Abstract
Mot1 is an essential yeast Snf2/Swi2-related ATPase that exerts both positive and negative effects on gene expression. In vitro, Mot1 can disrupt TATA-binding protein–DNA complexes in an ATP-dependent reaction. This activity can explain Mot1-mediated transcriptional repression, but how Mot1 activates transcription is unknown. We demonstrate that, remarkably, Mot1 is localized in vivo to promoters for both Mot1-repressed and Mot1-activated genes. Moreover, Mot1 ATPase activity is required for both activation and repression of gene activity. These findings suggest a novel function for the Mot1 ATPase at activated genes, perhaps involving ATP-driven reorganization of the preinitiation complex. Mot1 regulates the expression of ≈3% of yeast genes in cells grown in rich medium. Most of these genes are repressed by Mot1, consistent with Mot1's ATP-dependent TATA-binding protein–DNA dissociating activity. Additionally, ≈77% of the Mot1-repressed genes are involved in the diauxic shift, stress response, mating, or sporulation. The gene sets controlled by NC2 and Srb10 are strongly correlated with the Mot1-controlled set, suggesting that these factors cooperate in transcriptional control on a global scale.
A critical step in RNA polymerase II transcription involves the association of TATA-binding protein (TBP) with promoter DNA (1). TBP binding to promoters is often a rate-limiting step in transcription, and TBP or TBP-containing complexes are well-characterized targets of transcriptional regulators (2, 3). Mot1 was isolated genetically as a repressor of weak promoters in vivo (4–6). Consistent with its behavior as a transcriptional repressor, Mot1 can disrupt TBP–DNA complexes in vitro in an ATP-dependent reaction (7).
While Mot1's ATP-dependent TBP–DNA dissociating activity correlates well with its genetic isolation as a repressor and its function as a repressor of transcription in vitro (7), Mot1 also activates the expression of some genes in vivo (6, 8, 9). The transcriptional activation function of Mot1 may be caused by its ability to redistribute limiting TBP among different TBP binding sites in genomic DNA (8–10). On the other hand, Mot1 may have another activity that allows it to function as a transcriptional activator at specific promoters, depending on the promoter sequence, chromatin context, or association of TBP with other factors. Mot1's ATPase activity is essential for yeast cell viability (7), but a role for the ATPase activity in activation or repression of specific genes has not been established. Here we define an in vivo role for Mot1 as largely a repressor of transcription, although a subset of genes require Mot1 for their full expression. Remarkably, Mot1 is associated with promoters that are Mot1-activated, and transcriptional activation by Mot1 requires Mot1's ATPase activity. These results suggest an additional activity for this essential, conserved regulator of TBP function.
Materials and Methods
Yeast strains, growth conditions, RNA isolation, Northern blotting, chromatin immunoprecipitation, microarray hybridizations, and data analysis are described in detail with Figs. 5–7 and Tables 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org.
Yeast Strains and Growth Conditions.
In brief, for microarray analysis and Northern blots, wild-type and temperature-sensitive congenic mot1–14 and mot1–42 yeast strains were grown in rich medium (yeast extract/peptone/dextrose) at 30°C to an OD600 ≈1.0, then shifted to 35oC for 45 min, and cells were harvested. RNA was also obtained from WCS132 (tsm1, ref. 11), JR374 (taf145–869, ref. 12; Joe Reese, personal communication), SHY258 (toa2–3; ref. 13), JS306 (BNA1+) and JS663 (bna1Δ, ref. 14) cells grown in the same way. Experiments (see Fig. 3) were performed by using haploid yeast cells with a deletion of the chromosomal copy of MOT1, mot1–42 carried on pRS313 (15) and MOT1+, mot1-K1303A (both pRS315-derived) or pRS315 (15) plasmid vector alone. The cells were grown in synthetic media to maintain plasmid selection and heat-shocked, and RNA was harvested and analyzed as in Fig. 1. Alternatively, mot1Δ cells carrying mot1–42 on pRS313 and GAL1-driven mot1-K1303A or wild-type MOT1 on pRS315 were grown in synthetic media without leucine or histidine and containing 2% galactose plus 0.5% sucrose. Steady-state message levels were analyzed from these strains in the same way.
Figure 3.
