Abstract
Ribonuclease P (RNase P) is a ubiquitous endoribonuclease that cleaves precursor tRNAs to generate mature 5′ termini. Although RNase P from all kingdoms of life have been found to have essential RNA subunits, the number and size of the protein subunits ranges from one small protein in bacteria to at least nine proteins of up to 100 kDa. In Saccharomyces cerevisiae nuclear RNase P, the enzyme is composed of ten subunits: a single RNA and nine essential proteins. The spatial organization of these components within the enzyme is not yet understood. In this study we examine the likely binary protein–protein and protein–RNA subunit interactions by using directed two- and three-hybrid tests in yeast. Only two protein subunits, Pop1p and Pop4p, specifically bind the RNA subunit. Pop4p also interacted with seven of the other eight protein subunits. The remaining protein subunits all showed one or more specific protein–protein interactions with the other integral protein subunits. Of particular interest was the behavior of Rpr2p, the only protein subunit found in RNase P but not in the closely related enzyme, RNase MRP. Rpr2p interacts strongly with itself as well as with Pop4p. Similar interactions with self and Pop4p were also detected for Snm1p, the only unique protein subunit so far identified in RNase MRP. This observation is consistent with Snm1p and Rpr2p serving analogous functions in the two enzymes. This study provides a low-resolution map of the multisubunit architecture of the ribonucleoprotein enzyme, nuclear RNase P from S. cerevisiae.
Ribonuclease P (RNase P) is an essential endoribonuclease that acts early in tRNA biogenesis to remove the 5′ leader sequences of precursor tRNAs (pretRNAs) (1–3). The enzyme has been identified in every organism tested, in all kingdoms of life. In most cases, the enzyme is composed of a single RNA subunit and one or more protein subunits (1, 4). The RNA subunit forms the catalytic core of the enzyme, and the bacterial and some archaeal RNA subunits alone are catalytic in vitro (5–7). In contrast, no eukaryotic RNase P RNA subunits have been shown to be catalytic in the absence of protein. In bacteria, RNase P is composed of a catalytic RNA subunit and a single small protein subunit. Studies on the bacterial RNase P protein suggest that the protein plays a role in substrate recognition (8–11). Recent data show that at least one form of archaeal RNase P consists of four or more proteins and a single RNA subunit (12). Moreover, the identified archaeal proteins appear to be homologs of the eukaryotic RNase P proteins and not the bacterial proteins (T. A. Hall and J. W. Brown, personal communication).
Eukaryotic nuclear RNase P contains an RNA subunit similar in size to its bacterial and archaeal counterparts, containing all of the most conserved “critical regions” from the bacterial consensus structure (13). However, the protein content is far more complex and is absolutely required for activity. Human nuclear RNase P appears to contain at least ten proteins (14–19). At least six of these proteins are homologs of integral RNase P subunits identified in Saccharomyces cerevisiae (2, 16–19). The nuclear enzyme has been purified to homogeneity from S. cerevisiae (20), and shown to contain nine tightly associated proteins that are essential for RNase P activity and for life (20–24). The proteins range in size from 15.5 to 100 kDa and seven of the nine are highly basic (Table 1), with varied levels of nonspecific RNA binding activity in vitro (unpublished observations). Interestingly, eight of the nine protein subunits appear also to be required subunits of RNase MRP, an endoribonuclease that participates in the major preribosomal RNA maturation pathway (20–27). Further evidence of the close evolutionary relationship between RNase MRP and RNase P is that MRP has an RNA subunit, NME1 RNA, that is structurally related to the RPR1 RNA subunit of RNase P (28–32). Both RNase P and RNase MRP RNAs are found in the yeast nucleolus (26, 33–35). In human cells, RNase MRP is clearly nucleolar, whereas association of RNase P with the nucleolus might occur primarily during enzyme assembly (36–38). Despite the extensive protein overlap between RNases P and MRP and the shared subcellular location, the two enzymes appear to exist in separate complexes (39).
Table 1.
Subunit composition of S. cerevisiae RNase P and RNase MRP
| Yeast
gene
|
Subunit type | Molecular mass,* kDa | Isoelectric point,* pI | Ref. | |
|---|---|---|---|---|---|
| RNase P | RNase MRP | ||||
| RPR1 | — | RNA | 120 | — | 61 |
| — | NME1 | RNA | 112 | — | 25, 27 |
| POP1 | POP1 | Protein | 100.5 | 9.84 | 21 |
| POP3 | POP3 | Protein | 22.6 | 9.57 | 22 |
| POP4 | POP4 | Protein | 32.9 | 9.26 | 24 |
| POP5 | POP5 | Protein | 19.6 | 7.79 | 20 |
| POP6 | POP6 | Protein | 18.2 | 9.28 | 20 |
| POP7 | POP7 | Protein | 15.8 | 9.34 | 20, 62 |
| POP8 | POP8 | Protein | 15.5 | 4.57 | 20 |
| RPP1 | RPP1 | Protein | 32.2 | 9.76 | 23 |
| RPR2 | — | Protein | 16.3 | 9.99 | 20 |
| — | SNM1 | Protein | 22.5 | 9.81 | 63 |
Predicted molecular masses and isoelectric points based on amino acid composition.
