Abstract
Cellular metabolism of glucose is required for stimulation of insulin secretion from pancreatic β cells, but the precise metabolic coupling factors involved in this process are not known. In an effort to better understand mechanisms of fuel-mediated insulin secretion, we have adapted 13C NMR and isotopomer methods to measure influx of metabolic fuels into the tricarboxylic acid (TCA) cycle in insulinoma cells. Mitochondrial metabolism of [U-13C3]pyruvate, derived from [U-13C6]glucose, was compared in four clonal rat insulinoma cell 1-derived cell lines with varying degrees of glucose responsiveness. A 13C isotopomer analysis of glutamate isolated from these cells showed that the fraction of acetyl-CoA derived from [U-13C6]glucose was the same in all four cell lines (44 ± 5%, 70 ± 3%, and 84 ± 4% with 3, 6, or 12 mM glucose, respectively). The 13C NMR spectra also demonstrated the existence of two compartmental pools of pyruvate, one that exchanges with TCA cycle intermediates and a second pool derived from [U-13C6]glucose that feeds acetyl-CoA into the TCA cycle. The 13C NMR spectra were consistent with a metabolic model where the two pyruvate pools do not randomly mix. Flux between the mitochondrial intermediates and the first pool of pyruvate (pyruvate cycling) varied in proportion to glucose responsiveness in the four cell lines. Furthermore, stimulation of pyruvate cycling with dimethylmalate or its inhibition with phenylacetic acid led to proportional changes in insulin secretion. These findings indicate that exchange of pyruvate with TCA cycle intermediates, rather than oxidation of pyruvate via acetyl-CoA, correlates with glucose-stimulated insulin secretion.
Glucose stimulates insulin secretion by means of its metabolism in pancreatic islet β cells, but the coupling factors that relate metabolism of the hexose to exocytosis of insulin have not been fully delineated. A minimal working model holds that an increase in glucose concentration causes a rise in the [ATP]/[ADP] ratio in β cells, resulting in closure of ATP-regulated K+ (KATP) channels, membrane depolarization, influx of Ca2+, and subsequent exocytosis of insulin (1, 2). Glucose oxidation increases in proportion to the external glucose concentration in β cells, and this has been taken as evidence for a direct link between fuel oxidation, ATP production, and insulin secretion (3). However, β cells also have high levels of pyruvate carboxylase (PC) activity (4), which is remarkable in light of their lack of phosphoenolpyruvatecarboxykinase (PEPCK) expression (5) and low lipogenic capacity (6). Radioisotopic methods have been used to estimate that 40% of the pyruvate generated during glucose stimulation of β cells enters mitochondrial metabolism via PC-catalyzed conversion to oxaloacetate (OAA), with most of the remainder metabolized to acetyl-CoA via the pyruvate dehydrogenase (PDH) reaction (7–10). It has been further proposed that PC-catalyzed anaplerotic influx of pyruvate into the tricarboxylic acid (TCA) cycle is linked to efflux of other intermediates from the mitochondria, including malate (11) or citrate (12), resulting in synthesis of important coupling factors. Cytosolic malate can be reconverted to pyruvate via the malic enzyme, completing a pyruvate-malate cycle. An alternate cycle occurs when citrate leaves the mitochondria to be cleaved to acetyl-CoA and OAA by citrate lyase. Acetyl-CoA so formed can be converted to malonyl-CoA, which has been proposed as a coupling factor (13, 14), although evidence against this idea has also been presented (15, 16). The OAA formed by means of citrate cleavage can in turn be converted to malate via cytosolic malate dehydrogenase activity, and then back to pyruvate via malic enzyme to complete a pyruvate-citrate cycle. A cofactor common to both the pyruvate-malate and pyruvate-citrate cycles is NADPH produced as a byproduct of the malic enzyme (7).
Although the foregoing studies clearly establish that pyruvate enters mitochondrial metabolism both through PDH and PC in islet cells, direct evidence linking either of these pathways to insulin secretion is lacking. Indirect evidence for a role of PC-catalyzed anaplerosis (formation of OAA from pyruvate) comes from the following studies. (i) Comparison of 14CO2 production from [3,4-14C]glucose and [6-14C]glucose reveals that pyruvate carboxylation is highly active in β cells but virtually lacking in α cells (10). (ii) Incubation of rat insulinoma cell (INS) 1 insulinoma cells or isolated rat islets with phenylacetic acid (PAA), an inhibitor of PC, impairs glucose-stimulated insulin secretion (GSIS) without affecting glycolytic rate (12). (iii) Engineering of INS-1 cells for expression of glycerol kinase confers glycerol-stimulated insulin secretion, such that glycerol and glucose have similar maximal potency for stimulation of insulin secretion, despite relatively low rates of glycerol oxidation (17). These findings suggest that it would be unwise to neglect the potential contribution of pyruvate cycling to signal generation in GSIS, and that further investigation is warranted.