Mot1 ATPase is required for activation and repression of transcription. (A) Northern blot analysis of the indicated messages is shown for indicated yeast strains. Each strain has a deletion of the chromosomal copy of MOT1 and one or two plasmid-borne alleles of MOT1 (shown above the lanes) under control of the MOT1 promoter. For lanes 1–4, cells were grown and harvested as in Fig. 1. Lane 1, RNA from a strain carrying mot1–42 and a plasmid vector without a MOT1 gene. Lane 2, RNA from a strain carrying mot1–42 as well as wild-type MOT1 on a second plasmid. Lanes 3 and 4, two independent RNA samples from a yeast strain carrying mot1–42 and a mot1 allele encoding K1303A that destroys Mot1 ATPase activity. RNA in lanes 5–8 was derived from mot1–42 cells containing plasmid vector, wild-type MOT1, or mot1(K1303A) under control of the GAL1 promoter. Cells were grown as in A (lanes 1–4) except that the medium contained 2% galactose and 0.5% sucrose to partially induce the GAL1 promoter. Lanes 7 and 8 represent two independent RNA preparations from the same strain. (B) Gel mobility-shift analysis using radiolabeled TATA-containing DNA, recombinant TBP, and purified Mot1 or Mot1-K1303A as indicated. Reactions were incubated at 35°C before loading on the gel. The position of Mot1-TBP-DNA complex is represented by 3°. (C) Western blot analysis of yeast whole-cell extracts was performed to detect epitope-tagged Mot1 as described (42). Lanes 1–5 contained 75 μg and lane 6 contained 37 μg whole-cell extract protein. In lanes 1–5 the indicated genes were expressed under control of the MOT1 promoter; in lane 6 mot1-K1303A was expressed under GAL1 control. The arrow indicates the position of Mot1.
Figure 1.
Mot1, TFIIA, and TAF dependence of selected genes. The indicated strains were grown at 30°C to an OD600 of ≈1.0, then heat-shocked for 45 min at 35°C, and total RNA was harvested. Twenty micrograms of RNA from each strain was then resolved by electrophoresis, transferred to nitrocellulose, and probed with radiolabeled DNAs encoding the indicated ORFs. (A) Mot1-repressed and activated genes identified by microarray analysis. (B) Mot1-independent genes. (C) Comparison of message levels in wild-type, mot1–14, and bna1Δ strains. Inactivation of Mot1 led to variable effects on transcript levels for some genes. For instance, BNA1 message levels were reduced 4-fold in A but only ≈2-fold in C. These effects fall roughly within the range defined by microarray analysis (3.7 ± 1.4-fold).
RNA Isolation and Northern Blots.
Total yeast RNA was isolated by using a hot acid phenol extraction protocol (16). Poly(A)+ RNA was prepared from total RNA by using a Qiagen (Chatsworth, CA) Oligotex Midi Kit according to the instructions supplied by the manufacturer. For Northern blots, 5–20 μg total RNA was separated by electrophoresis in formaldehyde agarose gels, transferred to nitrocellulose, and probed with random-primed DNA probes. Bands were detected by autoradiography, and band intensities were quantitated by using a PhosphorImager.
Microarray Hybridization Experiments.
cDNA microarray chips containing 6,024 yeast ORFs were prepared as described (17, 18). Poly(A)+ RNA (2–4 μg) was labeled with Cy3 and Cy5-conjugated dUTP (Amersham Pharmacia) by reverse transcription reaction and hybridized to the chips (17). cDNA chips were scanned by using an Axon Scanner (Axon Instruments, Foster City, CA), and images were analyzed by using microarray suite software (Scanalytics, Fairfax, VA). The relative fluorescence intensity was measured for each labeled RNA, and a ratio of the values for the intensity of each fluor bound to each probe was calculated. The amount of autofluorescence generated in the Cy3 channel was measured, and a minimum intensity cut-off was set just above this value. The distribution of the ratio of all of the genes was calculated, and intensity ratio values that differed from the median with a confidence interval of more than 95.0% (19) were scored as significant changes. The same RNA was labeled and hybridized in three independent reactions. The data for each array was normalized by using the mean of all of the targets on the array, and the coefficient of variance for each hybridization was less than 0.3. A database tool, maps (20), was used to compile the overall list of consistent, significantly changed genes across the multiple hybridizations. The complete Mot1 microarray data set is available at http://dir.niehs.nih.gov/microarray/datasets.
