Abstract
Biochemical experiments and genomic sequence analysis showed that Deinococcus radiodurans and Thermus thermophilus do not possess asparagine synthetase (encoded by asnA or asnB), the enzyme forming asparagine from aspartate. Instead these organisms derive asparagine from asparaginyl-tRNA, which is made from aspartate in the tRNA-dependent transamidation pathway [Becker, H. D. & Kern, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12832–12837; and Curnow, A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Söll, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12838–12843]. A genetic knockout disrupting this pathway deprives D. radiodurans of the ability to synthesize asparagine and confers asparagine auxotrophy. The organism's capacity to make asparagine could be restored by transformation with Escherichia coli asnB. This result demonstrates that in Deinococcus, the only route to asparagine is via asparaginyl-tRNA. Analysis of the completed genomes of many bacteria reveal that, barring the existence of an unknown pathway of asparagine biosynthesis, a wide spectrum of bacteria rely on the tRNA-dependent transamidation pathway as the sole route to asparagine.
Asparagine, one of the 21 cotranslationally inserted amino acids that make up proteins, is known to be synthesized from aspartate in an ATP-dependent amidation reaction (1). Two mechanistically distinct asparagine synthetases are known (2–4). The one encoded by asnA utilizes ammonia as amide donor, whereas the asnB-derived protein works with glutamine. These enzymes are present in organisms of all domains, the major one being asparagine synthetase B, which is encoded in different organisms by a small number of related genes. Both enzymes are well studied biochemically (5), and their crystal structures are known (6, 7). Until recently they were assumed to be the sole biosynthetic route to asparagine. However, Thermus thermophilus and Deinococcus radiodurans lack these enzymes; instead they employ a tRNA-dependent transamidation mechanism for conversion of aspartate to asparagine (8, 9).
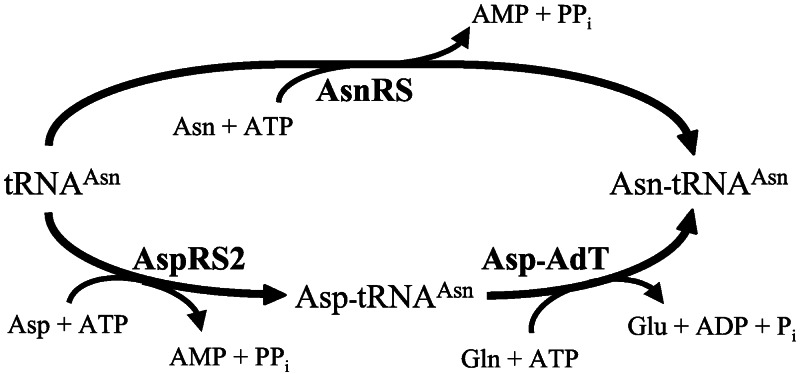
Two routes of Asn-tRNA synthesis exist in D. radiodurans (Fig. 1). Similar to many bacteria, Deinococcus contains a tRNA-dependent two-step pathway of Asn-tRNA formation. In the first step a nondiscriminating aspartyl-tRNA synthetase (AspRS)2 generates the misacylated Asp-tRNAAsn species, which then is amidated to the correctly charged Asn-tRNAAsn by the heterotrimeric Asp-tRNAAsn amidotransferase (Asp-AdT; encoded by the gatCAB genes) with glutamine serving as the amide donor (9). In addition, the organism also contains asparaginyl-tRNA synthetase (AsnRS; ref. 10), which is active and produces Asn-tRNA in the canonical aminoacylation reaction (9). The close Deinococcus relative T. thermophilus has similar enzymes and presumably uses the same asparagine biosynthetic routes (8, 11, 12). It was suggested earlier (8, 9) that the role of Asp-AdT in D. radiodurans and T. thermophilus is to synthesize the cell's entire supply of asparagine, because no asnA or asnB orthologs are present in the genome (10), and because biochemical analysis of crude cell extracts did not reveal the presence of any tRNA-independent asparagine synthetase activity (8, 9). Here we present data from D. radiodurans that prove this role to be correct and propose that tRNA-dependent asparagine synthesis occurs in many bacteria as the sole synthetic route to this essential amino acid.
Figure 1.
Redundant pathways of Asn-tRNAAsn synthesis in D. radiodurans. Shown are the direct pathway (top half of diagram) and the transamidation pathway (bottom half of diagram).
Materials and Methods
Chemicals and Enzymes.
Oligonucleotides were synthesized, and PCR products were sequenced by the Keck Foundation Biotechnology Resource Laboratory at Yale University. Uniformly labeled [14C]asparagine [228.4 mCi/mmol (1 Ci = 37 GBq)] was from NEN Life Science Products. Uniformly labeled [14C]aspartate (213 mCi/mmol) and [3H]aspartate (28 mCi/mmol) were from Amersham Pharmacia.
Strains, Plasmids, and Bacterial Growth.
D. radiodurans strain R1 was obtained from John Battista (Louisiana State University, Baton Rouge, LA). Plasmids pMD66 (13) and pMD405 were kindly provided by Michael J. Daly (Uniformed Services University of the Health Sciences, Bethesda, MD). pMD66 replicates autonomously in both Escherichia coli and D. radiodurans. pMD405 contains ampicillin-resistance and kanamycin-resistance markers and an origin of replication for E. coli but no replication capacity for D. radiodurans. D. radiodurans was grown at 30°C with vigorous shaking or on medium containing 1.5% agar. Complex medium for D. radiodurans was TGY (0.8% tryptone/0.4% yeast extract/0.1% glucose). Minimal medium for D. radiodurans (14) was 20 mM potassium phosphate (pH 8.0)/0.2 mM MgCl2/0.1 mM CaCl2/5 μM manganese acetate/5 μM (NH4)Mo7O24/5 μM FeSO4/10 μg/ml methionine/25 μg/ml histidine/30 μg/ml cysteine/1 μg/ml nicotinic acid/2 mg/ml fructose. Where necessary, the medium was supplemented with 10 μg/ml kanamycin/2.5 μg/ml tetracycline/3 μg/ml chloramphenicol/20 μg/ml asparagine. E. coli strain DH5α was grown at 37°C on LB medium (1% tryptone/0.5% yeast extract/0.5% NaCl/1.5% agar) supplemented where necessary with 50 μg/ml ampicillin and 30 μg/ml tetracycline. E. coli strain JF448 (15), obtained from the E. coli Genetic Stock Center at Yale University, has an asparagine auxotrophic phenotype (asnA and asnB). It was grown for 3 days at 37°C on M9 minimal agar plates containing 1 μg/ml thiamine and 30 μg/ml asparagine.
