Abstract
The Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) functions in splicing group I introns by promoting the formation of the catalytically active RNA structure. Previous work suggested that CYT-18 recognizes a conserved tRNA-like structure of the group I intron catalytic core. Here, directed hydroxyl-radical cleavage assays show that the nucleotide-binding fold and C-terminal domains of CYT-18 interact with the expected group I intron cognates of the aminoacyl-acceptor stem and D-anticodon arms, respectively. Further, three-dimensional graphic modeling, supported by biochemical data, shows that conserved regions of group I introns can be superimposed over interacting regions of the tRNA in a Thermus thermophilus TyrRS/tRNATyr cocrystal structure. Our results support the hypothesis that CYT-18 and other aminoacyl-tRNA synthetases interact with group I introns by recognizing conserved tRNA-like structural features of the intron RNAs.
The Neurospora crassa mitochondrial (mt) tyrosyl-tRNA synthetase (TyrRS), or CYT-18 protein, functions in both tRNATyr aminoacylation and group I intron splicing (1). The splicing function reflects that CYT-18 recognizes conserved structural features of group I intron and promotes the formation of the catalytically active RNA structure (2–5). Group I introns, similar to tRNAs, have minimal sequence conservation but share a conserved three-dimensional structure consisting of two double-helical domains (6, 7). The group I intron's P4–P6 domain is formed by the coaxial stacking of secondary structure elements P5, P4, P6, and P6a, and the P3–P9 domain is formed by P3, P8, P7, and P9, with the juxtaposition of the two domains creating a cleft that contains the intron's active site. RNA-footprinting experiments with the CYT-18-dependent N. crassa mt large subunit rRNA (LSU) and ND1 introns showed that the protein interacts with the phosphodiester backbone on the side opposite the active-site cleft, with most of the potential contact sites in the P4–P6 domain and a few additional sites in the P3–P9 domain (4, 5). Biochemical and genetic studies led to a model in which CYT-18 binds first to the P4–P6 domain to promote its assembly and then makes additional contacts with the P3–P9 domain to stabilize the two domains in the correct relative orientation to form the intron's active site (4, 5, 8–10).
Remarkably, comparison of the CYT-18 binding sites in the N. crassa mt LSU and ND1 introns with that in the N. crassa mt tRNATyr by graphic modeling revealed an extended three-dimensional overlap between the tRNA and highly conserved regions of the group I intron catalytic core (5). In this overlap, the group I intron's P4–P6 stacked helices almost completely superimpose on the tRNA's D-anticodon arm stacked helices, P7 overlaps the variable arm, and P9 largely parallels the acceptor stem, with the discriminator base directly overlapping L9-1, a putative protein contact site in both RNAs. These structural similarities suggest that aminoacyl-tRNA synthetases (aaRSs) and other tRNA-binding proteins may be predisposed to function in group I intron splicing. The yeast mt leucyl-tRNA synthetase (LeuRS), a second example, acts in concert with an intron-encoded maturase to function in splicing two closely related group I introns in yeast mitochondria (11, 12).
Class I aaRSs such as TyrRSs consist of a structurally conserved N-terminal nucleotide-binding fold domain that catalyzes amino acid activation and tRNA charging followed by an aaRS-specific C-terminal RNA-binding domain, which interacts with the anticodon arm of the tRNA (13). The nucleotide-binding fold interacts with the tRNA's acceptor stem and typically contains an insertion denoted connective peptide 1, which contributes to determining acceptor-helix identity (14, 15). In TyrRSs, the nucleotide-binding fold is followed by an α-helical intermediate domain linked via a flexible hinge to a C-terminal RNA-binding domain, containing a region homologous to ribosomal protein S-4 (16). In agreement with previous models, a cocrystal structure of the Thermus thermophilus TyrRS complexed with tRNATyr showed that the tRNA binds across the surface of the two subunits of the α2 homodimer, with the tRNA's acceptor stem interacting with the nucleotide-binding fold of one subunit and the anticodon arm interacting with the C-terminal domain of the other subunit (A. Yaremchuck, I. Kriklivyi, M. Tukalo, and S.C., unpublished information). The x-ray crystal structure also confirmed that TyrRS is unique among class I aaRSs in approaching the acceptor stem from the major groove side, as is characteristic of class II aaRSs (17).
The CYT-18 protein is homologous to bacterial TyrRSs (Fig. 1) and also functions as an α2 homodimer, with each dimer binding one group I intron RNA (20). Despite these structural similarities, however, only the N. crassa mt TyrRS and that of the related fungus P. anserina function in group I intron splicing, whereas the Escherichia coli and yeast mt TyrRSs do not function in splicing (21). Thus, the two fungal mt TyrRSs must have some adaptation of the canonical structure that confers splicing activity. Mutational analysis showed that regions of CYT-18 required for splicing overlap those that function in tRNA binding (22). An apparent exception was an idiosyncratic N-terminal extension, the deletion of which specifically inhibited group I intron splicing (23), but recent findings suggest that this region functions indirectly by stabilizing the structure of another region that contacts the intron RNA directly (18). Otherwise, most mutations in different protein regions have parallel effects on splicing and TyrRS activity, and a group I intron RNA was found to be a competitive inhibitor of aminoacylation, providing direct evidence that the two RNAs compete for the same or an overlapping binding site (2, 22). Together, these findings suggest that CYT-18 forms similar but not identical complexes with its tRNATyr and group I intron RNA substrates.