Although much progress has been made on determining the composition of RNase P, very little detail is available on the organization of the RNA and protein subunits in the enzyme complex. Even in bacteria, where the structure of the protein subunit has been solved (40, 41) and three-dimensional models of the RNA subunit exist (42, 43), the spatial organization the RNA and protein is still unclear. Our attempts to study these questions for S. cerevisiae nuclear RNase P by in vitro reconstitution from purified subunits were foiled by insolubility and aggregation of the individually expressed proteins. As an alternative, we have used a directed “two-hybrid” system to test for specific protein–protein interactions between subunits expressed inside yeast (44–46), and the “three-hybrid” system to detect specific interactions between the RNase P RNA and protein subunits (47). These data have yielded a low-resolution map of the spatial organization of S. cerevisiae RNase P subunits in the enzyme.
Materials and Methods
Media, Strains, and Plasmids.
Standard yeast genetic techniques and media were used (48, 49). All two-hybrid tests were performed in the L40 reporter strain [Mata, his3Δ200, trp1–901, leu2–3,112, ade2, LYS2∷(lexAop)4-HIS3, URA3∷(lexAop)8-lacZGAL4] (50, 51). The ORFs of all nine RNase P protein subunits and the MRP subunit, Snm1p (see Table 1), were individually cloned into the pBTM116-ADE2 (pLexA) plasmid (bait) between the XmaI and SalI sites, which produces a fusion protein linked to the entire coding region of the Escherichia coli LexA protein (52). The ORFs were also cloned into the pACTII plasmid (a gift of S. Elledge, Baylor College of Medicine, Houston) (prey) between the XmaI and SacI restriction sites, which produces a fusion protein linked to the GAL4 activation domain. All fusion junctions were sequenced. The L40 strain was transformed with all 109 combinations of the bait and prey plasmids, including those lacking ORF insertions (negative controls) and the pLexA-Ras/pVP16-Rip51 (positive control; ref. 53). Double transformants containing bait and prey plasmids were selected on SDC-trp-ura-leu-lys media. Fusion protein expression was checked by Western blot using monoclonal antibodies against the LexA protein and Gal4 activation domain (Santa Cruz Biotechnology; data not shown).
All three-hybrid tests were performed using the pIIIA-MS2–2 RNA expression plasmid (a gift of M. Wickens, University of Wisconsin, Madison) in the L40-coat reporter strain [Mata, ura3–52, leu2–3,112, his3Δ200, trp1Δ1, ade2, LYS2∷(lexAop)4-HIS3, ura3∷(lexAop)8-lacZ, LexA-MS2 coat(TRP1)] (47). The sense and antisense forms of RPR1 RNA were cloned into the SmaI restriction site of pIIIA-MS2–2. The resulting RNA polymerase III transcripts encode a hybrid RNA containing the RPR1 leader, the inserted RNA sequence, two tandem MS2 sites, and the RPR1 terminator. The pACTII fusion protein plasmids of the nine RNase P protein subunits and controls were transformed pairwise with pIIIA-MS2–2 RNA bait expression plasmids. pACTII/pIIIA-MS2-2 transformants were selected on SDC-trp-ura-leu-lys media. The control plasmids for the three-hybrid study, pIII-IRE and pAD-IRP, have been described (47) and were gifts from M. Wickens.
Identification of Interacting Subunits.
The L40 and L40-coat yeast strains each contain two integrated reporter genes: the yeast HIS3 gene and the bacterial lacZ gene. Two isolates from each double transformation were tested for their ability to grow on SDC-trp-ura-leu-lys-his plates in the absence or presence of 1 mM, 5 mM, 10 mM, and 20 mM 3-amino-1,2,4-triazole (3-AT, Sigma). Colonies growing after 2–3 days at 30°C were initially classified as positive for protein–protein or protein–RNA interactions. The same two transformation isolates were also tested for their ability to produce β-galactosidase, using a filter assay (54). Colonies were grown 2–4 days on SDC-trp-ura-leu-lys at 30°C, transferred to nitrocellulose filters and lysed by freeze/thaw by using liquid nitrogen. The filters were placed in Petri dishes containing 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-Gal). The reaction was incubated at 30°C up to 24 h and stopped by addition of 100 mM EDTA. Colony color was evaluated qualitatively relative to positive and negative controls.
Results
Identification of Protein–Protein Interactions Among RNase P Protein Subunits.
After extensive purification, nuclear RNase P from S. cerevisiae contains a single RNA subunit and nine essential protein subunits (Table 1; ref. 20). Although we have cloned and overexpressed all of the individual subunits, reconstitution of the ribonucleoprotein enzyme and study of binary subunit–subunit interactions in vitro have been unsuccessful. Most of the recombinant proteins are largely insoluble and the largest subunit, Pop1p, is particularly unstable. To identify potential protein–protein interactions among the protein subunits of yeast RNase P, we therefore performed an in vivo two-hybrid test in which all of the possible combinations of the individual subunits were tested against each other as bait and prey. The results of the two- and three-hybrid tests are shown in Figs. 1 and 2, and summarized schematically in Fig. 3.