Analysis of Microarray Data.
The alpha-factor time series (21) and diauxic shift expression pattern (17) were examined for each Mot1-regulated gene. To obtain quantitative estimates of the degree of overlap between the Mot1 microarray data and other data sets, comparisons were performed in Microsoft excel between the Mot1 data set and published data sets available from http://cmgm.stanford.edu/pbrown/explore/index.html (diauxic shift and Tup1) and http://web.wi.mit.edu/young/pub/expressionanalysis.html (TAF145, Tsm1, Gcn5, Bur6, and Spt3). The Mot1 data set was also compared with the collection of environmental and stress response data sets reported in ref. 22 (http://genome-www.stanford.edu/yeast_stress/). The expression tree (Fig. 2C) was constructed by using cluster and treeview programs (http://rana.lbl.gov/EisenSoftware.htm).
Figure 2.
Summary of microarray analysis results. Most Mot1-regulated genes fall into known regulatory groups. (A) Venn diagram of Mot1-regulated genes, showing fractions of Mot1-regulated genes induced by diauxic shift, mating pheromone, or stress. (B) Venn diagrams showing fractions of genome activated by diauxic shift or repressed by Mot1, Srb10, and the Bur6 component of NC2, and relative sizes of overlaps. (C) Clustering of effects of Mot1, NC2, or Taf145 mutations with representative stress responses: Mot1 and NC2 have a similar relationship with stress response gene overexpression. (D) Pie chart showing fractions of Mot1-regulated genes that are also induced by the diauxic shift, or repressed by Srb10, or NC2. Categories are not inclusive, i.e., the yellow piece does not represent all genes repressed by Mot1 and induced in the diauxic shift, but only the fraction of such genes that are not additionally regulated by NC2 or Srb10, so far as is known.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation was performed as described (23) with minor modifications using strains containing tandem affinity purification-tagged (24) or untagged Mot1. The normalized PCR signal obtained by using primers for Mot1-controlled promoters was roughly 3-fold greater than the signal obtained by using primers for other promoters or ORFs (except the ACT1 ORF), and the analysis was performed at least three times with two independently prepared batches of chromatin.
Gel Mobility-Shift Analysis.
Gel mobility-shift assays were performed by using purified recombinant Mot1 or mot1(K1303A), yeast TBP core domain, and a 110-bp fragment of the adenovirus major late promoter as described (25) with the exception that the reactions were incubated for 20 min at 35°C before loading on the gel.
Results
Global Analysis of Genes Controlled by Mot1.
The set of genes whose expression is affected by mutation of MOT1 was determined by cohybridization of glass slide microarrays containing probes for 6,024 yeast ORFs. Three independent hybridizations were performed by using RNA from wild-type MOT1+ cells and cells carrying a temperature-sensitive allele of MOT1, mot1–14. The distribution of the ratios of the intensities of the two RNAs was used to identify those genes whose expression was significantly different from the mean with 99% confidence (19). In rich medium, Mot1 regulates the expression of ≈182 genes, or about 3% of yeast genes. Among this set, expression of 176 genes was induced by mutation of MOT1, suggesting that Mot1 normally functions to repress transcription of these genes. Expression of six genes was decreased in the mot1 strain, indicating that Mot1 also activates transcription of some genes.
The MOT1 dependence of 15 genes was confirmed by Northern blot analysis (Fig. 1 A and B). Message levels were compared in two strains, each encoding a different mutation in MOT1. All genes examined by Northern analysis were similarly affected in the two mot1 strains, suggesting that very similar or identical sets of genes are affected by these two mot1 alleles. Mutation of MOT1 resulted in derepression of INO1 transcription 5- to 6-fold, derepression of THI5 about 4.5-fold, and derepression of HSP26 transcription 3.8- to 4.5-fold, for example. In contrast, transcription of BNA1, URA1, and YDR539W was decreased 2.5- to 4-fold, and RPS5, RPL5, RAD16, SAN1, and ACT1 message levels were affected less than 30% by mutation of MOT1. Quantitation of all message levels is reported in Tables 1 and 2 (see Materials and Methods). The apparent sizes of some messages are different in the MOT1+ and mot1 strains (e.g., HSP26, Fig. 1A), perhaps reflecting a role for Mot1 in regulating start site selection in vivo (8).
Correlations with Other Transcriptional Regulators and the Diauxic Shift.