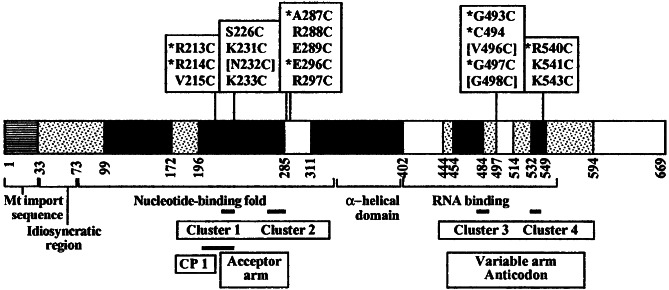
Figure 1.
Map of the CYT-18 protein showing the location of single cysteine substitutions for incorporation of (EDTA-2-aminoethyl)-2-pyridyl disulfide-Fe(III) (EPD-Fe). Black boxes indicate regions strongly conserved among bacterial TyrRSs, and stippled boxes indicate regions conserved only between the N. crassa and Podospora anserina mt TyrRSs, which function in group I intron splicing (18). CYT-18 variants having single cysteine substitutions for conjugation of EPD-Fe at different positions are shown above. Asterisks indicate variant proteins, the EPD-Fe derivatives of which gave specific cleavages in the intron RNAs, and brackets indicate proteins that were largely insoluble. The remaining proteins were soluble and active but did not give specific cleavages. The boundaries of the nucleotide-binding fold, α-helical intermediate domain, and C-terminal RNA-binding domain based on homology to other TyrRSs are shown below (18). Black bars indicate boundaries of connective peptide 1 and clusters 1–4, which contain conserved amino acid residues involved in tRNA binding in bacterial TyrRSs, with their predicted tRNATyr interaction sites indicated below (17, 19).
Here we used directed hydroxyl-radical cleavage assays to show that the nucleotide-binding fold and C-terminal domains of CYT-18 are positioned to interact with the expected cognates of the tRNA's acceptor and D-anticodon arms, respectively. Further, three-dimensional graphic modeling, supported by biochemical data, shows that key regions of the group I intron can be aligned with the tRNA in the T. thermophilus TyrRS/tRNATyr cocrystal structure. Our results support the hypothesis that CYT-18 interacts with group I introns, because the introns resemble tRNAs.
Materials and Methods
E. coli Strains and Growth Media.
E. coli strain DH5αF′ was used for cloning, and HMS174(DE3) plysS (Novagen) was used for CYT-18 protein expression. Bacteria were grown in LB medium supplemented with ampicillin (100 mg/liter) or chloramphenicol (25 mg/liter) to maintain plasmids.
Construction of CYT-18 Protein Mutants.
Wild-type CYT-18 protein was expressed from plasmid pEX560, which contains a 638-aa CYT-18 ORF lacking the mt targeting sequence cloned downstream of the phage T7 promoter in pET3a (22). CYT-18 proteins lacking one or both endogenous cysteine residues or containing single-cysteine substitutions at different positions were constructed by generating two partially overlapping PCR products that then were ligated together into the parent plasmid to replace the desired region of the CYT-18 ORF. The 5′- and 3′-PCR products each contained a unique restriction site for cloning and an overlap region, which contained both the desired mutation and a common restriction site, introduced via silent mutations, for joining the fragments. The regions generated by PCR were sequenced.
Synthesis of Intron-Containing RNAs.
In vitro transcripts containing ΔORF derivatives of the N. crassa mt LSU and ND1 introns and flanking exons were synthesized from pLSUcam and pND10, respectively. pLSUcam is a derivative of pBD5a (24) in which a T7 promoter was swapped for the T3 promoter. pLSUcam is digested with BamHI and transcribed with T7 RNA polymerase to yield a 440-nt RNA containing a 19-nt 5′ exon, the 388-nt intron, and a 33-nt 3′ exon. pND10 contains the ND1 intron and flanking exons from pND1m (25) cloned between the BamHI and EcoRI sites of pET5 (Novagen), with hammerhead and hepatitis δ virus ribozymes appended to the 5′ and 3′ ends, respectively (26). Transcription of EcoRI-digested pND10 accompanied by ribozyme cleavage yields a 210-nt RNA containing a 6-nt 5′ exon, the 197-nt ND1 intron, and a 7-nt 3′ exon. In vitro transcription and 5′ labeling with phage T4 polynucleotide kinase and gel purification of labeled RNAs were performed as described (27). Before use, RNAs were renatured by incubating in splicing buffer (100 mM KCl/5 mM MgCl2,/25 mM Tris⋅HCl, pH 7.5/10% glycerol) for 20 min at 55°C and then cooled to room temperature (4).
Protein Purification and Biochemical Assays.
Wild-type and mutant CYT-18 proteins were purified by the polyethyleneimine-precipitation procedure (20) except that DTT and EDTA were omitted from all solutions, and the protein was eluted from heparin-Sepharose columns by using a step gradient (27). Solutions were treated with Chelex-100 (Sigma) to remove trace amounts of metals. Tyrosyl-adenylation, TyrRS, and RNA splicing assays were performed as described (18). The RNA splicing assays used 10 nM CYT-18 protein and 20 nM 32P-labeled RNA substrate. Wild-type and mutant CYT-18 preparations were >90% pure and contained 80–95% active protein, as judged by tyrosyl-adenylation assays. CYT-18 concentrations refer to the homodimer.
Directed Hydroxyl-Radical Probing.