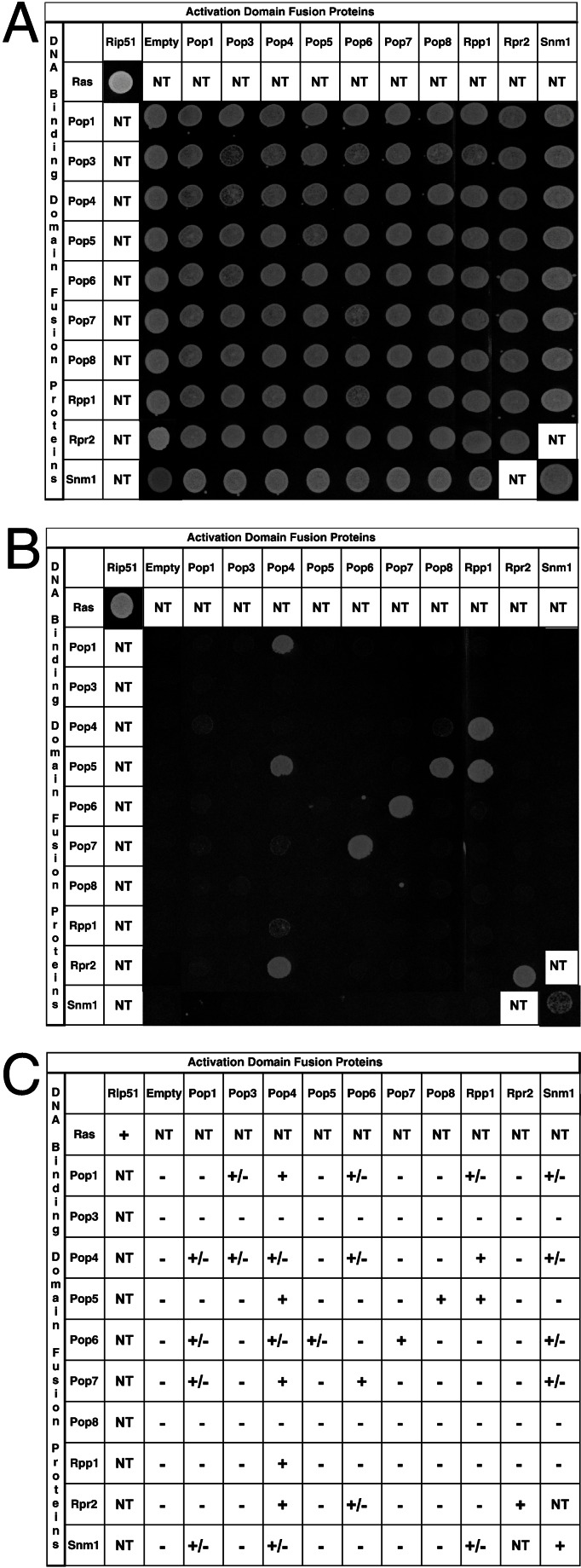
Figure 1.
Identification of protein–protein interactions among RNase P protein subunits by using two-hybrid analysis. Yeast strains transformed with the indicated plasmids were tested for their ability to grow on synthetic media containing histidine (A) and synthetic media lacking histidine plus 1 mM 3-aminotriazole (B). A summary of the results from the β-galactosidase assays performed on the yeast strains is shown in C. A positive protein–protein interaction is indicated by the ability of the yeast strains to grow in the absence of histidine and their ability to produce β-galactosidase. − Indicates no blue color was observed relative to positive control strain transformed with pLexA-Ras and pVP16-Rip51; +/− indicates a light blue color relative to positive control, and + indicates a dark blue color similar to the positive control. NT indicates that a combination was not tested because the proteins were known not to be part of the same complex in vivo.
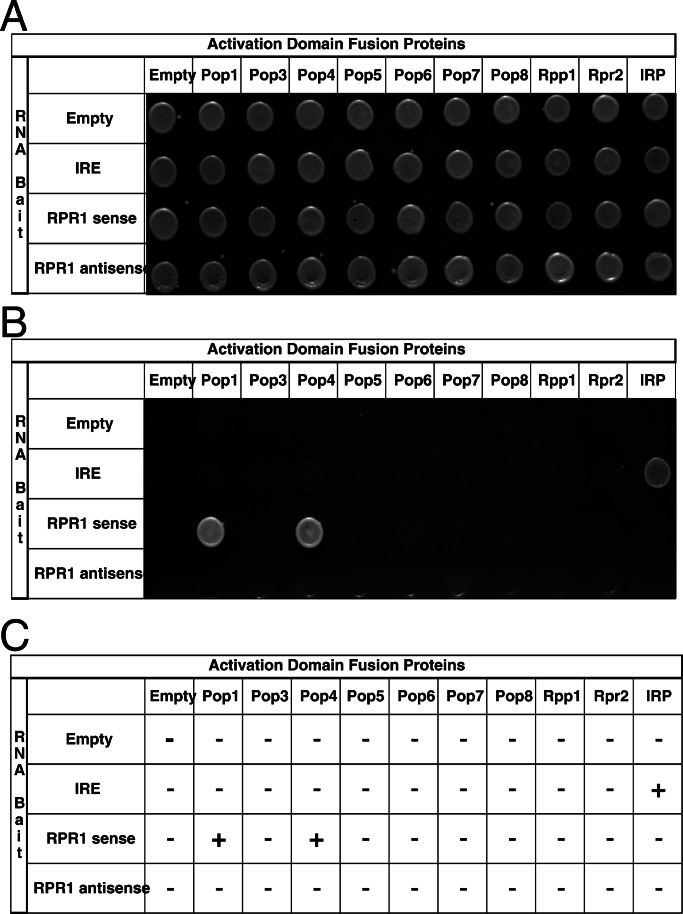
Figure 2.
