethered to a GAL4 DBD (data not shown). Next, we tested whether this mutant affects PPARα and PPARγ transcriptional activity. As shown in Fig. 4 D and E, the PPARδ DBDm that does not bind PPRE loses most of its ability to repress both PPARα and PPARγ transcription, albeit a small amount of repression is retained. These results support the notion that competition for DNA binding constitutes an important part of PPARδ-mediated isoform repression.
Figure 4.
Mutation of PPARδ DNA binding domain (C90A,C93A) relieves its repression of PPARα and γ activity. (A) Mutation C90A,C93A disrupts PPARδ DNA binding activity. Gel shift analysis of PPAR isoforms and the PPARδ DNA binding domain mutant (PPARδ DBDm) are shown. (B) The expression level of PPAR isoforms and the PPARδ DBDm. One microliter of each 35S-labeled TNT product was analyzed by SDS/PAGE. (C) The PPARδ DBDm interacts with SMRT in a mammalian two-hybrid assay. (D and E) The PPARδ DBD mutant relieves repression on PPARα (D) and PPARγ (E) transcriptional activity. CV-1 cells were transfected as indicated and treated with solvent, Wy (D), or 15d-PGJ2 (E) for 24 h.
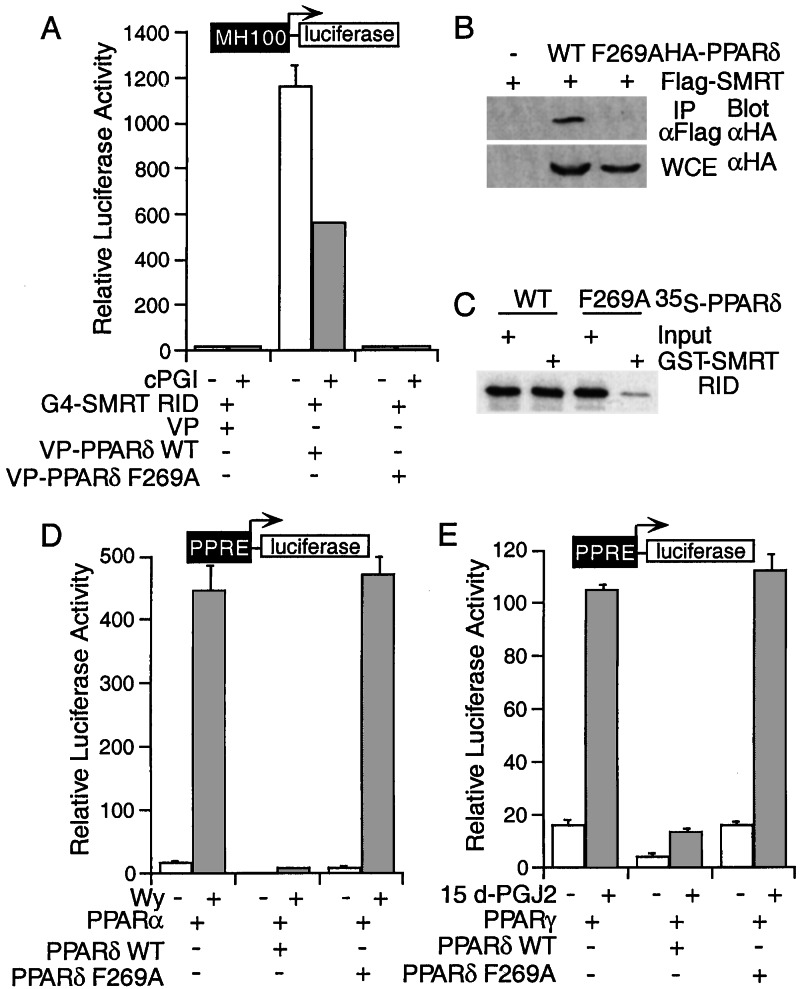
While searching for mutations that affect the PPARδ–corepressor interaction, we identified a phenylalanine (F) to alanine (A) mutation at amino acid 269 in the PPARδ LBD that abolishes its ability to interact with SMRT. This was demonstrated by a mammalian two-hybrid assay (Fig. 5A) and further confirmed by an immunoprecipitation analysis. The PPARδ F269A mutant was barely detectable in the Flag-SMRT immunocomplex, whereas wild-type PPARδ is readily coimmunoprecipitated with Flag-tagged SMRT (Fig. 5B). The inability of the F269A mutant to interact with SMRT was also demonstrated by in vitro GST-pull-down assay (Fig. 5C). To determine whether the interaction with SMRT is critical for the repression of PPARα and PPARγ activity by PPARδ, we examined whether the F269A mutant could alter PPARα and PPARγ transcriptional activation. As expected, coexpression of the PPARδ F269A mutant had little effect on PPARα and PPARγ transcriptional activity (Fig. 5 D and E), further demonstrating that a functional ligand binding domain competent to recruit the corepressor SMRT is important for the cross-repression activity of PPARδ.