RNA from congenic strains encoding mutations in TAF145, Tsm1, and the small subunit of TFIIA (Toa2) also was analyzed by Northern blot (Fig. 1 A and B). Transcription of BNA1, URA1, and YDR539W each was observed to depend on both TAF145 and Mot1. In contrast, Mot1 dependence was not observed to correlate simply with dependence on Tsm1. Likewise, genes repressed by Mot1 displayed differing degrees of dependence on Toa2, the small subunit of TFIIA (Fig. 1). Three genes activated by Mot1 also depended on TFIIA, suggesting that these factors function as coactivators of some genes. Investigation of the Mot1-controlled genes by manually recording annotated features available from public databases indicated that ≈77% of the Mot1-repressed genes are either induced during the diauxic shift, as part of the general response to environmental stress, and/or during mating and sporulation (Fig. 2A). For instance, among the genes analyzed in Fig. 1, AGA1 and SGA1 are alpha-factor induced; INO1, GND2, YDR533C, YGR043C, and YHR087W are all induced by alpha-factor and during the diauxic shift, TSL1 is a stress-induced gene also activated during the diauxic shift, and HSP26 is activated by stress, alpha-factor, and during the diauxic shift (17, 21). Interestingly, BNA1 encodes a component of the de novo synthesis pathway for nicotinic acid; altered levels of nicotinic acid could influence the levels of SIR2's NAD-dependent histone deacetylase activity (26, 27) and thereby activate expression of genes involved in mating and sporulation (27). To test this idea, message levels in the mot1–14 strain were compared with those in wild-type BNA1 and bna1Δ strains. As shown in Fig. 1C, deletion of BNA1 does not lead to induction of alpha factor-induced genes, indicating that derepression of these genes in the mot1 strains is not an indirect effect of decreased BNA1 expression. There are also no known or predicted transcription factors in the Mot1-controlled set with activities or expression profiles linking them to the expression of other genes in the set (not shown). Thus, the effects of Mot1 on transcription reported here appear to be direct, a conclusion supported by chromatin immunoprecipitation results presented below. Other indirect effects of Mot1 on genes not examined in detail cannot be ruled out, however.
To see whether the set of Mot1-regulated genes was correlated with gene sets regulated by some other factor, the complete Mot1 data set was aligned with other available data sets, and genes induced or repressed more than 2-fold were counted. (The 2-fold cut-off criterion was required to compare the Mot1 data with data sets from other investigators; this criterion defines a somewhat larger number of Mot1-controlled genes than the number of genes defined by the 99% confidence criterion.) A strong correlation was observed between Mot1 repression and diauxic shift activation (17) (correlation coefficient 0.4; Fig. 2). Of 258 Mot1-repressed genes, 109 are induced more than 2-fold in the diauxic shift program. The cyclin kinase Srb10 has also been reported to repress diauxic shift-induced genes (28). There are 78 genes repressed more than 2-fold by Mot1 and Srb10. Of these, 56 are diauxic shift-induced (Fig. 2B; correlation coefficient 0.2).
The Mot1 microarray data set was compared with the data set for the TFIID component TAF145 (28). This comparison revealed that of 21 genes that are activated by Mot1 (by the 2-fold cut-off criterion), 15 are TAF145-dependent (correlation coefficient 0.3). Comparison of the data sets for Mot1 and the TBP-binding factor NC2 (Bur6 component; ref. 29) was also informative: 155 of 247 Mot1-repressed genes are also NC2-repressed, and 10 genes are activated by both Mot1 and NC2 (correlation coefficient 0.3, Fig. 2). Nine of these 10 genes are TAF145-dependent. Furthermore, 51 of the 56 genes that are diauxic shift-induced, Mot1-repressed, and Srb10-repressed are also NC2-repressed. NC2 represses over 85% of all genes repressed by both Mot1 and Srb10 (Fig. 2D). NC2 represses over 83% of all Mot1-repressed genes induced during the diauxic shift (Fig. 2 B and D). There was no correlation, however, between the genes controlled by Mot1 and those controlled by the TFIID component, Tsm1, or between the gene sets controlled by Mot1 and the other transcriptional regulators Spt3, Gcn5, Srb5, Rpd3, Tup1, and Snf2/Swi2 (not shown). The sets of Mot1-, NC2-, and TAF145-dependent genes were compared with expression patterns during a large variety of stress responses (22) by using the programs cluster and treeview (Fig. 2C; see Materials and Methods). This analysis substantiated the strong and specific correlation between the Mot1 and NC2 data sets and also demonstrated a correlation between genes controlled by Mot1 and those affected by overexpression of the stress response transcription factors Msn2 or Msn4.