EPD-Fe-conjugated CYT-18 proteins were prepared by incubating 10 μM protein with 20 μM EPD-Fe (obtained from Robert Fox, University of Texas Medical Branch, Galveston, TX) in 100 μl of 0.5 M KCl/25 mM Tris⋅HCl, pH 7.5/10% glycerol for 60 min at room temperature and then microdialyzing using a 0.45-μm Type VS filter (Whatman) against 250 mM KCl/25 mM Tris⋅HCl, pH 7.5/10% glycerol for 60 min at room temperature to remove unincorporated EPD-Fe. The conjugated proteins were stored at 4°C. Accessibility of cysteine residues to modification was determined by using 5,5′-dithiobis(2-nitrobenzoic acid) (28). For hydroxyl-radical cleavage, 5′-labeled RNAs (50 nM) were complexed with equimolar EPD-Fe-conjugated protein in splicing buffer for 20 min at room temperature. Cleavage reactions were initiated by adding sodium ascorbate to 30 mM, incubated for 60 min at room temperature, and terminated by adding 1 μl of 10 M thiourea and 10 μg of E. coli tRNA carrier followed by extraction with phenol/chloroform/isoamyl alcohol (25:24:1). The cleavage products were analyzed in denaturing 6 or 9% polyacrylamide gels against RNA sequencing ladders generated by partial alkaline hydrolysis of the same 5′-labeled RNAs or iodine cleavage of 5′-labeled 10% α-S-ATP- or α-S-GTP-substituted RNAs.
Three-Dimensional Graphic Modeling.
Graphic modeling was done by using the INSIGHT II software package (Accelrys, San Diego). To construct the models, the previous group I intron RNA models were aligned with tRNATyr in the T. thermophilus cocrystal structure by using the overlaps between the CG pair at P4 bp-3 and D-arm bp-2, P7(5′)-3,4 and variable arm 47 and 47:1, and L9-1 and the discriminator base (5).
Results
Experimental Strategy.
The experimental strategy was to construct a series of CYT-18 protein mutants having single cysteine substitutions at different positions for conjugation of EPD-Fe (Fig. 1; ref. 29). In the presence of a reducing agent, the EPD-Fe conjugate produces short-lived hydroxyl radicals that cleave the phosphodiester backbone of nearby RNA. The conjugated protein is bound to 5′-labeled RNA, and the locations of the cleavage sites are determined by gel electrophoresis, yielding information about the proximity of specific sites in the protein and RNA. Because the Fe is tethered by a 14-Å flexible linker and generates hydroxyl radicals with a 10-Å cleavage radius, cleavages are expected within ≈24 Å of the conjugated cysteine residue (29).
Construction of a Cysteineless CYT-18 Protein.
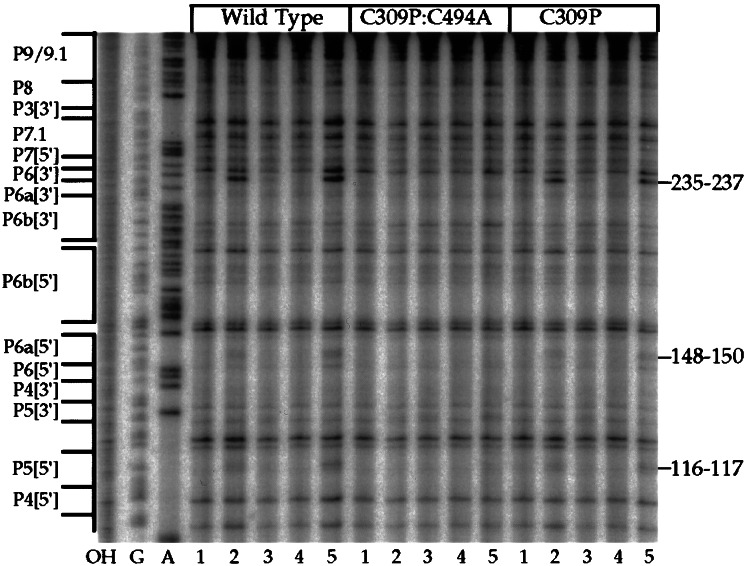
The CYT-18 protein is a favorable one for the incorporation of EPD-Fe, because it contains only two endogenous cysteine residues. To obtain a cysteineless CYT-18 protein as a starting point for further modifications, we replaced these endogenous cysteines with the corresponding amino acids from the P. anserina mt TyrRS, which also functions in group I intron splicing (21). Biochemical assays showed that the mutant proteins lacking one (C309P) or both (C309P:C494A) endogenous cysteines have wild-type tyrosyl-adenylation and TyrRS activity, and their EPD-Fe derivatives have wild-type splicing activity with both the N. crassa mt LSU and ND1 introns (data not shown).
Fig. 2 shows hydroxyl-radical cleavage assays for the wild-type and mutant proteins complexed with 5′-labeled N. crassa mt LSU intron RNA. The EPD-Fe derivatives of the wild-type and C309P proteins both gave specific cleavages that were not seen with the protein lacking the two endogenous cysteines. Group I intron structures are described by a convention in which conserved base-paired stems (P) are numbered sequentially from the 5′ end, with [5′] and [3′] indicating the 5′ and 3′ sides of the stem. The stems are separated by joining regions (J) and capped by loops (L). The specific cleavages observed in Fig. 2 map to three regions of the mt LSU intron: J6/6a-P6[3′] (235–237), P6a[5′] (148–150), and P5[5′] (116–117), with the cleavages in J6/6a-P6 by far the strongest. In control reactions, the specific cleavages were abolished in the presence of 0.5 M KCl, which dissociates CYT-18–intron RNA complexes (lanes 4), were competed by unmodified protein (lanes 3), and were not competed by nonspecific RNA (lanes 5). The finding that the same cleavages were obtained with the wild-type and C309P proteins indicates that all the cleavages result from EPD-Fe modification at C494, which is located in the C-terminal RNA-binding domain.
Figure 2.