Identification of RNase P protein subunits that interact specifically with the RPR1 RNA, using three-hybrid analysis. Yeast strains transformed with the indicated plasmids were tested for their ability to grow in the presence of histidine (A) and absence of histidine plus 20 mM 3-aminotriazole (B). A summary of the results from the β-galactosidase activity of three-hybrid strains is shown in C. A positive RNA–protein is indicated by the ability of the yeast strains to grow in the absence of histidine and their ability to produce β-galactosidase. − Indicates no blue color was observed relative to positive control strain transformed with pIRE and pIRP and + indicates a dark blue color similar to that observed with the positive control.
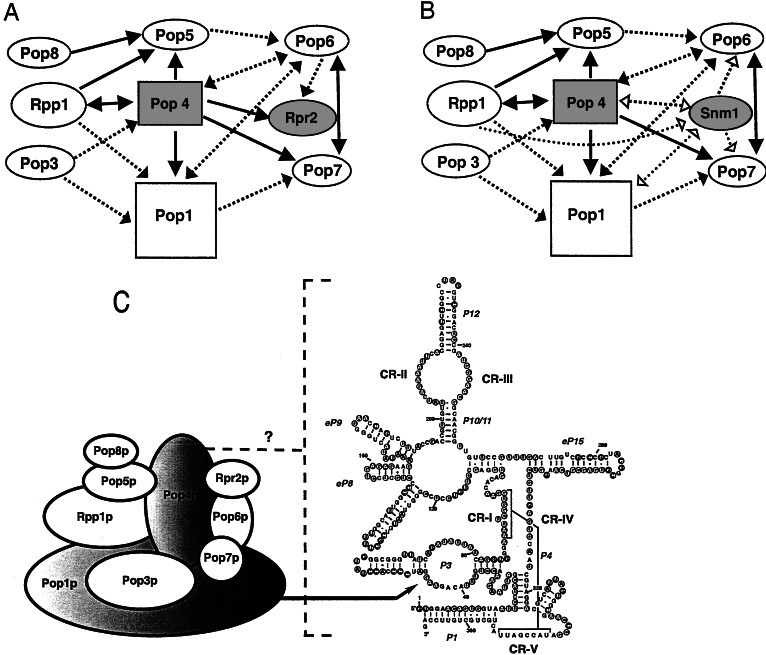
Figure 3.
Schematic representation of the protein–protein and protein–RNA interactions among RNases P and MRP subunits as derived from data in Figs. 1 and 2. (A) RNase P protein subunits that bind specifically to the RNase P RNA are drawn as rectangles. The direction of the arrow indicates which vector the protein subunit was expressed from when the protein–protein interaction was observed; the protein subunit at the beginning of the arrow was fused to the activation domain and the protein subunit at the end of the arrow was fused to the DNA binding domain (pACTII → pLexA). Arrowheads at both ends (↔) indicates that the interaction is reciprocal—i.e., observed in both directions. A dashed line (– – –) indicates that the potential protein–protein interaction was only observed in the β-galactosidase assay. Protein subunits that interact with themselves are shown shaded. (B) A summary of the protein–protein interactions that include Snm1p, rather than Rpr2p. Additional protein–protein interactions observed for Snm1p, but not seen for the Rpr2p subunit of RNase P, are indicated by dashed lines with open arrowheads. Other conventions are the same as used in A. (C) A spatial model of the results for the two-hybrid and three-hybrid test results of S. cerevisiae nuclear RNase P is shown. Ovals represent the protein subunits of yeast nuclear RNase P. The shaded ovals of Pop1p and Pop4p indicate that they bind specifically to the RPR1 RNA in the three-hybrid test. One of the recognition sites in the RPR1 RNA for Pop1p binding is the P3 loop, as indicated by the arrow (55). The binding site(s) for Pop4p are currently unknown, which is indicated by the dotted bracket and a question mark. The predicted secondary structure of RPR1 RNA is adapted from the model proposed by Frank et al. (64). The five critical regions, CR-I to CR-V, are numbered based on the Chen and Pace nomenclature (13). “P” represents helical regions, with numbers assigned according to the bacterial structure (65). “eP” indicates the eukaryotic paired regions whose homology to particular bacterial structures is uncertain, but which occupy the same positions as in the bacterial structure (64). Nucleotides in filled circles show protection from chemical modification and nuclease attack in the holoenzyme, whereas the nucleotides in open circles indicate exposure to solution (66).