Figure 5.
Mutation of PPARδ F269A abolishes its interaction with SMRT and the repression of PPAR isoform activity. (A) Mutation of Phe-269 disrupts PPARδ-SMRT interaction in mammalian two-hybrid assay. (B) The PPARδ F269A is not associated with SMRT in a coimmunoprecipitation assay. Two hundred ninety-three cells were transfected as indicated. The cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA antibody. An aliquot of the lysate input was assayed in parallel by Western blot with HA-specific antibody. (C) The PPARδ F269A does not interact with SMRT in a GST-pull-down assay. (D and E) The PPARδ F269A mutant loses its ability to repress PPARα (D) and PPARγ (E) transcriptional activity. CV-1 cells were transfected as indicated and treated with solvent, Wy (D), or 15d-PGJ2 (E) for 24 h.
Because all three PPARs activate transcription through dimerization with RXR, another possible mechanism for this isoform repression might be a competition for a limited amount of RXR. However, the repression fold by PPARδ remained the same with increased expression of RXR (data not shown), suggesting that competition for RXR is likely not a major factor involved in this repression. In summary, the results together demonstrate that binding of PPARδ to PPRE DNA elements and recruitment of corepressors underlie the mechanisms for PPARδ-mediated isoform repression.
PPARδ Represses PPARα Target Gene Expression.
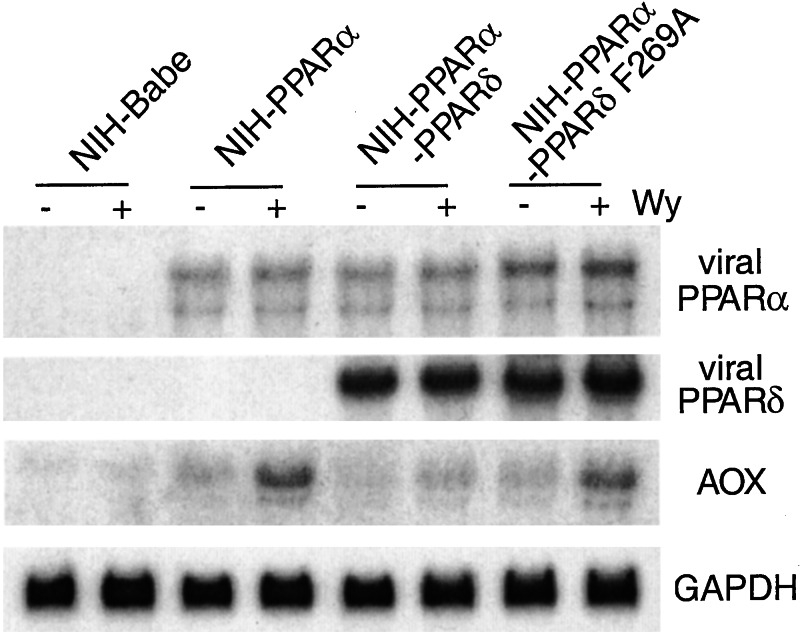
The repression of PPARα-mediated transcription by PPARδ in transient transfection assays prompted us to assess whether expression of PPARδ could repress endogenous PPARα target gene expression. An NIH-PPARα-PPARδ cell line was generated by infecting NIH 3T3 cells with a PPARα- and a PPARδ-expressing retrovirus under double drug selection. The NIH-PPARα-PPARδ cells express significant levels of both transduced genes (Fig. 6). To assess the effect of PPARδ on PPARα activity, the expression level of AOX, a known PPARα target gene, was examined in NIH-PPARα-PPARδ cells treated with solvent or the PPARα-specific ligand Wy14,643. Both basal and Wy-induced expression of AOX is repressed in the NIH-PPARα-PPARδ cells, in contrast to the induction of AOX expression in the NIH-PPARα cells. No expression of AOX is seen in 3T3 babe cells because of the lack of PPARα expression as expected. This result demonstrates that PPARδ indeed represses both basal and ligand-induced PPARα target gene expression.