Mot1 ATPase Activity Is Required for Both Activation and Repression of Transcription.
Mot1 ATPase activity is required for its essential function in vivo (7), but the ATPase requirement for regulation of specific genes has not been tested. Strains were constructed with a deletion of the chromosomal copy of MOT1, and plasmid-borne copies of the conditional mot1–42 allele and the mot1(K1303A) allele that encodes catalytically dead Mot1 (7). After heat shock to inactivate mot1–42, RNA was prepared, and RNA levels from selected genes were analyzed by Northern blot. As shown in Fig. 3A (lanes 1–4), INO1 transcription was induced by mutation of MOT1, whereas BNA1, URA1, and YDR539W transcription was barely detectable in the strain without a functional allele of MOT1 (Fig. 3A, compare lanes 2 and 1). Transcription of the ACT1 gene is unaffected by mutation of MOT1 as expected. Message levels for INO1, BNA1, URA1, and YDR539W in the mot1–42 strain were unaffected by introduction of the mot1 (K1303A) allele (Fig. 3A, compare lanes 3 and 4 to lane 1), indicating that ATPase-defective mot1 cannot support either Mot1-dependent activation or repression of transcription. As shown in Fig. 3B, recombinant mot1(K1303A) formed ternary complexes with TBP and DNA in vitro at 35°C as well as wild-type Mot1, indicating that the TBP–DNA binding activity of mot1(K1303A) is not temperature-sensitive. The levels of mot1(K1303A) in whole-cell extracts were equivalent to wild-type Mot1 levels as judged by immunoblotting (Fig. 3C, lanes 1 and 2), although in some extracts we observed somewhat less of the K1303A mutant protein compared with wild-type Mot1 (e.g., Fig. 3C, lane 4 versus 5). To ensure that the failure of mot1(K1303A) to support Mot1 function in transcription in vivo was not caused by reduced levels of the mutant protein, we expressed mot1-K1303A under control of the GAL1 promoter. When cells were grown in media containing 2% galactose plus 0.5% sucrose, levels of mot1-K1303A were 2- to 4-fold higher than the level of wild-type Mot1 (Fig. 3C, lanes 4 and 6). RNA from cells containing the GAL1-driven mot1(K1303A) gene was analyzed by Northern blotting as above. As shown in Fig. 3A (lanes 5–8), message levels for BNA1 and HSP26 were equivalent in mot1–42 cells whether or not mot1-K1303A was overexpressed compared with wild-type Mot1. Thus, Mot1 control of these genes requires functional ATPase activity.
Mot1 Localization on Chromatin in Vivo.
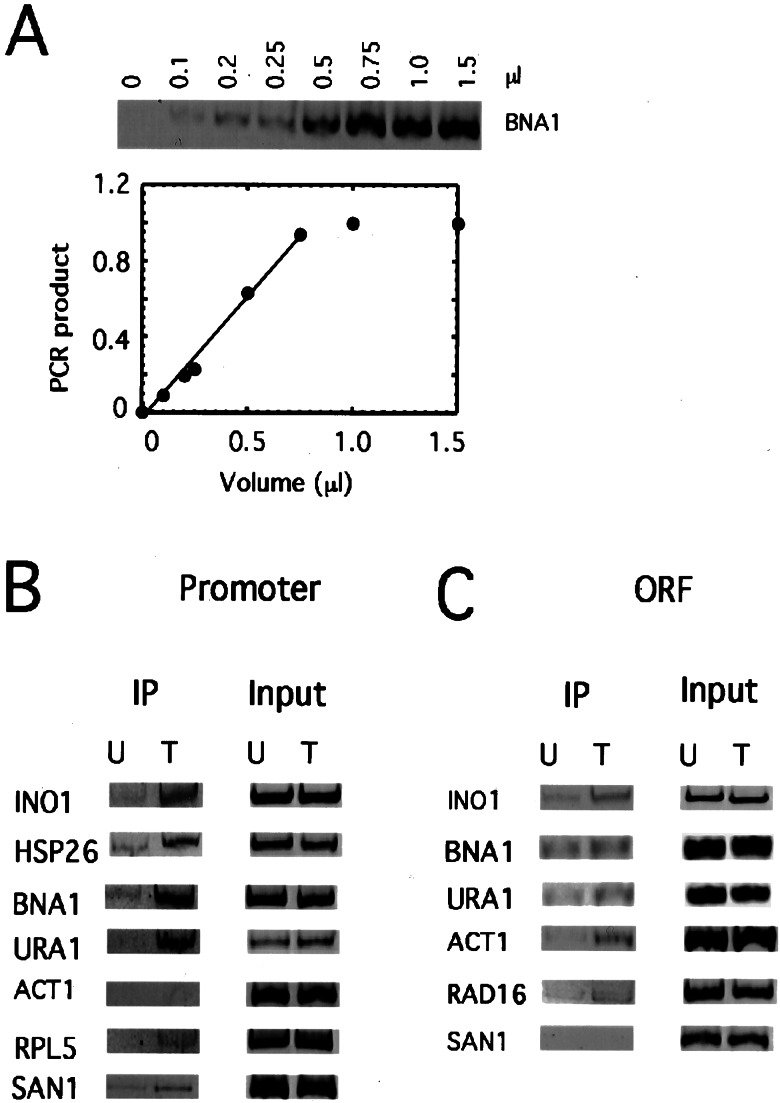