Preparation of tRNA Transcripts.
tRNAAsp and tRNAAsn genes were identified in the preliminary D. radiodurans genomic DNA sequence by the program TRNASCAN (16). The tRNAAsp gene was produced by PCR using the Pfu Turbo polymerase (Stratagene) and subcloned into pUC119. The tRNAAsn gene was constructed in pUC119 from six overlapping oligonucleotides. To enable in vitro transcription of tRNAAsn with T7 RNA polymerase, the first base pair was changed to G1-C72. The 5′ end of each construct contained the T7 promoter, and the 3′ end contained a BstNI site that, after BstNI digestion, allowed generation of the correct 3′ end of the DNA template for in vitro transcription. Reactions were performed as described (17); the resulting RNA was ethanol-precipitated, resuspended in loading buffer [10 mM Hepes (pH 7.3)/1 mM Na2-EDTA/7 M urea], and heated to 85°C for 10 min before loading onto a Q Sepharose column (Amersham Pharmacia). Transcripts were eluted from the column at 1.3 M NaCl. Fractions were concentrated, desalted, ethanol-precipitated, and resuspended in 10 mM Hepes (pH 7.2).
Preparation of Unfractionated and in Vivo Expressed tRNA.
Unfractionated D. radiodurans tRNA was prepared as described (9). For generation of in vivo expressed D. radiodurans tRNAAsn, the tRNAAsn gene was constructed from two overlapping oligonucleotides and cloned into the pGFIB vector (18). The plasmid was transformed into E. coli DH5α, and unfractionated tRNA was prepared from cultures of these cells as described above (9). Comparison of aminoacylation reactions with this tRNA and unfractionated tRNA from DH5α showed that the D. radiodurans tRNAAsn comprised 10–15% of the tRNA.
Preparation of D. radiodurans Cell Extract.
D. radiodurans wild-type strain R1 and the deletion mutant strains were grown in TGY medium including kanamycin for the mutant strains. Cells (5 g) from logarithmic-phase growth were harvested by centrifugation at 4,000 × g for 10 min at 4°C and resuspended in 100 ml of ice-cold 95% ethanol to remove the cells' outer membrane. The ethanol-stripped cells were harvested immediately by centrifugation at 4,000 × g for 5 min at 4°C. The resulting pellets were resuspended in 5 ml of 25 mM Hepes (pH 7.2)/1 mM MgCl2/30 mM KCl/5 mM DTT/4 mM 2-mercaptoethanol/10% glycerol. Lysozyme (2 mg/ml) was added to the suspension, and the mixture was incubated on ice for 30 min. The cells were disrupted by sonication with a Branson Sonifier 250 at 60% output for 15 sec plus cooling on ice for 3 min, repeated 10–15 times. The disrupted cells were centrifuged at 100,000 × g for 2 h at 4°C. The extracts were dialyzed against the above buffer containing 50% glycerol and stored at −20°C.
Preparation of D. radiodurans AspRS1 and AspRS2.
As described earlier (9), AspRS1 was overexpressed as a His-tagged enzyme from pET15b (Novagen) and purified on nickel-nitrilotriacetic acid resin (Qiagen, Chatsworth, CA). AspRS2 was overexpressed from pCYB2 (New England Biolabs) as a self-cleaving chitin-binding construct and purified on chitin agarose resin (New England Biolabs). To obtain cleavage of the intein fusion product, a glycine residue was added to the C terminus of AspRS2. The enzyme preparations were >95% pure as judged by Coomassie-stained polyacrylamide gel.
Aminoacyl-tRNA Synthetase Assays.
Reactions (100 μl) were performed as described (9) at 30°C and contained 100 mM Hepes (pH 7.2), 50 mM KCl, 10 mM MgCl2, 5 mM DTT, 6 μM [3H]aspartate, 2.5 mM unlabeled aspartate, 2 mM ATP, and 100 nM AspRS1 or 200 nM AspRS2. Aliquots (10 μl) of the reaction were removed at various time intervals and spotted onto a 3MM filter disk (Whatman) that was immersed in 10% trichloroacetic acid for 10 min and then washed twice with 5% trichloroacetic acid. The filters were rinsed in ethanol, dried-and counted in 3 ml of Ultima Gold scintillation mixture fluid (Packard).
For kinetic analysis, a higher specific activity aspartate mix was composed of 16 μM [3H]aspartate and 2.5 mM unlabeled aspartate. Aliquots were taken every 15 sec, and initial rates were determined from a minimum of five time points in the linear range of duplications of the assay. The enzyme concentrations were held at 40 nM AspRS1 and 100 nM AspRS2. The concentrations of ATP and aspartate were at least 5-fold above their determined apparent Km values (2 mM ATP and 0.5 mM aspartate in the AspRS1 reactions, and 2 mM ATP and 1.25 mM to 2.5 mM aspartate for AspRS2). When tRNAAsn was used, the Km of AspRS2 for aspartate was 2-fold higher than that when tRNAAsp was used. Therefore, for AspRS2, aspartate was kept at 1.25 mM for tRNAAsp and 2.5 mM for tRNAAsn. Transcript concentrations were at 3 μM for AspRS1 and 20 μM for AspRS2. For AspRS1, the concentration of aspartate was varied from 2 to 250 μM, ATP was varied from 0.05 to 5 mM, and tRNAAsp transcript was varied from 0.01 to 0.5 μM. For AspRS2, aspartate was varied from 0.02 to 1.25 mM for tRNAAsp and 0.02 to 2.5 mM for tRNAAsn; ATP was varied from 13 μM to 1.6 mM; tRNAAsp transcript was varied from 0.04 to 5 μM; and tRNAAsn transcript was varied from 0.08 to 20 μM.