The complete ORF of each protein subunit was fused independently to the LexA protein in the pBTM116 vector (bait) or to the Gal-4 activation domain in the pACTII vector (prey). All combinations of bait and prey plasmids were transformed into the tester strain L40, which has two reporter genes for two-hybrid interactions, HIS3 and lacZ. Thus, transformants were assayed for cell growth in the absence of histidine (plus 3-AT) and for β-galactosidase activity. The growth test for the HIS3 reporter gene is shown in Fig. 1. Fig. 1A is a control that shows that all strains grow on media selecting only for the presence of the bait and prey plasmids. In Fig. 1B, expression of the HIS3 reporter is also required for growth, and only a subset of bait/prey combinations are viable. When a positive interaction was noted by growth in the absence of histidine (plus 3-AT), the transformants grew well up to 20 mM 3-AT, the highest concentration tested. The results of the β-galactosidase assays for the two-hybrid test are shown in Fig. 1C. All potential interactions were compared with the positive control strain, L40 transformed with pLexA-Ras and pVP16-Rip51. This positive control strain grows well in the presence of 20 mM 3-AT and gives a strong β-galactosidase signal in the filter assay. All binary combinations that gave a strong positive signal for HIS3 expression, also gave a strong β-galactosidase signal and are denoted by solid lines in Fig. 3. In contrast, there were several instances where we observed clearly reproducible β-galactosidase signals, but there was little or no growth in the absence of histidine. It is possible that the more sensitive β-galactosidase assays were detecting more transient interactions between bait and prey subunits in these cases. These presumably weaker interactions are noted by dashed lines in Fig. 3, although they are interpreted with less confidence than cases in which both reporter genes were expressed.
Not all interactions were reciprocal. For example, Pop4p interacted strongly with Pop5p only when Pop4p was expressed from the prey plasmid; no interaction was noted when Pop5p was expressed from the prey plasmid and Pop4p expressed from the bait plasmid. In contrast, Pop4p interacted strongly with Rpp1p whether it was expressed from the bait or prey plasmids. This lack of reciprocity is not unusual when testing two-hybrid interactions and several explanations have been suggested, including poor folding of some protein fusions or failure of a particular fusion protein to enter the nucleus (51).
It is worth noting that no interacting partners were observed for Pop3p and Pop8p when these ORFs were fused to pLexA (bait). This lack of interaction was not due to the absence of protein expression as determined by Western blot analysis (data not shown). In addition, expression of these proteins on these high copy plasmids was not toxic to the cells, because transformants containing these pLexA fusions grew well if we did not select for HIS3 expression. Thus, we cannot rule out that these fusion proteins might have a problem such as misfolding. Except for Pop8p, all subunits tested interacted with at least two other protein subunits when used as bait and prey. Pop4p, Rpp1p, and Pop5p were each involved in multiple strong protein–protein interactions as represented in Fig. 3. Pop4p binds seven of the eight other protein subunits, in addition to its ability to interact strongly with itself. Rpr2p, the unique RNase P protein subunit, bound strongly to Pop4p and itself, but interacted only weakly with Pop6p.
Interactions with the Unique Protein Subunit of RNase MRP, Snm1p.
Although RNase MRP has not been biochemically purified, genetic and immunoprecipitation studies have suggested that eight subunits of yeast nuclear RNase P are also associated with RNase MRP. Snm1p is the only unique protein subunit of RNase MRP identified to date (Table 1). To compare protein–protein interactions of Snm1p with those of Rpr2p, we tested the complete ORF of Snm1p as bait and prey in the two-hybrid system against the eight shared subunits of RNases MRP and P (Fig. 1). Like Rpr2p, Snm1p interacts strongly with itself and weakly with Pop6p. In contrast, the interaction of Snm1p with Pop4p is weaker than the interaction of Rpr2p with Pop4p. Snm1p also interacts with several other shared subunits, Pop1p, Pop7p, and Rpp1p, that do not give a signal with Rpr2p. The Snm1p protein–protein interactions are summarized in Fig. 3B.
Identification of Protein–RNA Interactions.
At least one of the nine RNase P protein subunits must bind directly to the RPR1 RNA. However, specific RNA–protein interactions between in vitro transcribed RPR1 RNA and purified recombinant proteins have been difficult to interpret. Results from gel shift mobility assays have demonstrated that the majority of the protein subunits exhibit a high degree of nonspecific RNA binding activity.