Directed hydroxyl-radical cleavage of the N. crassa mt LSU intron with EPD-Fe derivatives of wild-type and variant CYT-18 proteins lacking one or both endogenous cysteine residues. A 5′-labeled in vitro transcript containing the N. crassa mt LSU intron was complexed with the indicated EPD-Fe-conjugated CYT-18 proteins, and cleavage sites were mapped in a denaturing 6% polyacrylamide gel. OH−, alkaline hydrolysis ladder; G and A, RNA sequencing ladders; lanes 1, EPD-Fe-modified protein incubated with RNA in the absence of ascorbate to initiate hydroxyl-radical cleavage; lanes 2, cleavage reactions after the addition of ascorbate; lanes 3, cleavage reactions in the presence of a 2-fold molar excess of unmodified wild-type CYT-18 protein; lanes 4, cleavage reactions in 0.5 M KCl; lanes 5, cleavage reactions in the presence of a 5-fold molar excess of nonspecific competitor RNA (pTRSE15/NsiI; ref. 30). Regions of the mt LSU intron are shown to the left, and the cleavages sites are indicated to the right.
Directed Hydroxyl-Radical Cleavage Assays.
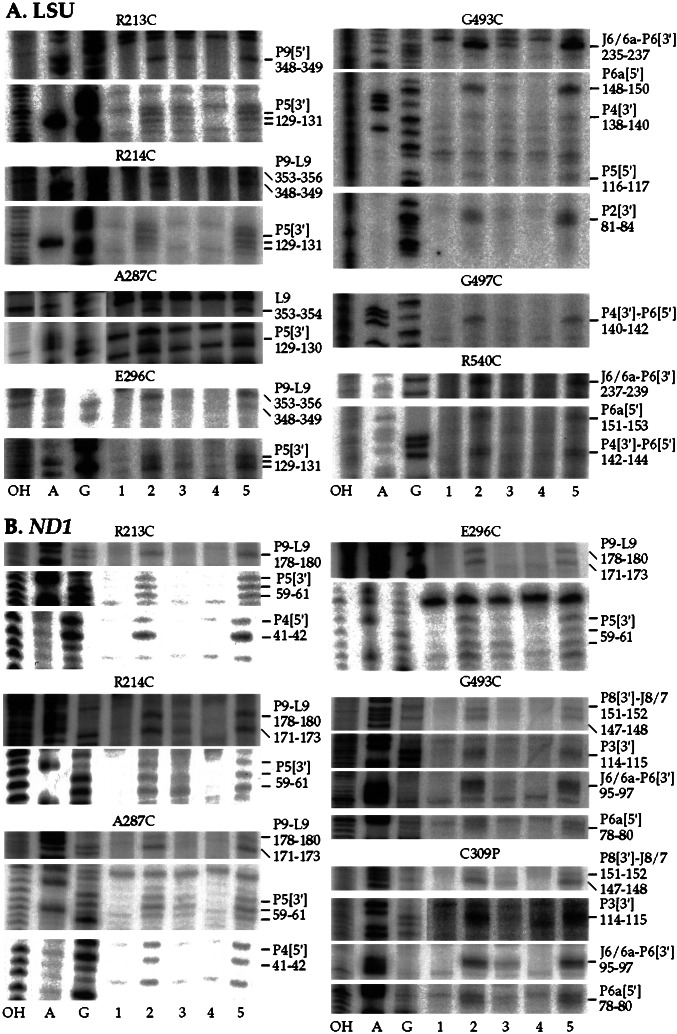
The CYT-18 protein lacking both endogenous cysteine residues was used as a starting point for constructing a series of single-cysteine substitutions in CYT-18. The cysteines were engineered in or near four regions of CYT-18 that contain clusters of conserved amino acid residues involved in tRNA binding in bacterial TyrRSs (clusters 1–4; see Fig. 1; refs. 17 and 19). Clusters 1 and 2 in the nucleotide-binding fold region interact with the acceptor stem of tRNATyr, and clusters 3 and 4 in the C-terminal domain interact with the variable and anticodon arms. The endogenous cysteine, C494, lies in cluster 3. Typically, we replaced nonconserved amino acids neighboring conserved basic amino acids identified as being involved in tRNA binding. In some cases, however, we replaced the conserved amino acid itself, the assumption being that loss of a single phosphate-backbone contact would not impair the very tight binding of CYT-18 to the intron RNA substantially (apparent Kd measured kinetically = <0.3 pM; ref. 20). In all, we constructed a total of 20 single-cysteine substitutions of which 17 gave active protein and 8 gave specific cleavages (see Fig. 1). All the active proteins have wild-type tyrosyl-adenylation activity, and all but one retain 80–100% wild-type splicing activity with both the LSU and ND1 introns after conjugation of EPD-Fe (data not shown). The single exception, EPD-Fe R540C, had 20–50% wild-type splicing activity with both introns. Representative hydroxyl-radical cleavage assays are shown in Fig. 3, and the locations of the cleavages in the intron RNAs are summarized in Fig. 4.
Figure 3.
Directed hydroxyl-radical probing of the N. crassa mt LSU and ND1 introns with variant CYT-18 proteins containing EPD-Fe conjugated to different positions. A and B show cleavage reactions with the mt LSU and ND1 introns, respectively. The reactions and gel lanes are as described for Fig. 2. Mutant proteins are indicated above, and cleavage sites are indicated to the right.