Preparation of Genomic DNA.
D. radiodurans genomic DNA was prepared as described (19) with modifications. Cells from a 500-ml culture were collected by centrifugation at 4°C for 15 min at 5,000 × g. The cell pellet was resuspended in 2 ml of 100% ethanol to facilitate the removal of lipid-rich outer membranes. The stripped cells were collected by centrifugation and resuspended in water. Lysozyme (4 mg) was added and the suspension incubated at 37°C for 30 min. SDS (2%), Na2-EDTA [0.1 M (pH 8.0)], and Pronase E (4 mg) were added, and the lysate was incubated at 50°C for 3 h. After removal of proteins by multiple extractions with phenol/chloroform/isoamyl alcohol (25:24:1), chromosomal DNA was recovered from the aqueous layer by ethanol precipitation. The DNA was resuspended in 10 mM Tris⋅HCl (pH 8.0).
Construction of an Artificial Operon for Asparagine Synthesis with D. radiodurans aspS2 and gatCAB genes.
To construct an artificial operon for asparagine synthesis, the NdeI/BamHI fragment containing the D. radiodurans gatCAB genes from the pET20b clone (9) was ligated into the pCYB1 vector (New England Biolabs) such that there was no protein fusion with the intein of pCYB1. This process yielded the vector pCYB1-DRgatCAB. The D. radiodurans aspS2 gene subsequently was cloned into this vector by PCR-amplifying aspS2 with a forward primer containing the NdeI recognition site and a reverse primer containing the ribosome binding sequence for the gatC gene plus an AseI recognition site. The amplified aspS2 gene was cloned into the pCR-TOPO vector (Invitrogen), and the NdeI/AseI fragment containing aspS2 was subcloned into the NdeI site of pCYB1-DRgatCAB, yielding pCYB1-DRaspS2gatCAB.
Construction of Deletion Mutants.
D. radiodurans strains with deletions of asnS (encoding AsnRS) or aspS2 (encoding AspRS2) were constructed by an inversion overlap extension PCR knockout method. By using the Expand Long Template PCR system (Roche), for each gene deletion two PCR products were generated from D. radiodurans genomic DNA based on the known sequence (10). PCR with primers A and B yielded the upstream region of the gene including the 5′ end of asnS or aspS2, generating products of 3.0 or 2.7 kb, respectively. PCR with primers C and D similarly yielded the downstream region including the 3′ end of asnS or aspS2, generating products of 3.0 or 2.7 kb, respectively. The 5′ ends of primers A and D additionally contained complementary forms of the sequence 5′-GCGGCCGCGTTTAAACGGCGCGCC-3′, which is comprised of the adjacent 8-bp restriction sites NotI, PmeI, and AscI, to aid in subsequent clone identification by restriction analysis. This sequence also allowed overlap extension PCR using the two PCR products above as template plus primers E and F. Primers E and F contained 5′-XbaI sites for cloning, overlapped the genomic sequence immediately outside of the gene, and were on the 3′ side of primers B and C, respectively. The PCR products generated with primers E and F contained 54 or 71 bp of the 5′ end of asnS or aspS2, respectively, and 57 or 72 bp of the 3′ end of asnS or aspS2, respectively. Overlap PCR generated an ≈6-kb fragment simultaneously linking and inverting the upstream and downstream regions of the gene. The resulting fragment was cloned into the XbaI site of plasmid pMD405 for amplification in E. coli. D. radiodurans was transformed (20) with these plasmids and plated on TGY medium plus kanamycin. A double-crossover recombination into the chromosome is expected to delete the gene without disrupting its flanking regions. The crossover event was verified in kanamycin-resistant transformants by PCR analysis. The AsnRS activity was verified in D. radiodurans cell extracts.
Complementations of D. radiodurans with E. coli asnB and E. coli JF448 with the Artificial Operon for Asparagine Synthesis.
The asnB gene encoding asparagine synthetase was amplified from E. coli W3110 genomic DNA and cloned into the ScaI site of pMD66. This shuttle vector confers ampicillin resistance in E. coli and kanamycin and tetracycline resistance in D. radiodurans. Transformation of D. radiodurans was carried out as described (20). pCYB1, pCYB1-DRaspS2, pCYB1-DRgatCAB, and pCYB1-DRaspS2gatCAB were transformed into JF448. Ampicillin-resistant colonies were streaked onto M9 minimal agar plates containing 1 μg/ml thiamine and 1 mM isopropyl β-D-thiogalactoside with or without 30 μg/ml asparagine and incubated at 37°C for 3 days.
Phylogenetic Analysis.