In the current study, we have combined novel cell line models with analysis of metabolism by 13C NMR to provide a better understanding of the potential role of PC-catalyzed pyruvate metabolism in regulation of insulin secretion. We have shown that 13C-isotopomer analysis of glutamate spectra can provide detailed metabolic information about pathways intersecting with the TCA cycle (18–20). In the current study, this technology has been applied to a unique set of cell lines derived from rat INS-1 insulinoma cells. These cell lines were procured by stable transfection of parental INS-1 cells with a plasmid containing an antibiotic-resistance marker (21, 22). From among 60 cell lines screened, 70% exhibited weak GSIS (≤2-fold), 17% had moderate responses (2–5-fold), and a few clones had increases in insulin secretion of 10–15-fold as glucose was raised from 3 to 15 mM. Importantly, the response characteristics of the robustly and poorly responsive clones were stable over long periods of continuous tissue culture (21). This stability has allowed us, in the current study, to use 13C NMR to measure the relative contributions of PC- and PDH-gated pathways of pyruvate metabolism in clones with weak, moderate, or robust glucose responsiveness. We find that PC-catalyzed pyruvate cycling, but not the fractional contribution of glucose to acetyl-CoA formation (PDH-catalyzed pyruvate metabolism), is directly correlated with the degree of glucose responsiveness in our INS-1-derived cell lines. Furthermore, PAA impairs, whereas malate, given as the cell-permeable methyl ester, potentiates GSIS in glucose-responsive clones in close correlation with changes in PC-catalyzed cycling of pyruvate. These experiments provide evidence for a key role of pyruvate cycling in control of insulin secretion.
Methods
Cell Lines.
Four clonal cell lines (832/1, 832/2, 832/13, and 834/40), derived from INS-1 insulinoma β cells using a transfection-selection strategy (21, 22), were used in these studies. Cells were grown to confluence in RPMI medium 1640 containing 11.1 mM glucose and supplemented with 10% FBS/100 units/ml penicillin/100 μg/ml streptomycin/10 mM Hepes/2 mM glutamine/1 mM sodium pyruvate/50 μM β-mercaptoethanol in 15-cm plates at 37°C in a humidified atmosphere containing 5% CO2.
Oxygen Consumption.
O2 consumption by cells was measured with a YSI model 5300 biological oxygen monitor equipped with a Clarke (Yellow Springs, Ohio) oxygen electrode. The electrode was calibrated by assuming an oxygen concentration of 221 μM for air-saturated water, 37°C, and zero after addition of excess sodium dithionite. Actively respiring cells were added to an air-saturated solution in a water-jacketed airtight chamber, the chamber was sealed, and disappearance of O2 was recorded over a 15–30-min period. Total protein in the chamber was measured, and O2 consumption was reported as nmol/min/mg of cell protein.
NMR Spectroscopy.
After cells were grown to confluence, tissue culture medium was removed, and the cells were washed once with PBS. The cells were preincubated in Hepes-buffered saline solution (HBSS;114 mM NaCl/4.7 mmol KCl/1.2 mM KH2PO4/1.16 mM MgSO4/20 mM Hepes/2.5 mM CaCl2/25.5 mM NaHCO3, pH 7.2) containing 3 mM glucose for 1 h then incubated with 3, 6, or 12 mM [U-13C6]glucose (Cambridge Isotope Laboratories, Cambridge, MA) in the same Hepes-buffered saline for 4 h at 37°C. In addition, dimethylmalate (DMM) and PAA (both from Sigma) were added to the HBSS in certain experiments at the concentrations indicated in the respective legends. Samples from the assay buffer were removed before preparation of the cell extracts for NMR and used for insulin radioimmunoassays as described (21). Then, the cells were washed once with ice-cold PBS and extracted with ice-cold 3.5% perchloric acid. Extracts from 3 plates were pooled, neutralized, and lyophilized. In a few samples, cellular glutamate was purified by reconstituting the freeze-dried extract in water, applying the sample to a Dowex-50 column (H+ form), and eluting the amino acid mixture with 2N ammonium hydroxide. The eluant was freeze-dried and dissolved in 0.5 ml of 2H2O for analysis by 13C NMR. Proton-decoupled 13C NMR spectra were recorded on either a 500 (11.75T) or 600 MHz (14T) Varian INOVA NMR spectrometer by using a 45° pulse and a 3-s repetition time on a 5-mm tunable broadband probe. The areas of the multiplets arising from 13C-13C spin-spin coupling in the glutamate C2, C3, and C4 resonances were determined by using the line-fitting routine in the PC-based NMR program, nuts (Acorn NMR, Fremont, CA). These multiplet areas were used to perform a 13C-isotopomer analysis (18–20) to determine a metabolic profile for metabolism of [U-13C6]glucose in the TCA cycle. Preliminary experiments showed that cellular glutamate became highly enriched in 13C in these cells as a result of oxidation of [U-13C6]glucose in the TCA cycle and that cellular glutamate was readily detected by 13C NMR in extracts of ≈1.5 × 108 cells. Preliminary experiments on cells incubated with [U-13C6]glucose for various times indicated that isotopic steady-state was reached by ≈3 h.