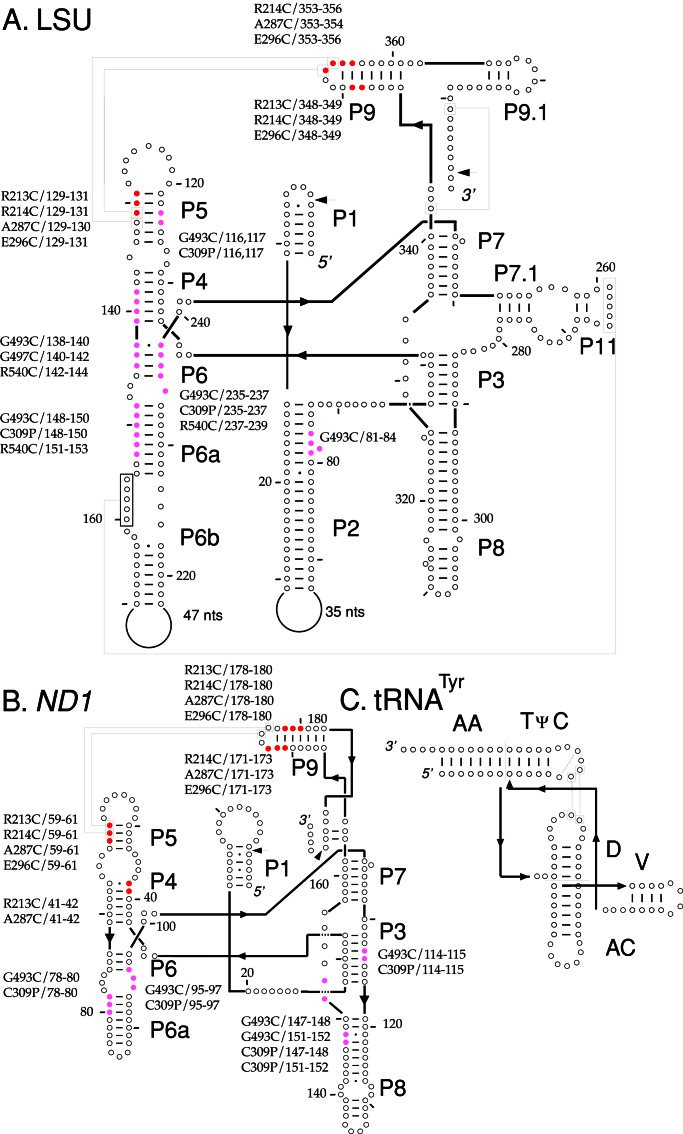
Figure 4.
Summary of directed hydroxyl-radical cleavages in the N. crassa mt LSU and ND1 introns. (A and B) The predicted secondary structures of the 388-nt mt LSU and 197-nt ND1 introns used in this study. Cleavages from EPD-Fe conjugated to different positions in the N- and C-terminal domains of CYT-18 are indicated in red and magenta, respectively. Tertiary interactions are indicated by boxed positions connected by lines, and splice sites are indicated by arrows. (C) The secondary structure of N. crassa mt tRNATyr drawn in a similar orientation for comparison.
Clusters 1 and 2 are expected to interact with the group I intron structure P9/L9, which is the cognate of the tRNA's acceptor stem. Four cluster 1 and 2 mutants gave specific EPD-Fe cleavages: R213C and R214C near the end of connective peptide 1 in cluster 1, A287C in cluster 2, and E296C just downstream of cluster 2 in a 16-aa CYT-18-specific insertion. All four proteins cleaved both introns in P9/L9, as expected, and more weakly in P5, an adjacent structure that is a docking site for the L9 tetraloop. EPD-Fe R213C and A287C also cleaved a neighboring region of P4 in the ND1 intron. Other cluster 1 and 2 mutants, EPD-Fe V215C, S226C, K231C, K233C, R288C, E289C, and R297C, gave no specific cleavages even though their cysteine residues were accessible for modification, and their EPD-Fe derivatives were fully active for splicing both the LSU and ND1 intron. Although EPD-Fe cleavages in the N. crassa mt RNATyr could not be mapped precisely because of the relatively weak binding and small size of the RNA, additional experiments showed that all four cluster 1 and 2 mutants that cleaved the intron RNAs also cleaved the tRNA, and those that failed to cleave the intron RNAs also failed to cleave the tRNA (data not shown). Thus, all the tested cluster 1 and 2 positions are disposed similarly to cleave or not cleave both the intron and tRNA substrates, and all four mutants that cleaved the intron did so in P9/L9, the cognate of the acceptor stem, and to a lesser extent in the adjacent structure P5.
Clusters 3 and 4 in the C-terminal RNA-binding domain are expected to interact with the group I intron's P4–P6 stacked helices, the cognate of the tRNA's D-anticodon arm stacked helices, and with P7, the cognate of the variable arm. In addition to the endogenous cysteine C494 in the C309P mutant (Fig. 2), three other cluster 3 and 4 mutants gave specific cleavages. EPD-Fe–G493C, which is adjacent to the endogenous C494 in cluster 3, cleaved the LSU intron at the same locations as C309P in J6/6a-P6[3′], P6a[5′], and P5[5′] as well as additional sites in P4[3′] and P2[3′]. Both C309P and G493C cleaved the ND1 intron in P8[3′]-J8/7, P3[3′], J6/6a-P6[3′], and P6a[5′]. The remaining two mutants, G497C in cluster 3 and R540C, the only active variant in cluster 4, cleaved the LSU intron but not the ND1 intron, likely reflecting more extensive interaction of the C-terminal domain with the LSU intron (see Discussion). Both mutants cleaved the LSU intron around the junction of P4[3′] and P6[5′], and EPD-Fe R540C also cleaved the LSU intron in P6a[5′] and P6[3′]. Thus, as expected, the cluster 3 and 4 mutants cleaved primarily in the P4–P6 domain, the cognate of the D-anticodon arm stacked helices of tRNATyr, with a few, mostly weaker cleavages in other regions.
Structure Modeling.