For the protein sequence alignments the following amino acid sequences for AsnB proteins and their homologs were obtained from the nonredundant protein database at the National Center for Biotechnology Information: Aedes aegypti (gb|AAB95197.1), Aeropyrum pernix (dbj|BAA81051.1), Arabidopsis thaliana (sp|P49078), Archaeoglobus fulgidus (gb|AAB89808.1), Asparagus officinalis (sp|P31752), Bacillus halodurans (dbj|BAB05227.1), Bacillus subtilis (sp|P54420, sp|P42113, and sp|O05272), Bordetella bronchiseptica (emb|CAA07658.1), Caenorhabditis elegans (gi|7438080 and emb|CAA92825.1), Corynebacterium glutamicum (dbj|BAA89484.1), Desulfovibrio gigas (gb|AAF34252.1), Drosophila melanogaster (gb|AAF45462.1), E. coli (sp|P22106), Halobacterium sp. (gi|10580431), Homo sapiens (sp|P08243 and ref|NP_061921.1), Methanothermobacter thermautotrophicus (gb|AAB84920.1), Methanococcus jannaschii (sp|Q58516), Mycobacterium tuberculosis (sp|Q10374), Plasmodium falciparum (gi|7494204), Pseudomonas aeruginosa (gb|AAG05472.1, gb|AAG03441.1, and gb|AAG06847.1), Pyrobaculum aerophilum(gb|AAL62750.1), Pyrococcus abyssi (emb|CAB50044.1 and emb|CAB50067.1), Pyrococcus horikoshii OT3 (dbj|BAA30201.1), Saccharomyces cerevisiae (sp|P49089, sp|P49090, and sp|Q04489), Schizosaccharomyces pombe (gi|7492408), Thermoplasma acidophilum (emb|CAC11460.1), Vibrio cholerae (gb|AAC46243.1), and Xylella fastidiosa (gb|AAF82931.1). Data from partial genome sequences were obtained from: Bacillus stearothermophilus, www.genome.ou.edu/bstearo.html; Giardia lamblia, hermes.mbl.edu/baypaul/Giardia-HTML; Acidithiobacillus ferrooxidans, Geobacter sulfurreducens, and Shewanella putrefaciens, www.tigr.org; Pyrococcus furiosus, www.genome.utah.edu; and Clostridium difficile, Streptomyces coelicolor, and Candida albicans, www.sanger.ac.uk. Sequence data from Prosthecobacter sp. and Bifidobacterium longum were provided by R. Overbeek (Integrated Genomics, Chicago) and F. Arigoni (Nestlé Research Center, Lausanne, Switzerland), respectively.
Phylogenetic Inference.
From the alignment of 50 AsnB proteins, 408 positions were deemed to be aligned confidently. These data were analyzed as described (21) by protein maximum parsimony methods using a heuristic search algorithm (PAUP* 4.0 BETA 2, D. Swofford, Sinauer, Sunderland, MA). The 1,000 shortest trees were evaluated by maximum likelihood criteria, using the PROTML program (Version 2.2) in the MOLPHY package (22) with the JTT model for amino acid substitutions. Bootstrap percentages for each node in the tree were estimated by the resampling estimated log-likelihood method (23) using the PROTML program to compare the 1,000 most parsimonious trees. The CONSENSE program [PHYLIP (phylogeny inference package), Version 3.5c; J. Felsenstein, Department of Genetics, University of Washington, Seattle] was used to construct a consensus tree from the resampling estimated log-likelihood weightings. Phylogenetic trees were viewed and edited with the TREEVIEW program (Version 1.5.2; ref. 24).
Results
D. radiodurans Contains Two AspRSs.
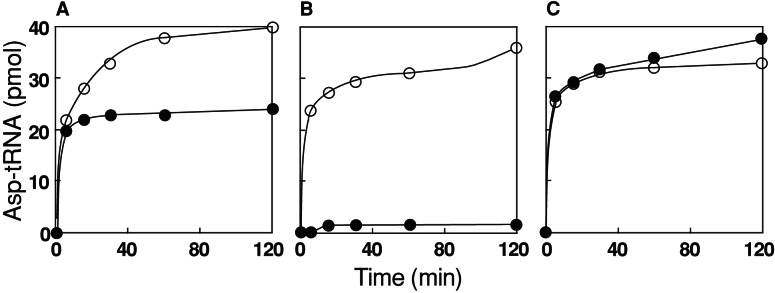
As discussed earlier (9) Deinococcus employs two routes of Asn-tRNA synthesis: canonical aminoacylation and tRNA-dependent transamidation (depicted in Fig. 1). A key enzyme for the latter pathway is a nondiscriminating AspRS capable of generating the misacylated Asp-tRNAAsn transamidation substrate. Actually, D. radiodurans contains two AspRS enzymes (9, 10). AspRS1 is a discriminating synthetase only capable of acylating the tRNAAsp isoacceptors, whereas the smaller, archaeal genre AspRS2 enzyme has relaxed tRNA specificity and aspartylates both tRNAAsp and tRNAAsn (9, 11, 12, 25, 26). To investigate the biochemical properties of these enzymes, they were purified from heterologously overexpressed cloned genes, and their activity was tested with D. radiodurans tRNAAsp and tRNAAsn species (Fig. 2). AspRS1 aminoacylated 3.4% of unfractionated tRNA from D. radiodurans. In contrast, AspRS2 aminoacylated a higher proportion (≈5%) of this tRNA (Fig. 2A), which is consistent with an ability to aminoacylate additional tRNA species. To determine whether the difference was caused by misaminoacylation of tRNAAsn, we assayed the ability of each AspRS to aminoacylate specific tRNA gene transcripts. Fig. 2B shows that only AspRS2 could aminoacylate the tRNAAsn, whereas both AspRS enzymes were fully capable of aminoacylating the tRNAAsp transcript (Fig. 2C).
Figure 2.
Asp-tRNA formation by the two D. radiodurans AspRS enzymes. AspRS1 (●) and AspRS2 (○) were used to aminoacylate, per reaction, unfractionated D. radiodurans tRNA (A, 800 pmol), D. radiodurans tRNAAsn transcript (B, 40 pmol), and D. radiodurans tRNAAsp transcript (C, 40 pmol).
Detailed steady-state kinetic analysis with the tRNAAsn and tRNAAsp transcripts showed that the Km of AspRS1 for the tRNAAsp transcript was 0.22 μM, which is similar to the value of the E. coli enzyme for the homologous substrate of 0.6 μM (27). The Km of AspRS2 for each transcript was higher than for AspRS1 at 3.1 μM for tRNAAsp and 1.8 μM for tRNAAsn. The comparable Km values of AspRS2 for both tRNAAsp and tRNAAsn demonstrates that AspRS2 recognizes both tRNAs to a similar degree. The Km values for the other substrates are ATP (AspRS1), 216 μM; ATP (AspRS2), 171 μM (with tRNAAsp) and 24 μM (with tRNAAsn); Asp (AspRS1), 69 μM; and Asp (AspRS2) 247 μM (with tRNAAsp) and 442 μM (with tRNAAsn). These data are in general agreement with values for the two T. thermophilus AspRS enzymes (12, 28).
Two Functionally Redundant Routes of Asn-tRNA Synthesis in D. radiodurans.