To identify specific protein–RNA interactions between RPR1 RNA and the nine protein subunits, we therefore performed an in vivo three-hybrid test. The vectors expressing the activation domain fusion proteins (prey) used in the two-hybrid test were cotransformed into the L40-coat reporter strain with pIIIA-MS2–2 RNA hybrid plasmid containing no RNA insert, a negative control IRE RNA, RPR1 RNA in the sense orientation, or RPR1 RNA in the antisense orientation as an additional negative control. Northern analysis was performed to confirm that the RNA hybrids were expressed in roughly comparable steady-state amounts (data not shown). Double transformants were tested for HIS3 expression (Fig. 2B) and assayed for β-galactosidase activity (Fig. 2C). Pop1p and Pop4p were the only subunits that bound the RPR1 RNA in the sense orientation but not the antisense orientation. None of the proteins were shown to interact with RNA from control plasmids containing no RNA insert or the IRE RNA. It should be noted that Pop4p tends to give HIS3 positives with several RNA baits (such as the P3 subdomain of RNase P RNA and NME1 RNA in both the sense and antisense direction) at 3-AT concentrations lower than 5 mM; thus, the three-hybrid tests are shown at 20 mM 3-AT. This tendency of Pop4p to bind several different RNA baits, in addition to the sense RPR1 RNA, suggests that Pop4p has greater nonspecific RNA binding tendencies than Pop1p, even though it shows specificity in the sense versus antisense test. In the β-galactosidase filter assay, Pop1p and Pop4p were the only proteins that gave a positive test (Fig. 3C).
We did not detect specific binding of any of these proteins, including Snm1p, when the RNA subunit of RNase MRP, NME1 RNA, was used as bait (data not shown). This was somewhat surprising given that Pop1p was expected to bind to the conserved P3 region of the NME1 RNA (31, 55). Northern analysis revealed that the NME1 hybrid RNA was being expressed (data not shown). The negative result is possibly due to misfolding of the NME1 RNA in this context, and such negative results in this test are considered inconclusive.
We conclude that the three-hybrid test allowed us to identify the proteins that bound RPR1 RNA specifically in the absence of stoichiometric levels of the other subunits. These results leave open the possibility that other protein subunits also contact the RNA, but that the protein–RNA binary interaction is insufficiently stable to obtain a signal in the absence of other subunits. This caveat also applies to binary protein–protein interactions that were not observed in the two-hybrid test.
Discussion
Despite the progress that has been made in the identification of the protein and RNA components of eukaryotic RNase P, little is known about the architecture of the enzyme. Nuclear RNase P purified from S. cerevisiae contains a single RNA subunit and nine protein subunits. The relatively complex subunit composition of the yeast RNase P and the extensive protein subunit overlap between RNases P and MRP raises an obvious question: What is the spatial organization of the protein subunits within the RNases P and MRP complexes? Here we describe the in vivo binary interactions between subunits of the yeast nuclear RNase P enzyme. We emphasize at the beginning of this discussion that many interactions among subunits could be overlooked in this approach, especially if the interaction requires three or more subunits for stability. It is also technically possible that the observed subunit–subunit interactions are really indirect, with the interaction mediated by additional subunits that are recruited from the yeast nucleus. It seems unlikely that this is generally the case, because both the bait and the prey fusion proteins are expressed at much higher levels than the other subunits and only a very limited number of subunit–subunit contacts give positives relative to the 100 possible binary interactions.
The two-hybrid analysis allowed us to identify extensive protein–protein interactions among protein subunits of S. cerevisiae RNases P and MRP. A summary of these protein–protein interactions is shown in Fig. 3 A and B with a spatial model shown in C. Several strong protein–protein interactions were identified for Pop1p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, Rpp1p, and Rpr2p. Data presented here suggest that Pop4p plays a central role in the RNases P and MRP, interacting with both the shared and unique subunits of each enzyme as well as the RNA subunit of RNase P.
A recent two-hybrid analysis of human RNase P with a partial set of the protein components did not identify any strong interactions among the protein subunits (57). Several of the weak interactions were consistent with some of the observations in this study and inconsistent with others. It should be noted when considering differing two-hybrid results that not all yeast protein subunits have recognizable human homologs and at least four human proteins have not been found in the yeast enzyme. The protein–protein and/or protein–RNA interactions within the individual complexes might be influenced by proteins that are found in one organism and not the other.
Our data indicate that Pop4p, Rpr2p, and Snm1p can strongly interact with themselves as well as with other proteins. This suggests that these proteins may be present in more than on copy per enzyme. The question of protein stoichiometry in the RNase P complex has not been determined because of the miniscule quantities obtained in biochemical purification. The only information available concerning subunit stoichiometry comes from a recent study showing that there is one RNase P RNA subunit per RNase P complex and that RNases P and MRP are not part of the same ribonucleoprotein particles in the cell (39).