The positional information from the EPD-Fe cleavage experiments enabled us to orient the protein on the RNA for structure modeling. Fig. 5 shows models of the complexes between CYT-18 and the mt LSU and ND1 introns, respectively, based on the cocrystal structure of the T. thermophilus TyrRS–tRNATyr complex. The homologous regions of the T. thermophilus TyrRS and CYT-18 are expected to have similar three-dimensional structures, although CYT-18 has several small insertions and a longer C-terminal domain, which are not represented in the models. The models of the complexes were constructed without reference to the biochemical data by simply aligning the group I intron RNA with the tRNATyr in the T. thermophilus cocrystal structure using the previously described three-dimensional overlaps between group I introns and tRNAs (ref. 5; see Materials and Methods). Auspiciously, the orientation of the tRNA's long variable arm in the x-ray crystal structure more closely parallels P7 than it did in the previous models, which were based on tRNASer.
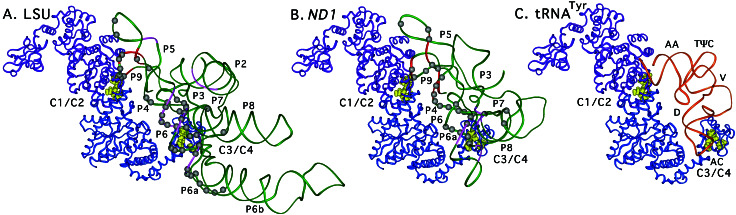
Figure 5.
Models of the N. crassa mt LSU and ND1 introns complexed with the T. thermophilus TyrRS. The models are shown in A and B, with a drawing of the T. thermophilus TyrRS/tRNATyr cocrystal structure shown in C for comparison (A. Yaremchuk, I. Kriklivyi, M. Tukalo, and S.C., unpublished data). The two TyrRS subunits are blue and purple, and the RNA phosphodiester backbone is green for the introns and orange for the tRNA. EPD-Fe-conjugated amino acid residues that gave specific cleavages in the intron RNAs are highlighted in yellow, and the cleaved regions of the phosphodiester backbone are shown in red and magenta for N- and C-terminal cleavages, respectively. E296C, which is in a CYT-18-specific insertion, is represented by yellow highlighting of the closest amino acid residue, S290. Gray balls indicate putative protein-protection sites on the phosphodiester backbone detected in iodine-footprinting experiments (4, 5). Protection sites likely caused by protein-induced RNA structural changes in P3[5′] in both introns and J4/5, P7[3′], P7.1, and P6b in the LSU intron are not shown. C1/C2, clusters 1 and 2; C3/C4, clusters 3 and 4.
The models show that the group I introns, similar to tRNATyr, bind across the surface of the two subunits of the homodimer, interacting with the N-terminal domain of one subunit and the C-terminal domain of the other. In agreement with the predicted group I intron–tRNATyr structural homologies, the C-terminal domain interacts primarily with the group I introns' P4–P6 stacked helices, the cognate of the tRNA's D-anticodon arm stacked helices, and the nucleotide-binding fold interacts with P9/L9, the cognate of the tRNA's acceptor stem, as well as P5, which interacts with L9. As noted previously, the discriminator base and L9-1 are putative contact sites in both RNA substrates and overlap in the alignments. The group I introns of course lack the 3′ CCA, which extends into the TyrRS active site. The models and biochemical data suggest that there also could be interactions between the C-terminal domain and P7, the cognate of the long variable arm, and between the α-helical intermediate domain and J6/6a, the cognate of the anticodon loop. The extended C-terminal domain of CYT-18 also may interact with P2, P3, and P8, which do not have cognates in the tRNA.
Although the models are based on the group I intron–tRNATyr alignment, there nevertheless is good agreement with biochemical data. Thus, 35/40 EPD-Fe cleavages (Fig. 5, red and magenta regions) fall within 33 Å of the derivatized amino acid (Fig. 5, yellow), and all strong cleavages fall within 28 Å. The only cleavages that cannot be explained by minor adjustments in the model are the weak cleavages from two cluster 3 positions (G493C and the endogenous C494 in C309P) in P5[5′] in the LSU intron. These cleavages are not observed in the more compact ND1 intron and could reflect the conformational mobility of the flexibly hinged C-terminal domain in either subunit of the homodimer. If not, they require either a conformational change in the intron RNA or that the interacting C-terminal domain in the model be shifted up from the position in the cocrystal structure. At least some shift in the C-terminal domain seems necessary in any case to accommodate the bulkier intron RNA in this region and to account for the lack of observed cleavages in P7, the cognate of the variable arm.
Also in agreement with the models, most of the phosphate-backbone protections mapped previously in iodine-footprinting experiments (gray spheres) are in regions protected by the protein in the model. Importantly, the phosphate-backbone protections around the junction of the P4–P6 stacked helices basically are superimposable with those at the junction of the D-anticodon arm stacked helices, with the protein facing the minor groove of P4 and the major groove of P6, as for the cognate structures in the tRNA (5, 27). These findings strongly suggest a very similar disposition of the α-helical and C-terminal domains with respect to both the intron and tRNA substrates in this region. Except for two P5 protections in the ND1 intron, the only unaccounted for phosphate protections in the models are in P8 in both introns and in P6a in the LSU intron and can be explained by additional interactions with CYT-18's larger C-terminal domain and/or some shift in the position of the C-terminal domain in the complex. We note that the biochemical data show more extensive C-terminal contacts with the LSU intron than with the ND1 intron, which is consistent with the observation that the C-terminal domain plays a greater role in splicing the LSU intron than the ND1 intron (18).