To demonstrate that two routes to Asn-tRNA formation exist in D. radiodurans, we attempted to create independent genetic knockouts of each pathway. The asnS gene of the published D. radiodurans R1 genomic sequence was reported to have an authentic frameshift (10); the predicted protein of 320 aa is not expected to be active. However, when we sequenced the asnS gene amplified from our isolate of D. radiodurans R1 DNA, we found an intact ORF of 521 aa in length. In agreement with this finding, we previously demonstrated AsnRS activity in a cell extract from this Deinococcus isolate (9).
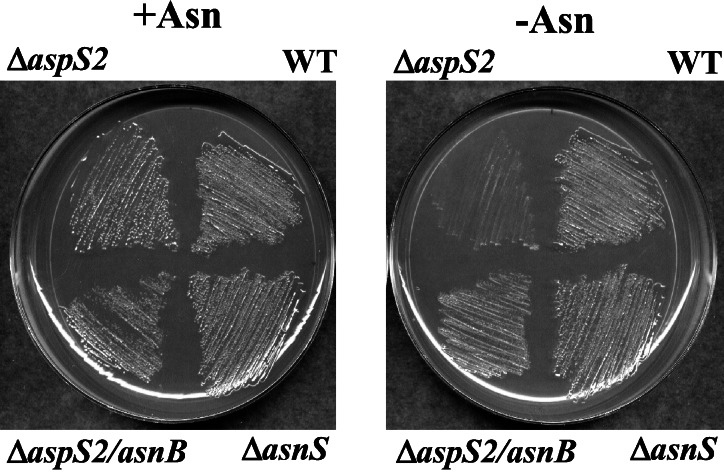
We then proceeded to make two D. radiodurans strains with gene disruptions in either asnS (blocking aminoacylation, Fig. 1) or aspS2 (blocking the transamidation pathway, Fig. 1). Both strains were found to be viable (Fig. 3 Left). Therefore, D. radiodurans possesses redundant routes to Asn-tRNA formation, and each can support the organism's growth independently.
Figure 3.
Growth properties of D. radiodurans strain constructs. ΔaspS2, aspS2 deletion strain; WT, wild-type parent strain; ΔaspS2/asnB, aspS2 deletion strain complemented in trans with E. coli asnB; ΔasnS, asnS deletion strain. Growth on minimal agar plates in the presence (+Asn) or absence (−Asn) of asparagine is shown.
tRNA-Dependent Transamidation Is the Sole Route to Asparagine Formation in D. radiodurans.
The D. radiodurans genomic sequence has no apparent homologs of the asnA or asnB genes that encode the two types of asparagine synthetase (10). In accordance with this absence, asparagine synthetase activity could not be detected in cell extracts of D. radiodurans (9). Because there are no other known biosynthetic routes to asparagine (29), we reasoned that the tRNA-mediated transamidation pathway of Asn-tRNA synthesis is the route to asparagine formation in this organism. Thus, if this pathway is blocked by the aspS2 deletion, the resulting D. radiodurans strain should be auxotrophic for asparagine. This phenotype of asparagine-dependent growth was verified on minimal medium in which the mutant did not grow without asparagine (Fig. 3 Right) but did grow in the presence of asparagine (Fig. 3 Left). In contrast, both the wild type and the asnS deletion mutant strains grew in the presence or absence of asparagine (Fig. 3). The asnS mutant does not require asparagine, because it forms the amino acid in a tRNA-dependent manner via the AspRS2/Asp-AdT transamidation pathway (Fig. 1). To demonstrate further that the aspS2 deletion mutant requires asparagine, the strain was complemented in trans with the E. coli asnB gene (carried on a pMD66 plasmid). Similar to the wild type and the asnS deletion mutant, the aspS2 strain complemented with asnB did not require added asparagine for growth (Fig. 3). These results show that D. radiodurans forms asparagine only in a tRNA-dependent manner.
The end-product of the transamidation pathway is Asn-tRNA. Thus, if free asparagine is needed by the cell, Asn-tRNA must be hydrolyzed. Enzymes carrying out this function may exist, analogous to the protein that releases d-tyrosine from charged tRNA (30). Alternatively, AsnRS may fill this role, because synthetase-catalyzed hydrolysis of aminoacyl-tRNA in the absence of the other substrates (AMP and PPi) is known (31). The fact that the D. radiodurans asnS deletion grows in the absence of asparagine (Fig. 3 Right) shows that AsnRS does not assume this role in Deinococcus.
The Deinococcus Transamidation Pathway Enzymes Rescue an E. coli Asparagine Auxotroph.
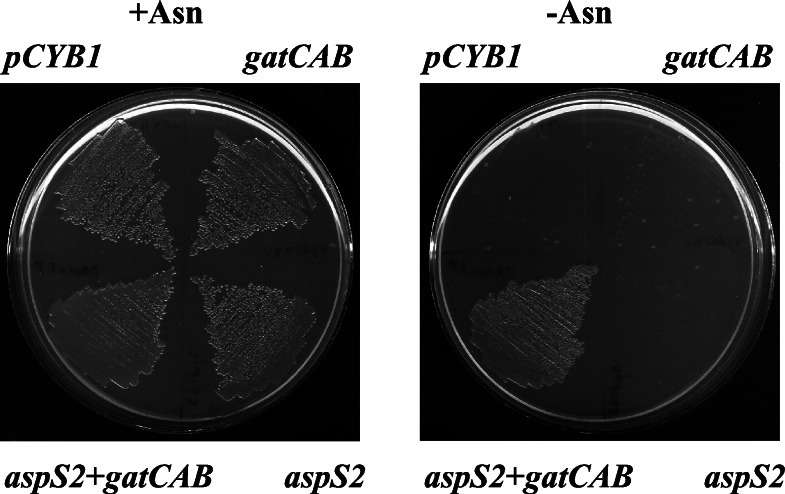
To demonstrate in another way that the transamidation pathway provides asparagine, we complemented the asparagine auxotrophic E. coli strain JF448 (asnA,asnB) with the D. radiodurans aspS2, gatC, gatA, and gatB genes. The transformed strain did not require asparagine for growth, unlike JF448 transformed with the empty vector or the aspS2 or gatCAB genes (Fig. 4). Thus, the Deinococcus nondiscriminating AspRS2 and the Asp-AdT enzymes are functional in asparagine synthesis in E. coli.