Figure 6.
PPARδ represses PPARα target gene expression. Total RNA was isolated from NIH-Babe, NIH-PPARα, NIH-PPARα-PPARδ, or NIH-PPARα-PPARδ F269A cells. The RNA blot was probed with PPARα, PPARδ, AOX, or glyceraldehyde-3-phosphate dehydrogenase sequentially.
PPARδ Inhibits PPARγ Signaling.
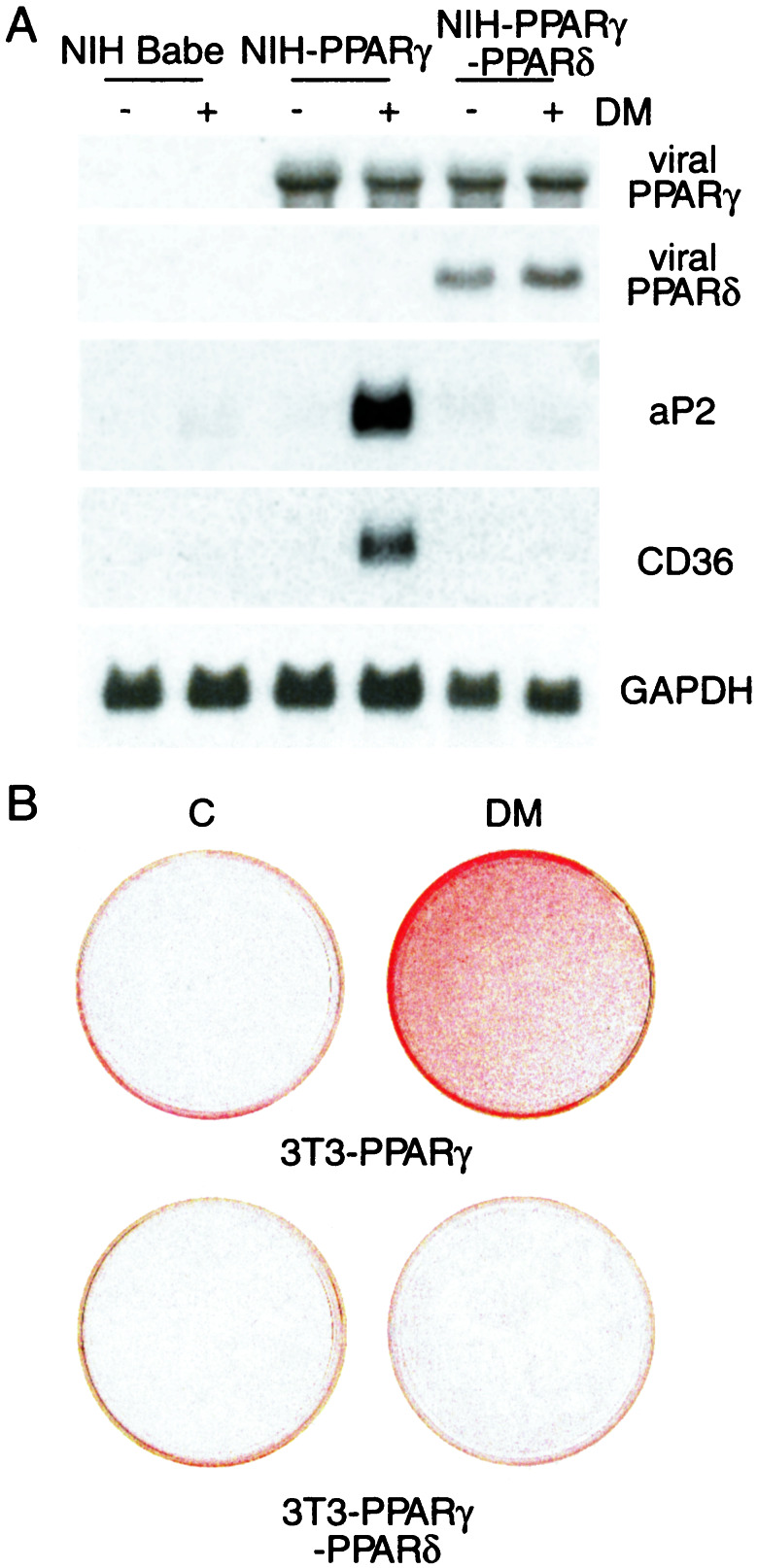
It has been shown that retrovirally expressed PPARγ2 promotes adipose differentiation of NIH 3T3 cells when exposed to differentiation media containing PPARγ ligands (5). The repression of PPARγ-mediated transcription by PPARδ prompted us to assess whether coexpression of PPARδ could repress the adipogenic function of PPARγ. An NIH-PPARγ-PPARδ cell line was generated by infecting NIH-PPARγ2 cells with a PPARδ-expressing retrovirus under double drug selection. The NIH-PPARγ-PPARδ cells express significant levels of both transduced genes (Fig. 7A).
Figure 7.
PPARδ represses PPARγ signaling. (A) PPARδ represses PPARγ target gene expression. Total RNA was isolated from NIH 3T3-Babe, NIH-PPARγ, or NIH-PPARγ-PPARδ cells. The RNA blot was probed with PPARδ, PPARγ, aP2, CD36, or glyceraldehyde-3-phosphate dehydrogenase sequentially. (B) PPARδ inhibits PPARγ-mediated adipogenesis. NIH-PPARγ or NIH-PPARγ-PPARδ cells were cultured in standard (C) or differentiation media (DM). After 7 days of differentiation, cells were fixed and stained with oil red O.
Northern blot analysis was performed to characterize the induction profile of PPARγ target genes in the NIH-PPARγ-PPARδ cells under the differentiation conditions. When cultured in differentiation media containing troglitazone, a PPARγ-specific ligand, the NIH-PPARγ2 cells activated expression of a number of typical PPARγ target genes, such as aP2 and CD36. However, these genes were dramatically inhibited in the NIH-PPARγ-PPARδ cells under the same conditions (Fig. 7A), further suggesting that PPARδ represses PPARγ-mediated transcription.
When cultured in differentiation media plus troglitazone, 20–30% of the NIH-PPARγ2 cells were differentiated into lipid-containing adipocytes as revealed by Oil-red O staining (5). However, lipid-containing cells were barely detectable in the NIH-PPARγ-PPARδ cells under the same conditions (Fig. 7B), indicating that PPARδ indeed can suppress PPARγ-mediated adipogenesis.