Discussion
Our results support the hypothesis that CYT-18 functions in splicing by recognizing highly conserved structural features of group I introns that resemble those in tRNAs. The hydroxyl-radical cleavage data identify putative interaction sites between the nucleotide-binding fold and the P9/P5 junction region and between the C-terminal domain and the P4–P6 stacked helices. With these putative interacting regions fixed, the structural models based on the T. thermophilus TyrRS/tRNATyr cocrystal structure indicate a decidedly tRNA-like recognition of the group I intron catalytic core, with the acceptor-arm cognate (P9/L9) interacting with the nucleotide-binding fold of one subunit, and the D-anticodon arm cognate (P4–P6) interacting with the C-terminal domain of the other subunit. The relatively large interface between the protein and intron RNA affords the potential for multiple contacts, as in tRNA binding, and explains how CYT-18 can suppress structural mutations throughout the group I intron catalytic core (3, 9, 10). The ability of CYT-18 to function in splicing many different group I introns likely reflects that the tRNA-like structural features with which it interacts are highly conserved in group I introns, presumably because they are required for the catalytic activity of the intron RNAs. Within this framework of conserved interactions, additional intron-specific and non-tRNA-like interactions also may contribute to CYT-18-dependent splicing.
With respect to splicing mechanism, our previous biochemical analysis suggested that CYT-18 interacts first with the P4–P6 domain to promote its assembly and then makes secondary contacts with the P3–P9 domain to bring the two major domains into the correct relative orientation to form the intron's active site (4, 5). Regarding the first step, the models show that the region of the P4–P6 domain around the junction of the P4–P6 stacked helices interacts with a protein scaffold formed by the C-terminal and α-helical domains, which evolved to recognize the D-anticodon arm of the tRNA. The use of this protein scaffold readily explains how CYT-18 can induce assembly of the P4–P6 domain as well as compensate for a variety of structural mutations that impair base stacking, base pairing, or base triple interactions around the junction of the P4–P6 stacked helices (9, 10). For the second step, the models suggest that CYT-18 could help establish the correct relative orientation of the two intron domains by stabilizing or substituting for critical interdomain interactions including the P5/L9 tetraloop/receptor interaction, J3/4 and J6/7 interdomain contacts, and the minor groove interdigitation between P3 and J6/J6a (6, 7). Consistent with this hypothesis, genetic assays with the phage T4 td intron show that CYT-18 can suppress mutations at all positions in J3/4 and J6/7 (9) as well as mutations that disrupt the P5-L9 interaction (X. Chen and A.M.L., unpublished data).
The models also explain previous findings that the N- and C-terminal domains of CYT-18 contribute differently to splicing different group I introns and in particular that deletion of the C-terminal RNA-binding domain abolished splicing and stable binding of the LSU intron but left substantial splicing activity with the ND1 intron and other group I introns (18). The models show that this truncated protein still can interact extensively with the ND1 intron via the nucleotide-binding fold and α-helical domains, with the latter responsible for many of the phosphate-backbone protections in P4-J6/6a. The greater interaction of the LSU intron with the C-terminal domain is reflected by the additional EPD-Fe cleavages from positions in clusters 3 and 4 and by the larger number of phosphate-backbone interactions extending farther down the P6 helix into P6a. We note that even though the C-terminal truncation did not abolish splicing of the ND1 intron, it increased koff by ≈5-fold, suggesting that the C-terminal domain makes a noncritical contribution to binding (18). Further, group I introns that ordinarily do not depend on the C-terminal domain readily acquire such dependence by mutations in certain regions of the catalytic core (X. Chen and A.M.L., unpublished data). Together, these findings suggest that all group I introns form fundamentally similar complexes involving the N- and C-terminal domains of CYT-18 and that dependence on the C-terminal domain reflects primarily the nature of the specific structural deficiencies that impair self-splicing in different introns.
We suggested previously that group I introns initially were self-splicing and became dependent on cellular proteins as a result of mutations that impaired self-splicing (31, 32). Although it is not clear yet to what extent other aaRSs function in group I intron splicing, it is possible that TyrRS has structural features that make it uniquely suited for such adaptation. Such features may include the ability to recognize a tRNA with a long variable arm in a specific orientation that matches P7 and the flexibly hinged C-terminal domain, which may facilitate the binding of some non-tRNA substrates. The latter may include not only group I introns but also the 3′-terminal tRNA-like structures of RNA viruses (33). In addition, CYT-18 contains several idiosyncratic insertions and an extended C-terminal domain, which may contribute to splicing via unique interactions with group I introns.
The yeast mt LeuRS, which functions in splicing the closely related mt group I introns bI4 and aI4, also may recognize tRNA-like structural features of these introns (11, 12). Further, the ability to splice these introns is inherent in other mt or bacterial LeuRSs, which can fully complement a yeast null mutant lacking the mt LeuRS (12). Unlike CYT-18, however, which functions in splicing many different group I introns, the LeuRS functions in splicing only the two closely related yeast mt DNA introns and does so by acting in concert with a maturase encoded by one of the introns. Thus, the LeuRS must recognize either idiosyncratic features of bI4 and aI4 or a subset of the conserved tRNA-like features in a manner not sufficient to promote splicing in the absence of the maturase.
Finally, the alignment with the crystal structure further supports the conclusion that group I introns have structural similarities to tRNAs. As discussed previously, these structural similarities may be coincidental, or they may reflect an evolutionary relationship between group I introns and tRNAs. One possibility is that group I introns evolved in the RNA world and gave rise to tRNAs during the evolution of protein synthesis (2). Another possibility is that a tRNA or tRNA-like structure evolved into a group I intron by acquiring catalytic activity, which enabled it to propagate as a mobile element by reverse splicing into other RNA sites (5). Both possibilities are remarkable.