Figure 4.
Coexpression of D. radiodurans aspS2 and gatCAB genes rescues asparagine auxotrophy of E. coli strain JF448. Growth on minimal agar plates in the presence (+Asn) or absence (−Asn) of asparagine is shown. The complementing genes were introduced on the pCYB1 vector.
Two Pathways of Asparagine Formation Exist in Nature.
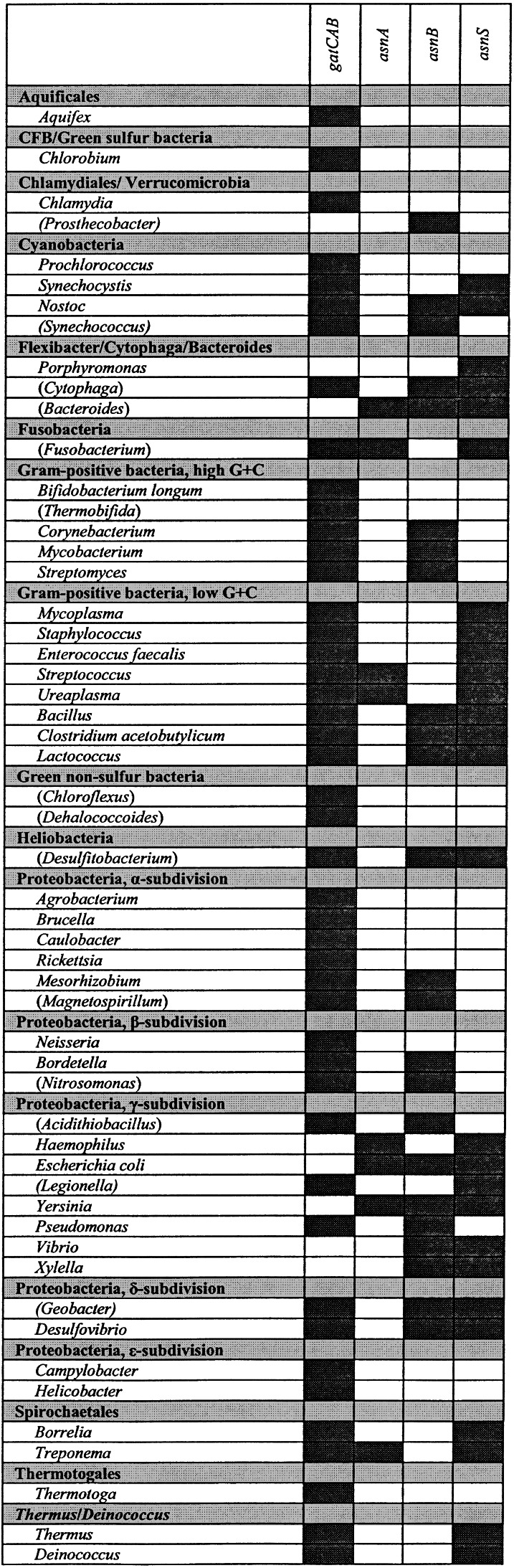
Based on the Deinococcus data presented above and assuming that there is no other still unknown pathway for asparagine synthesis, we examined bacteria with completed genome sequences for their possible asparagine biosynthetic mechanism. This analysis revealed that a large number of bacteria may use tRNA-dependent transamidation exclusively to generate asparagine. Table 1 shows that bacteria (with completed genome sequences) of 13 different genera, representing most of the main bacterial groups, have no apparent homologs for asparagine synthetase and AsnRS but contain the gatCAB genes encoding the bacterial Asp/Glu-tRNA amidotransferase. Therefore we propose that tRNA-dependent asparagine formation is widespread in the bacteria. The bacterial genomes also demonstrate that many other bacterial genera may make asparagine by the asparagine synthetase(s) of the AsnA or AsnB type (Table 1).
Table 1.
Presence (dark shadowed) of asparagine pathways among bacterial genera with complete (and incomplete) genomes
Discussion
Asparagine Biosynthesis Is Both tRNA-Dependent and tRNAIndependent.
The deletion experiments described above prove that D. radiodurans lacks a tRNA-independent route of asparagine biosynthesis. Unless there are yet undescribed biosynthetic routes to asparagine, one may conclude from existing genomic sequences and the established activities of the GatCAB enzyme that many bacteria synthesize asparagine only via this path (Table 1). Alternatively, some bacteria employ AsnRS to synthesize Asn-tRNAAsn with asparagine either imported or made from aspartate by the AsnA or AsnB enzymes (Table 1).
All the above pathways are distributed broadly among diverse bacteria (Table 1). However, some heterotrophic organisms such as Porphyromonas gingivalis apparently have no capacity for asparagine biosynthesis, relying solely on exogenous amino acids. The redundancy of asparagine biosynthetic pathways in bacteria epitomizes the modularity of both amino acid biosynthesis and tRNA-charging enzymes. As both evolution and genetics have demonstrated, these modular systems are interchangeable. Each system depends on compatible biosynthetic and tRNA-charging systems, emphasizing the unique role of aminoacyl-tRNA synthetases in connecting metabolism to translation.
Evolution and Structural Relationships of the Enzymes of Asparagine Biosynthesis.
Most bacteria have some capacity to synthesize asparagine. The asnA gene has been identified primarily in pathogenic, heterotrophic bacteria, many of which also have asnB genes (Table 1). A recent crystal structure of E. coli AsnA (6) supports the idea that asnA diverged from an AspRS gene (aspS). This model extends earlier sequence analyses (32) and site-directed mutagenesis experiments (33, 34) to explain how AsnA and AspRS form analogous aminoacyl-adenylate reaction intermediates (26, 35). Similarities between archaeal/eukaryal AspRS sequences and AsnA led to the suggestion that the asnA gene may have evolved specifically from an archaeal aspS gene (6).