Cells harboring epitope-tagged Mot1 expressed from the MOT1 promoter in the normal chromosomal locus, or the untagged control strain, were grown in rich medium, and formaldehyde crosslinked protein-DNA complexes were obtained by immunoprecipitation. PCR was used to determine whether specific promoters or ORFs were associated with Mot1 in the immunoprecipitates. The INO1 and HSP26 genes are repressed by Mot1, and Mot1 was found to be associated with promoters for both of these genes (Fig. 4). This is consistent with a role for Mot1 as a repressor by acting locally to remove TBP and prevent preinitiation complex assembly. Surprisingly, Mot1 was also associated with both the URA1 and BNA1 promoters, two genes whose expression is activated by Mot1. As Mot1 ATPase activity is required for activation of these genes, we suggest that Mot1 has a novel, ATP-dependent activity at activated promoters that facilitates some rate-limiting step in transcription initiation. By comparison, the association of Mot1 with the ACT1, RPL5, and SAN1 promoters, as well as the INO1 ORF, was barely detectable and there was no detectable association between Mot1 and the BNA1, URA1, RAD16, or SAN1 ORFs (Fig. 4). Of the six ORFs examined, Mot1 was clearly associated only with the ACT1 ORF, suggesting that fortuitous TBP binding does occur in vivo and may be monitored by Mot1. Thus, Mot1 is specifically associated with the promoters of Mot1-activated and repressed genes in vivo.
Figure 4.
Mot1 is localized to Mot1-activated and repressed promoters in vivo. (A) Dependence of PCR product yield on amount of input chromatin template. PCRs were performed with primers for the BNA1 promoter using increasing amounts of chromatin solution. The amounts of PCR product obtained versus input chromatin substrate are plotted below. All PCRs shown in B and C were performed by using a volume of chromatin solution yielding a product within the linear response range. (B) PCR was performed by using primers for the indicated promoters and chromatin solution derived from cells with epitope-tagged Mot1 (T) or untagged Mot1 (U). Chromatin template from reactions in the first two lanes (IP) was obtained from the immunoprecipitated material whereas the reactions in the second pair of lanes (Input) were performed by using purified chromatin that had not been subjected to immunoprecipitation. (C) PCR was performed as in B by using primers that amplify portions of the indicated ORFs.
Discussion
These results reveal a function for Mot1 in cells growing in rich medium as a repressor of genes induced during the diauxic shift, mating, sporulation, and as part of the response to environmental stress (Figs. 2A and 5). Mot1 also activates some genes, however, and the activation and repression functions of Mot1 both require Mot1 ATPase activity. Localization of Mot1 to both Mot1-activated and Mot1-repressed promoters in vivo suggests that these effects of Mot1 are direct, but that the consequences of having Mot1 associated with promoter DNA depend on the promoter context in which Mot1 is found. Below we suggest how Mot1 might exert these complex effects on gene expression.