Only Pop1p and Pop4p could be identified by the three-hybrid test as interacting specifically with the RNase P RNA subunit. In separate work, our lab has shown by mutational analysis of RNase P RNA that Pop1p interacts directly and specifically with the essential P3 subdomain of RNase P RNA (55). Pop4p bound specifically to the sense orientation of the RNase P RNA, but not to the antisense orientation (Fig. 2). At this time, the RNA determinants for Pop4p binding remain elusive, but they do not absolutely require the P3 subdomain of RNase P (unpublished data) and it might be that Pop4p is interacting with multiple sites in the RNA. A recent three-hybrid test of human RNase P RNA (H1 RNA) identified possible interactions with the human homologs of Pop4p (Rpp29p), Rpp1p (Rpp30p), and Rpr2p (Rpp21p) (56). The human results are in stark contrast to our results in yeast; the human three-hybrid study did not identify an interaction between H1 RNA and human Pop1p whereas we could not detect interactions between RPR1 RNA and either Rpp1p or Rpr2p. In line with the behavior of the yeast subunits, a three-hybrid test of the recently identified archaeal homologs of Rpp1p (MTH688p) and Rpr2p (MTH1618p) did not identify any interactions with the archaeal RNase P RNA (T. A. Hall and J. W. Brown, personal communication). Taking into account the low degree of identity between the protein homologs, it may be that specific binding of a protein to its cognate RNA in one organism requires additional subunit partners in another, thereby resulting in subtle differences in the spatial organization and assembly of RNase P enzymes among organisms or stability of the binary complex.
Currently, the functions of the individual protein subunits remains elusive. The yeast Pop3p protein has been shown to bind pretRNA and a variety of RNA molecules with high affinity while displaying a preference for single-stranded RNAs (58). In humans, Rpp21p (the homolog of yeast Rpr2p) has also been observed to bind precursor tRNA (19). It has been suggested that human Pop7p has ATPase activity (59). However, the significance of the latter observation is unclear with respect to S. cerevisiae Pop7p because the ATPase signature motif does not appear to be present in the yeast homolog. It is possible that only a subset of the protein subunits are necessary for RNase P catalytic activity while other proteins function in assembly and/or localization.
The low abundance of RNase P enzyme and our present inability to reconstitute RNase P from purified subunits precludes a meaningful biochemical test of the functional subunit interactions in the enzyme. The correlation of several protein–protein interactions in our yeast two-hybrid analysis with some of those observed in the archaeal and human two-hybrid tests supports the notion that these results recapitulate interactions in RNase P enzyme.
The subunit interaction map provided here provides several types of information that will prove useful in further investigations of the enzymes. First, it is clear that an extensive network of protein–protein contacts are possible in the absence of the RNA subunit—i.e., the RNA subunit may not necessarily be needed to nucleate protein complex formation and help bind each new protein. It is not yet known whether all, or a subset, of proteins can preassociate with each other before complexing with the RNA in the assembly of RNase P. This work can help guide interpretation of the investigation of enzyme assembly. The second line of experiments that will benefit from this map is the question of substrate recognition. It is clear that the nuclear enzyme shares some substrate recognition determinants with the bacterial enzymes, but that there are also one or more additional sites in the nuclear RNase P (60). A particularly interesting question for further study is which subunits in RNases P and MRP cause the substrate specificity to shift from pretRNAs to prerRNAs. It seems likely that the differences in either the RPR1/NME1 RNA subunits or the Rpr2p/Snm1p protein subunits, or both, are responsible. However, it is theoretically possible that the effects exerted by the changed subunits are indirect, for example by altering the geometry of RNA-binding common protein subunits in the RNase P and RNase MRP complexes. The predicted relationships of the proteins will facilitate spatial interpretation of various substrate interactions.
Acknowledgments
We are thankful to Ann Vojtek for helpful suggestions concerning this work. We are grateful to Marvin Wickens, Steve Elledge, and Ann Vojtek for many reagents and plasmids. Funding for this work was provided by National Institutes of Health Grant GM34869 (to D.R.E.), National Science Foundation Grant MCB-0077949 (to L.L. and J.M.Z.), and postdoctoral fellowships to F.H.-S. from United Negro College Fund/Pfizer and the Ford Foundation.
Abbreviation
- RNase P
ribonuclease P
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Frank D N, Pace N R. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Xiao S, Houser-Scott F, Engelke D. J Cell Physiol. 2001;187:11–21. doi: 10.1002/1097-4652(200104)187:1<11::AID-JCP1055>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman S. In: The RNA World. Gesteland R F, Cech T, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Press Lab. Press; 1999. pp. 351–380. [Google Scholar]
- 4.Schön A. FEMS Microbiol Rev. 1999;23:391–406. doi: 10.1111/j.1574-6976.1999.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 5.Pannucci J A, Haas E S, Hall T A, Harris J K, Brown J W. Proc Natl Acad Sci USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerrier-Takada C, Altman S. Science. 1984;223:285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- 7.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 8.Crary S M, Niranjanakumari S, Fierke C A. Biochemistry. 1998;37:9409–9416. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- 9.Kurz J C, Niranjanakumari S, Fierke C A. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 10.Niranjanakumari S, Stams T, Crary S M, Christianson D W, Fierke C A. Proc Natl Acad Sci USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gopalan V, Baxevanis A D, Landsman D, Altman S. J Mol Biol. 1997;267:818–829. doi: 10.1006/jmbi.1997.0906. [DOI] [PubMed] [Google Scholar]
- 12.Hall, T. A. & Brown, J. W. (2002) RNA, in press.