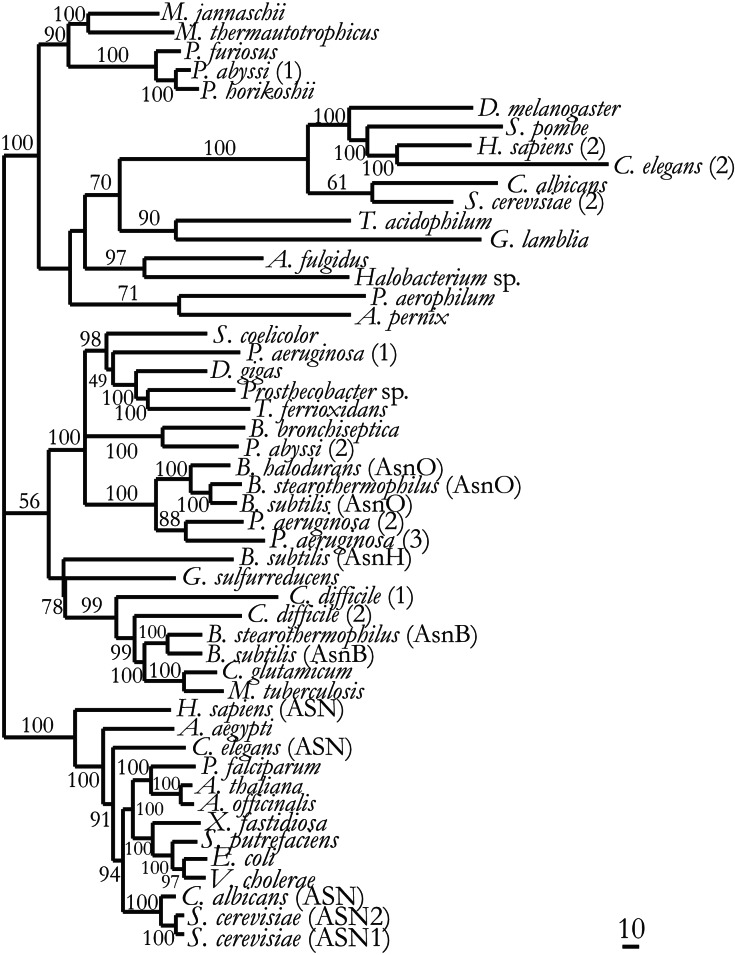
Phylogenetic analysis of AsnB, the glutamine-dependent asparagine synthetase, shows homologs segregating into three distinct lineages: bacterial, eukaryal, and archaeal (Fig. 5). These results are consistent with AsnB evolving before the segregation of organisms into the modern cell types. AsnB shows no similarity to either AsnA or aminoacyl-tRNA synthetase proteins. Rather, its N-terminal domain resembles glutamine phosphoribosylpyrophosphate amidotransferase, whereas its C-terminal domain resembles GMP synthetase (7). Although eukaryotes depend on asnB homologs for asparagine biosynthesis, this gene does not seem to have evolved in bacteria as a primary route to make asparagine, because few bacteria seem to rely exclusively on the AsnB+AsnRS system for asparagine biosynthesis (Table 1). Remarkably, asnB genes have been identified in genomes of proteobacteria and archaea that have no recognizable AsnRS.
Figure 5.
Phylogeny of AsnB homologs. Bootstrap percentages for each branch were estimated by the resampling estimated log-likelihood method. (Scale bar represents 10 substitutions per 100 aa positions.)
The aspartyl-tRNAAsn amidotransferase (GatCAB) is the most widespread route to asparagine synthesis in bacteria. Identification of tRNA-dependent asparagine biosynthesis by genomic analysis is complicated, however, by the fact that the aspartyl-tRNAAsn amidotransferase is one of two possible activities of the GatCAB enzyme. In most bacteria this amidotransferase also generates Gln-tRNAGln employing the glutamyl-tRNAGln activity. Whether an organism uses one or both of these activities is determined, in turn, by the presence of a nondiscriminating glutamyl-tRNA synthetase or AspRS in the genome (36). In any case, the absence of AsnRS in a genome suggests formation of Asn-tRNAAsn by the amidotransferase (e.g., Chlamydia; ref. 36). The amidotransferase pathway, including the nondiscriminating AspRS2, is therefore the sole route to translational asparagine biosynthesis in a majority of bacterial lineages (Table 1) and also in some archaeal lineages (37). As demonstrated here, GatCAB and AspRS2 are necessary and sufficient for asparagine biosynthesis in D. radiodurans. Furthermore, these genes are able to rescue asparagine auxotrophy (caused by asnA, asnB) in E. coli. It would be tempting to speculate that the amidotransferase pathway is the original “rational” route to asparagine, by forming directly Asn-tRNA, the required precursor for protein synthesis.
A Wider Role for tRNA-Dependent Amino Acid Transformations.
Pathways whereby aminoacylated tRNAs are modified further before their use in translation are well known. Selenocysteinyl-tRNA (reviewed in ref. 38) and formylmethionyl-tRNA formation (reviewed in ref. 39) are crucial for translation in many organisms. Although the transamidation route to Asn-tRNA and Gln-tRNA synthesis is known to be operating in most bacteria and archaea, its wider significance in being responsible for an organism's sole supply of asparagine has not been appreciated. Another example highlighting a close relationship between aminoacyl-tRNA synthesis and intermediary metabolism is the formation of 5-aminolevulinic acid, the universal precursor of porphyrins in many organisms. This metabolite is generated by reduction of glutamate attached to tRNA by a unique oxidoreductase with sequence-specific aminoacyl-tRNA recognition (40). These reactions suggest a closer than previously recognized tRNA-mediated connection between intermediary metabolism and protein synthesis.