Discussion
We provide evidence that PPARδ is capable of acting as an intrinsic transcriptional repressor and inhibiting PPARα- and γ-activated transcription. Consistent with its repression function, PPARδ is associated with the nuclear receptor corepressors SMRT, SHARP, and HDACs. Consequently, PPARδ acquires the ability to recruit histone deacetylase activity. The SMRT interaction domain lies in the LBD of PPARδ and a point mutation in this region (F269A) abolishes the interaction. Intriguingly, this mutant also loses its ability to repress PPAR target gene expression, further strengthening the notion that recruitment of corepressors is critical for PPARδ-mediated repression.
Although all three PPARs bind to nuclear receptor corepressors in solution (23, 35, 36), only PPARδ functions as a repressor in both GAL4 system and PPRE-mediated transcription. Possibly, PPARδ is the only PPAR isoform that associates with corepressors when bound to DNA. Indeed, supershift analysis revealed that SMRT only associates with DNA-bound PPARδ and not with DNA-bound PPARα or PPARγ (data not shown). This is consistent with previous observations that neither PPARα nor PPARγ was associated with corepressors when bound to DNA (23, 36) and provides a basis for the unique repression property of PPARδ.
Because all three PPARs bind to common DNA sequences, the isoform-specific repression by PPARδ is likely because of recruitment of corepressors to PPRE. Indeed, a PPARδ mutant that loses its DNA binding activity failed to repress PPARα and PPARγ activation, suggesting that competition for DNA binding represents at least one of the mechanisms for PPARδ-mediated repression. The lack of repression on ERE or TRE reporters further supports the notion that repression by PPARδ is PPRE-dependent. Because of its association with transcriptional corepressors and histone deacetylases, binding of PPARδ to a PPRE presumably results in histone deacetylation and chromatin remodeling.
The double-transduced stable cell lines expressing both PPARδ and PPARα/γ provide a useful tool for us to assess the repression function of PPARδ and its role on endogenous target genes. Consistent with the transient transfection analysis, in cells stably expressing PPARδ, both PPARα and PPARγ target gene expression was repressed, suggesting that PPARδ indeed functions as a transcriptional repressor of PPAR target genes.
It is intriguing that PPARδ represses both PPARα and PPARγ transcriptional activity. Given the widespread expression of PPARδ and the restricted expression of PPARα and PPARγ, and the differential developmental expression pattern of the PPAR isoforms, PPARδ could act as a tissue- and developmental-specific regulator of PPAR target genes. In this way, both the absolute levels of PPARs and the ratio of PPARδ versus PPARα and PPARγ will control the relative activity of each PPAR isoform. PPARα has been shown to play an important role in lipid metabolism, whereas PPARγ is mainly involved in adipogenesis. Accordingly, PPARδ acquires the ability to modulate both lipid metabolism and adipogenesis by increasing the dynamic range of both PPARα and PPARγ activity, which in turn provides a useful strategy for therapeutic control of PPAR-related disease such as diabetes and obesity.
Acknowledgments
We thank Dr. Peter Tontonoz for providing the 3T3-PPARγ2 cell line; Dr. Hung-ying Kao for the G4-ID1 mutant constructs; Drs. Helen Cho, Michael Downes, Richard Lin, Jun Sonoda, Wen Xie, and Ruth Yu for reading the manuscript; Geraldine Cabrera, Sambath Tiep, and Henry Juguilon for technical help; and Lita Ong and Elaine Stevens for administrative assistance. Y.S. is a fellow of the Susan G. Komen Breast Cancer Foundation. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by the Howard Hughes Medical Institute.