The strong codependence of genes on Mot1 and NC2 (Fig. 7) indicates that Mot1 and NC2 cooperate on a global scale. This relationship fits well with previous data linking the functions of these two factors (6, 30, 31). Most Mot1-activated promoters that we identified are also TAF145-dependent. Mot1 and NC2 are localized in vivo to promoters that they activate (Fig. 4; ref. 29), so the biochemical activity of Mot1 at Mot1-activated promoters may be a consequence of unique preinitiation complexes or subcomplexes containing TAF145, NC2, and Mot1 formed on these promoters. Srb10 and Mot1 repress an overlapping set of genes (Fig. 6), but it is unclear whether there is a mechanistic relationship between these factors. Srb10 kinase can inhibit transcription by means of C-terminal domain phosphorylation (32), and by interaction with the transcriptional repressor Tup1 (33). Thus, there may be features of core promoters repressed by both Mot1 and Srb10 that dictate their dependence on these two factors. However, Srb10 also affects transcription indirectly by regulating the cellular localization of the Msn2 stress response transcription factor (34).
Microarray analysis indicates that a selected subset of genes is repressed by Mot1, despite the fact that TBP is ubiquitously required for transcription. A direct effect of Mot1 as a TBP–DNA dissociating enzyme may be antagonized by other TBP-associated factors that prevent Mot1's interaction with TBP. On the basis of in vitro studies, TFIIA was proposed as one such antagonist of Mot1 function (35), but the dependence of genes on Mot1 and TFIIA (Fig. 1) indicate that TFIIA function cannot be simply described as a Mot1 antagonist. Likewise, TAF145 interacts with a surface on TBP that overlaps the Mot1 binding site (refs. 30 and 36; see below), but TAF145 and Mot1 are both required for activation of a subset of genes. Thus, TBP-associated factors that regulate Mot1 function are likely to be complex. Mot1 localization may also be dictated by a particular chromatin structure at Mot1-controlled promoters or Mot1 recruitment may be mediated by means of interactions with site-specific DNA binding proteins (37).
Mot1 might exert a kinetic effect on transcriptional control by, for instance, disassembling the transcription complex from an activated promoter when the activation signal is removed and transcription is returned to the nonactivated “ground state.” We found, however, that the rates of shut-off of the GAL1 and MET15 promoters were unaffected by a mutation in MOT1, indicating that Mot1 does not function in this way, at least at these two promoters (data not shown).
Our results suggest that Mot1 can act directly at activated promoters, and the ATP dependence of Mot1's transcriptional activation function suggests that Mot1 uses a novel enzymatic activity at activated promoters. One possibility is that Mot1 uses ATP hydrolysis at activated promoters to induce a conformational change in a TBP-containing complex that facilitates the loading or activity of preinitiation complex components at the promoter. Such a conformational change might be related to the conformational change induced in TFIID by TFIIA in the TFIIA–TFIID–DNA complex (38, 39). Recruitment of TBP to the promoter of polymerase II genes has been implicated as a rate-limiting step in transcription (reviewed in ref. 3) and the interplay between TFIID, NC2, Mot1, TFIIA, and other TBP-associated transcription factors that stabilize or destabilize TBP binding to DNA is not well understood. TAF145 interacts with the DNA binding surface of TBP (40) as well as the convex surface of TBP in a region that overlaps with the interaction surface for TFIIA and Mot1 (35, 36, 41). Surprisingly, although biochemical and structural studies indicate that Mot1, TAF145, and TFIIA bind competitively to TBP, BNA1, URA1, and YDR539W transcription each depend on all three of these factors for full expression. These factors may function in a pathway for assembly of a complex competent for transcription initiation. Sequential interaction with overlapping surfaces on TBP would then explain the requirement for three proteins that apparently cannot co-occupy the promoter. Another possibility is that Mot1, TAF145, and/or TFIIA can co-occupy the promoters that they activate, but that Mot1 ATPase activity is used to “remodel” the preinitiation complex, perhaps by moving TAFs with respect to each other and/or TBP to facilitate a rate-limiting conformational change in transcriptional activation.