Acknowledgments
We thank M. Ibba, C. R. Woese, and H. Zalkin for informative discussions, F. Arigoni, D. A. Bryant, and R. Overbeek for sharing unpublished genome data, and J. Battista and M. Daly for help with D. radiodurans manipulations. D.T.-H. was a National Institutes of Health Postdoctoral Fellow. This work was supported by grants from the Department of Energy and the National Institute of General Medical Sciences.
Abbreviations
- AspRS
aspartyl-tRNA synthetase
- Asp-AdT
Asp-tRNAAsn amidotransferase
- AsnRS
asparaginyl-tRNA synthetase
References
- 1.Milman H A, Cooney D A. Biochem J. 1979;181:51–59. doi: 10.1042/bj1810051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zalkin H, Truitt C D. J Biol Chem. 1977;252:5431–5436. [PubMed] [Google Scholar]
- 3.Nakamura M, Yamada M, Hirota Y, Sugimoto K, Oka A, Takanami M. Nucleic Acids Res. 1981;9:4669–4676. doi: 10.1093/nar/9.18.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scofield M A, Lewis W S, Schuster S M. J Biol Chem. 1990;265:12895–12902. [PubMed] [Google Scholar]
- 5.Richards N G, Schuster S M. Adv Enzymol Relat Areas Mol Biol. 1998;72:145–198. doi: 10.1002/9780470123188.ch5. [DOI] [PubMed] [Google Scholar]
- 6.Nakatsu T, Kato H, Oda J. Nat Struct Biol. 1998;5:15–19. doi: 10.1038/nsb0198-15. [DOI] [PubMed] [Google Scholar]
- 7.Larsen T M, Boehlein S K, Schuster S M, Richards N G J, Thoden J B, Holden H M, Rayment I. Biochemistry. 1999;38:16146–16157. doi: 10.1021/bi9915768. [DOI] [PubMed] [Google Scholar]
- 8.Becker H D, Kern D. Proc Natl Acad Sci USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curnow A W, Tumbula D L, Pelaschier J T, Min B, Söll D. Proc Natl Acad Sci USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, et al. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 12.Becker H D, Roy H, Moulinier L, Mazauric M H, Keith G, Kern D. Biochemistry. 2000;39:3216–3230. doi: 10.1021/bi992573y. [DOI] [PubMed] [Google Scholar]
- 13.Daly M J, Ouyang L, Fuchs P, Minton K W. J Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkateswaran A, McFarlan S C, Ghosal D, Minton K W, Vasilenko A, Makarova K, Wackett L P, Daly M J. Appl Environ Microbiol. 2000;66:2620–2626. doi: 10.1128/aem.66.6.2620-2626.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felton J, Michaelis S, Wright A. J Bacteriol. 1980;142:221–228. doi: 10.1128/jb.142.1.221-228.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe T M, Eddy S R. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampson J R, Uhlenbeck O C. Proc Natl Acad Sci USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Normanly J, Masson J M, Kleina L G, Abelson J, Miller J H. Proc Natl Acad Sci USA. 1986;83:6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattimore V, Udupa K S, Berne G A, Battista J R. J Bacteriol. 1995;177:5232–5237. doi: 10.1128/jb.177.18.5232-5237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masters C I, Smith M D, Gutman P D, Minton K W. J Bacteriol. 1991;173:6110–6117. doi: 10.1128/jb.173.19.6110-6117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woese C R, Olsen G, Ibba M, Söll D. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adachi J, Hasegawa M. Comput Sci Monogr. 1996;28:1–150. [Google Scholar]
- 23.Kishino H, Miyata T, Hasegawa M. J Mol Evol. 1990;31:151–160. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 24.Page R D M. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 25.Imanaka T, Lee S, Takagi M, Fujiwara S. Gene. 1995;164:153–156. doi: 10.1016/0378-1119(95)00491-n. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt E, Moulinier L, Fujiwara S, Imanaka T, Thierry J-C, Moras D. EMBO J. 1998;17:5227–5237. doi: 10.1093/emboj/17.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eriani G, Dirheimer G, Gangloff J. Nucleic Acids Res. 1990;18:7109–7118. doi: 10.1093/nar/18.23.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker H D, Reinbolt J, Kreutzer R, Giegé R, Kern D. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 29.Reitzer L J, Magasanik B. J Bacteriol. 1982;151:1299–1313. doi: 10.1128/jb.151.3.1299-1313.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soutourina J, Plateau P, Delort F, Peirotes A, Blanquet S. J Biol Chem. 1999;274:19109–19114. doi: 10.1074/jbc.274.27.19109. [DOI] [PubMed] [Google Scholar]
- 31.Ebel J P, Giegé R, Bonnet J, Kern D, Befort N, Bollack C, Fasiolo F, Gangloff J, Dirheimer G. Biochimie. 1973;55:547–557. doi: 10.1016/s0300-9084(73)80415-8. [DOI] [PubMed] [Google Scholar]
- 32.Wolf Y I, Aravind L, Grishin N V, Koonin E V. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 33.Gatti D L, Tzagoloff A. J Mol Biol. 1991;218:557–568. doi: 10.1016/0022-2836(91)90701-7. [DOI] [PubMed] [Google Scholar]
- 34.Hinchman S K, Henikoff S, Schuster S M. J Biol Chem. 1992;267:144–149. [PubMed] [Google Scholar]
- 35.Cedar H, Schwartz J H. J Biol Chem. 1969;244:4122–4127. [PubMed] [Google Scholar]
- 36.Raczniak G, Becker H D, Min B, Söll D. J Biol Chem. 2001;276:45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- 37.Tumbula D L, Becker H D, Chang W-Z, Söll D. Nature (London) 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 38.Blanquet S, Mechulam Y, Schmitt E. Curr Opin Struct Biol. 2000;10:95–101. doi: 10.1016/s0959-440x(99)00055-x. [DOI] [PubMed] [Google Scholar]
- 39.Böck A. Biofactors. 2000;11:77–78. doi: 10.1002/biof.5520110122. [DOI] [PubMed] [Google Scholar]
- 40.Moser J, Schubert W D, Beier V, Bringemeier I, Jahn D, Heinz D W. EMBO J. 2001;20:6583–6590. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]