Abbreviations
- PPAR
peroxisome proliferator-activated receptor
- PPRE
peroxisome proliferator response element
- SMRT
silencing mediator for retinoid and thyroid hormone receptors
- HDAC
histone deacetylase
- SHARP
SMRT and HDAC-associated repressor protein
- RXR
retinoid X receptor
- AOX
acyl-CoA oxidase
- GST
glutathione S-transferase
- DBD
DNA binding domain
- LBD
ligand binding domain
- RID
receptor interaction domain
- HA
hemagglutinin
References
- 1.Issemann I, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 2.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoonjans K, Staels B, Auwerx J. Biochim Biophys Acta. 1996;1302:93–109. doi: 10.1016/0005-2760(96)00066-5. [DOI] [PubMed] [Google Scholar]
- 4.Lee S S, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 6.Barak Y, Nelson M C, Ong E S, Jones Y Z, Ruiz-Lozano P, Chien K R, Koder A, Evans R M. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 7.Chawla A, Boisvert W A, Lee C H, Laffitte B A, Barak Y, Joseph S B, Liao D, Nagy L, Edwards P A, Curtiss L K, et al. Mol Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 8.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 9.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 10.Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et al. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 11.Rosen E D, Sarraf P, Troy A E, Bradwin G, Moore K, Milstone D S, Spiegelman B M, Mortensen R M. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 12.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amri E Z, Bonino F, Ailhaud G, Abumrad N A, Grimaldi P A. J Biol Chem. 1995;270:2367–2371. doi: 10.1074/jbc.270.5.2367. [DOI] [PubMed] [Google Scholar]
- 14.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 15.Oliver W R, Jr, Shenk J L, Snaith M R, Russell C S, Plunket K D, Bodkin N L, Lewis M C, Winegar D A, Sznaidman M L, Lambert M H, et al. Proc Natl Acad Sci USA. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He T C, Chan T A, Vogelstein B, Kinzler K W. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim H, Gupta R A, Ma W G, Paria B C, Moller D E, Morrow J D, DuBois R N, Trzaskos J M, Dey S K. Genes Dev. 1999;13:1561–1574. doi: 10.1101/gad.13.12.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J B, Zhang H, Rasmussen T H, Petersen R K, Flindt E N, Kristiansen K. J Biol Chem. 2001;276:3175–3182. doi: 10.1074/jbc.M005567200. [DOI] [PubMed] [Google Scholar]
- 19.Matsuura H, Adachi H, Smart R C, Xu X, Arata J, Jetten A M. Mol Cell Endocrinol. 1999;147:85–92. doi: 10.1016/s0303-7207(98)00214-7. [DOI] [PubMed] [Google Scholar]
- 20.Bastie C, Holst D, Gaillard D, Jehl-Pietri C, Grimaldi P A. J Biol Chem. 1999;274:21920–21925. doi: 10.1074/jbc.274.31.21920. [DOI] [PubMed] [Google Scholar]
- 21.Brun R P, Tontonoz P, Forman B M, Ellis R, Chen J, Evans R M, Spiegelman B M. Genes Dev. 1996;10:974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 22.Peters J M, Lee S S, Li W, Ward J M, Gavrilova O, Everett C, Reitman M L, Hudson L D, Gonzalez F J. Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamir I, Zhang J, Lazar M A. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 24.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 25.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 26.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 27.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Downes M, Xie W, Kao H Y, Ordentlich P, Tsai C C, Hon M, Evans R M. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Y, Mosser D D, Morimoto R I. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 31.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 32.Nagy L, Kao H Y, Love J D, Li C, Banayo E, Gooch J T, Krishna V, Chatterjee K, Evans R M, Schwabe J W. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X, Lazar M A. Nature (London) 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 34.Perissi V, Staszewski L M, McInerney E M, Kurokawa R, Krones A, Rose D W, Lambert M H, Milburn M V, Glass C K, Rosenfeld M G. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiRenzo J, Soderstrom M, Kurokawa R, Ogliastro M H, Ricote M, Ingrey S, Horlein A, Rosenfeld M G, Glass C K. Mol Cell Biol. 1997;17:2166–2176. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dowell P, Ishmael J E, Avram D, Peterson V J, Nevrivy D J, Leid M. J Biol Chem. 1999;274:15901–15907. doi: 10.1074/jbc.274.22.15901. [DOI] [PubMed] [Google Scholar]
- 37.Barak Y, Liao D, He W, Ong E S, Nelson M C, Olefsky J M, Boland R, Evans R M. Proc Natl Acad Sci USA. 2002;99:303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]