Supplementary Material
Acknowledgments
We are grateful to Lee Bennett and Pierre Bushel for their help in compiling the microarray data and Kate Johnson, Don Cox, and Pat Hurban for optimization/generation of yeast ORFs. We are also grateful to Marisol Santisteban, Mitch Smith, David Gross, Patrick Grant, Jeff Smith, Joe Reese, and Steve Hahn for yeast strains and reagents. Thanks to Joe Reese and Jeff Smith for discussion, Patrick Grant, Dan Engel, Sam Wilson and members of the Auble lab for critically reading the manuscript, and Jeff Smith for providing information before publication. This work was supported by National Institutes of Health Grant GM55763 to D.T.A.
Abbreviation
- TBP
TATA-binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lemon B, Tjian R. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 2.Maldonado E, Hampsey M, Reinberg D. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 3.Lee T I, Young R A. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 4.Davis J L, Kunisawa R, Thorner J. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piatti S, Tazzi R, Pizzagalli A, Plevani P, Lucchini G. Chromosoma. 1992;102:S107–S113. doi: 10.1007/BF02451793. [DOI] [PubMed] [Google Scholar]
- 6.Prelich G. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 8.Madison J M, Winston F. Mol Cell Biol. 1996;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collart M A. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muldrow T A, Campbell A M, Weil P A, Auble D T. Mol Cell Biol. 1999;19:2835–2845. doi: 10.1128/mcb.19.4.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee T I, Causton H C, Holstege F C P, Shen W-C, Hannett N, Jennings E G, Winston F, Green M R, Young R A. Nature (London) 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 12.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 13.Kang J J, Auble D T, Ranish J A, Hahn S. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandmeier, J. J., Celic, I., Boeke, J. D. & Smith, J. S. (2002) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 15.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt M E, Brown T A, Trumpower B L. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 18.Hauser N C, Vingron M, Scheideler M, Krems B, Hellmuth K, Entian K-D, Hoheisel J D. Yeast. 1998;14:1209–1221. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1209::AID-YEA311>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Dougherty E R, Bittner M L. J Biomed Optics. 1997;2:364–374. doi: 10.1117/12.281504. [DOI] [PubMed] [Google Scholar]
- 20.Bushel P, Hamadeh H, Bennett L, Sieber S, Martin K, Nuwaysir E F, Johnson K, Reynolds K, Paules R, Afshari C A. Bioinformatics. 2001;17:564–565. doi: 10.1093/bioinformatics/17.6.564. [DOI] [PubMed] [Google Scholar]
- 21.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y, Dai H, Walker W L, Hughes T R, et al. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 22.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo M H, Allis C D. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 24.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 25.Darst R P, Wang D, Auble D T. EMBO J. 2001;20:2028–2040. doi: 10.1093/emboj/20.8.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucharczyk R, Zagulski M, Rytka J, Herbert C J. FEBS Lett. 1998;424:127–130. doi: 10.1016/s0014-5793(98)00153-7. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein B E, Tong J K, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 29.Geisberg J V, Holstege F C, Young R A, Struhl K. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cang Y, Auble D T, Prelich G. EMBO J. 1999;18:6662–6671. doi: 10.1093/emboj/18.23.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire M, Xie J, Meisterernst M, Collart M A. Mol Microsc. 2000;36:163–173. doi: 10.1046/j.1365-2958.2000.01839.x. [DOI] [PubMed] [Google Scholar]
- 32.Hengartner C J, Myer V E, Liao S M, Wilson C J, Koh S S, Young R A. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 33.Zaman Z, Ansari A Z, Koh S S, Young R, Ptashne M. Proc Natl Acad Sci USA. 2001;98:2550–2554. doi: 10.1073/pnas.041611198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi Y, Huddleston M J, Zhang X, Young R A, Annan R S, Carr S A, Deshaies R J. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auble D T, Hahn S. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 36.Kokubo T, Swanson M J, Nishikawa J-I, Hinnebusch A G, Nakatani Y. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade P A, Jaehning J A. Mol Cell Biol. 1996;16:1641–1648. doi: 10.1128/mcb.16.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi T, Carey M. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 39.Oelgeschlager T, Chiang C-M, Roeder R G. Nature (London) 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 40.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 41.Bagby S, Mal T K, Ikura M. FEBS Lett. 2000;468:149–154. doi: 10.1016/s0014-5793(00)01213-8. [DOI] [PubMed] [Google Scholar]
- 42.Auble D T, Wang D, Post K W, Hahn S. Mol Cell Biol. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.