Acknowledgments
We thank Dr. Robert Fox (University of Texas Medical Branch, Galveston, TX) for EPD-Fe, Stacy Crain and Anthony Kaiser for technical assistance, and Georg Mohr and Paul Paukstelis for comments on the manuscript. This work was supported by National Institutes of Health Grant GM37951 and a collaborative grant from the Human Frontiers Science Program.
Abbreviations
- mt
mitochondrial
- TyrRS
tyrosyl-tRNA synthetase
- LSU
large subunit rRNA
- aaRS
aminoacyl-tRNA synthetase
- LeuRS
leucyl-tRNA synthetase
- EPD-Fe
(EDTA-2-aminoethyl)-2-pyridyl disulfide-Fe(III)
- P
base-paired stem
- J
joining region
- L
loop region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Akins R A, Lambowitz A M. Cell. 1987;50:331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- 2.Guo Q, Lambowitz A M. Genes Dev. 1992;6:1357–1372. doi: 10.1101/gad.6.8.1357. [DOI] [PubMed] [Google Scholar]
- 3.Mohr G, Zhang A, Gianelos J A, Belfort M, Lambowitz A M. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- 4.Caprara M G, Mohr G, Lambowitz A M. J Mol Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 5.Caprara M G, Lehnart V, Lambowitz A M, Westhof E. Cell. 1996;87:1135–1145. doi: 10.1016/s0092-8674(00)81807-3. [DOI] [PubMed] [Google Scholar]
- 6.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 7.Golden B L, Gooding A R, Podell E R, Cech T R. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 8.Saldanha R, Ellington A, Lambowitz A M. J Mol Biol. 1996;261:23–42. doi: 10.1006/jmbi.1996.0439. [DOI] [PubMed] [Google Scholar]
- 9.Myers C A, Wallweber G J, Rennard R, Kemel Y, Caprara M G, Mohr G, Lambowitz A M. J Mol Biol. 1996;262:87–104. doi: 10.1006/jmbi.1996.0501. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Gutell R R, Lambowitz A M. J Mol Biol. 2000;301:265–283. doi: 10.1006/jmbi.2000.3963. [DOI] [PubMed] [Google Scholar]
- 11.Herbert C J, Labouesse M, Dujardin G, Slonimski P P. EMBO J. 1988;7:473–483. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houman F, Rho S B, Zhang J, Shen X, Wang C-C, Schimmel P, Martinis S A. Proc Natl Acad Sci USA. 2000;97:13743–13748. doi: 10.1073/pnas.240465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnez J G, Moras D. Trends Biochem Sci. 1997;258:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 14.Rould M A, Perona J J, Söll D, Steitz T A. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 15.Wakasugi K, Quinee C L, Tao N, Schimmel P. EMBO J. 1998;17:297–305. doi: 10.1093/emboj/17.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillard C, Bedouelle H. Biochemistry. 2001;40:7192–7199. doi: 10.1021/bi010208c. [DOI] [PubMed] [Google Scholar]
- 17.Bedouelle H. Biochimie. 1990;72:589–598. doi: 10.1016/0300-9084(90)90122-w. [DOI] [PubMed] [Google Scholar]
- 18.Mohr G, Rennard R, Cherniack A D, Stryker J, Lambowitz A M. J Mol Biol. 2001;307:75–92. doi: 10.1006/jmbi.2000.4460. [DOI] [PubMed] [Google Scholar]
- 19.Nair S, Ribas de Pouplana L, Houman F, Avruch A, Shen X, Schimmel P. J Mol Biol. 1997;269:1–9. doi: 10.1006/jmbi.1997.1025. [DOI] [PubMed] [Google Scholar]
- 20.Saldanha R J, Patel S S, Surendran R, Lee J C, Lambowitz A M. Biochemistry. 1995;34:1275–1287. doi: 10.1021/bi00004a022. [DOI] [PubMed] [Google Scholar]
- 21.Kämper U, Kück U, Cherniack A D, Lambowitz A M. Mol Cell Biol. 1992;12:499–511. doi: 10.1128/mcb.12.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kittle J D, Jr, Mohr G, Gianelos J A, Wang H, Lambowitz A M. Genes Dev. 1991;5:1009–1021. doi: 10.1101/gad.5.6.1009. [DOI] [PubMed] [Google Scholar]
- 23.Cherniack A D, Garriga G, Kittle J D, Jr, Akins R A, Lambowitz A M. Cell. 1990;62:745–755. doi: 10.1016/0092-8674(90)90119-y. [DOI] [PubMed] [Google Scholar]
- 24.Guo Q, Akins R A, Garriga G, Lambowitz A M. J Biol Chem. 1991;266:1809–1819. [PubMed] [Google Scholar]
- 25.Wallweber G J, Mohr S, Rennard R, Caprara M G, Lambowitz A M. RNA. 1997;3:114–131. [PMC free article] [PubMed] [Google Scholar]
- 26.Ferré-D'Amaré A R, Doudna J A. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caprara M G, Myers C A, Lambowitz A M. J Mol Biol. 2001;308:165–190. doi: 10.1006/jmbi.2001.4581. [DOI] [PubMed] [Google Scholar]
- 28.Ellman G L. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 29.Hall K B, Fox R O. Methods. 1999;18:78–84. doi: 10.1006/meth.1999.0759. [DOI] [PubMed] [Google Scholar]
- 30.Chen B, Lambowitz A M. J Mol Biol. 1997;271:311–332. doi: 10.1006/jmbi.1997.1185. [DOI] [PubMed] [Google Scholar]
- 31.Lambowitz A M, Perlman P S. Trends Biochem Sci. 1990;15:440–444. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- 32.Lambowitz A M, Caprara M G, Zimmerly S, Perlman P S. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 451–485. [Google Scholar]
- 33.Mans M W, Pleij C W A, Bosch L. Eur J Biochem. 1991;201:303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]