- 13.Chen J L, Pace N R. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- 14.Jarrous N, Eder P S, Wesolowski D, Altman S. RNA. 1999;5:153–157. doi: 10.1017/s135583829800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarrous N, Altman S. Methods Enzymol. 2001;342:93–100. doi: 10.1016/s0076-6879(01)42538-9. [DOI] [PubMed] [Google Scholar]
- 16.van Eenennaam H, Pruijn G J, van Venrooij W J. Nucleic Acids Res. 1999;27:2465–2472. doi: 10.1093/nar/27.12.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lygerou Z, Pluk H, van Venrooij W J, Seraphin B. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- 18.van Eenennaam H, Lugtenberg D, Vogelzangs J H P, van Venrooij W J, Pruijn G J M. J Biol Chem. 2001;276:31635–31641. doi: 10.1074/jbc.M103399200. [DOI] [PubMed] [Google Scholar]
- 19.Jarrous N, Reiner R, Wesolowski D, Mann H, Guerrier-Takada C, Altman S. RNA. 2001;7:1153–1164. doi: 10.1017/s1355838201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 22.Dichtl B, Tollervey D. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolc V, Altman S. Genes Dev. 1997;11:2414–2425. doi: 10.1101/gad.11.18.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu S, Zengel J M, Lindahl L. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt M E, Clayton D A. Genes Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt M E, Clayton D A. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu S, Archer R H, Zengel J M, Lindahl L. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forster A C, Altman S. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt M E, Bennett J L, Dairaghi D J, Clayton D A. FASEB J. 1993;7:208–213. doi: 10.1096/fasebj.7.1.7678563. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt M E. J Mol Biol. 1999;292:827–836. doi: 10.1006/jmbi.1999.3116. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl L, Fretz S, Epps N, Zengel J M. RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrissey J P, Tollervey D. Trends Biochem Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- 33.Reimer G, Raska I, Scheer V, Tan E M. Exp Cell Res. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson M R, Cao L-G, Wang Y-L, Pederson T. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertrand E, Houser-Scott F, Kendall A, Singer R H, Engelke D R. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee B, Matera A G, Ward D C, Craft J. Proc Natl Acad Sci USA. 1996;93:11471–11476. doi: 10.1073/pnas.93.21.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matera A G, Frey M R, Margelot K, Wolin S L. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobson M R, Cao L-G, Taneja K, Singer R H, Wang Y L, Pederson T. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 39.Srisawat C, Engelke D R. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spitzfaden C, Nicholson N, Jones J J, Guth S, Lehr R, Prescott C D, Hegg L A, Eggleston D S. J Mol Biol. 2000;295:105–115. doi: 10.1006/jmbi.1999.3341. [DOI] [PubMed] [Google Scholar]
- 41.Stams T, Niranjanakumari S, Fierke C A, Christianson D W. Science. 1998;280:752–755. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- 42.Chen J L, Nolan J M, Harris M E, Pace N R. EMBO J. 1998;17:1515–1525. doi: 10.1093/emboj/17.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massire C, Jaeger L, Westhof E. J Mol Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 44.Walhout A J M, Boulton S J, Vidal M. Yeast. 2000;17:88–94. doi: 10.1002/1097-0061(20000630)17:2<88::AID-YEA20>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legrain P, Selig L. FEBS Lett. 2000;480:32–36. doi: 10.1016/s0014-5793(00)01774-9. [DOI] [PubMed] [Google Scholar]
- 46.Flores A, Briand J-F, Gadal O, Andrau J-C, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, et al. Proc Natl Acad Sci USA. 1999;96:7815–7520. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 49.Becker D M, Guarente L. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 50.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartel P L, Fields S. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 52.Bartel P L, Chien C-T, Sternglanz R, Fields S. In: Cellular Interactions in Development: A Practical Approach. Hartley D A, editor. Oxford: Oxford Univ. Press; 1993. pp. 153–179. [Google Scholar]
- 53.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 54.Breeden L, Nasmyth K. Cold Spring Harbor Symp Quant Biol. 1985;50:643–649. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 55.Ziehler W A, Morris J, Scott F H, Millikin C, Engelke D R. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang T, Guerrier-Takada C, Altman S. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang T, Altman S. Proc Natl Acad Sci USA. 2001;98:920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brusca E M, True H L, Celander D W. J Biol Chem. 2001;276:42543–42548. doi: 10.1074/jbc.M107293200. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Altman S. Proc Natl Acad Sci USA. 2001;98:441–444. doi: 10.1073/pnas.021555498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziehler W A, Day J J, Fierke C A, Engelke D R. Biochemistry. 2000;39:9909–9916. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]
- 61.Lee J Y, Rohlman C E, Moloney L A, Engelke D R. Mol Cell Biol. 1991;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stolc V, Katz A, Altman S. Proc Natl Acad Sci USA. 1998;95:6716–6721. doi: 10.1073/pnas.95.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitt M E, Clayton D A. Genes Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- 64.Frank D N, Adamidi C, Ehringer M A, Pitulle C, Pace N R. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haas E S, Brown J W, Pitulle C, Pace N R. Proc Natl Acad Sci USA. 1994;91:2527–2531. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tranguch A J, Kindelberger D W, Rohlman C E, Lee J Y, Engelke D R. Biochemistry. 1994;33:1778–1787. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]