13C Isotopomer Model.
Cellular uptake of [U-13C6]glucose results in the production of two equivalents of [U-13C3]pyruvate via glycolysis. The resulting [U-13C3]pyruvate has three possible metabolic fates: conversion to [U-13C3]lactate via lactate dehydrogenase (LDH), conversion to [U-13C2]acetyl-CoA via PDH, or conversion to [1,2,3-13C3]OAA via PC. If [U-13C3]pyruvate is converted exclusively to [U-13C2]acetyl-CoA and oxidized in the TCA cycle, then all intermediates involved in cycle reactions will become enriched with 13C in a predictable pattern. The five-carbon intermediate α-ketoglutarate (α-KG), for example, would consist of 16 distinct 13C isotopomers at isotopic steady state. If [U-13C3]pyruvate also enters the cycle via PC, then the same mix of 16 13C isotopomers of α-KG will be present at steady-state; however, the population of those 16 isotopomers will be heavily weighted by influx of [1,2,3-13C3]OAA formed via carboxylation of [U-13C3]pyruvate. These metabolic alternatives are easily distinguished by 13C NMR (Fig. 1). As cellular α-KG is too low to detect by NMR under most circumstances, the NMR analysis is based instead on tissue glutamate, a relatively abundant intermediate in rapid exchange with α-KG. Thus, one underlying assumption of 13C isotopomer analysis is that 13C NMR spectrum reported by glutamate at metabolic and isotopic steady-state mirrors the isotopomer distribution in α-KG.
Figure 1.
Modeling of pyruvate metabolism with13C isotopomer analysis. Two possible metabolic fates of [U-13C3]pyruvate (derived from [U-13C6]glucose) and the resultant modeled spectra of glutamate carbons 2 (C2) and 4 (C4) [simulated by using tcasim from (www2.swmed.edu/rogersmr/available_products.htm)]. (Top) We have assumed that [U-13C3]pyruvate enters the TCA cycle only via PDH, whereas in the Bottom, flux of [U-13C3]pyruvate through PDH and PC are equal. The 13C spectrum of glutamate clearly differentiates these two metabolic extremes.
Entry of any metabolite into the TCA cycle via a pathway other than acetyl-CoA is referred to as anaplerosis, a term originally coined by Kornberg (23) to represent any process that replenishes a TCA cycle intermediate. Kornberg's original definition applies only when the amount of one or more of a TCA cycle intermediate increases as, for example, during an altered metabolic state. However, flux through anaplerotic pathways can continue after the system reaches a new metabolic steady state by balancing flux into the cycle by a corresponding flux out. An example of this would be continual production of glucose from lactate by the liver. Such anaplerotic flux can be detected and quantified by isotopomer analysis of tissue glutamate collected at metabolic and isotopic steady-state (20). Fig. 1 illustrates the multiplet pattern anticipated for glutamates C2 and C4 for two different metabolic models, one where [U-13C3]pyruvate enters the TCA cycle exclusively via PDH and another where [U-13C3]pyruvate enters the cycle via both PDH and PC. Clearly, these two metabolic extremes are easily distinguished by a 13C NMR spectrum of glutamate. In this work, 13C isotopomer analysis of cell glutamate provided a detailed picture of metabolism in INS-1 cells after incubation with [U-13C6]glucose under “normal” tissue culture conditions. This analysis provided a direct comparison of mitochondrial metabolism in INS-1-derived cell lines with varying capacities for GSIS.
Results
NMR Spectra.
Fig. 2 shows representative 13C NMR spectra of the amino acid fraction of extracts of robustly glucose-responsive 832/13 cells (21) after incubation with 3, 6, or 12 mM [U-13C6]glucose for 4 h. Several features of these spectra deserve comment. First, it is clear that the amount of [U-13C6]glucose contributing to the acetyl-CoA pool in these cells increased dramatically as glucose in the incubation media was increased from 3 to 12 mM. This result is evidenced by the increase in area of the quartet (4Q) relative to the doublet (4D45) in the glutamate C4 resonance. Second, the multiplet pattern in the glutamate C2 resonance also changes, and the individual lines appear to become broader at the higher glucose concentration. This apparent line-broadening is the result of long-range coupling between glutamates C2 and C5 (24), reflecting an increase in [U-13C5]glutamate with increasing glucose concentration. However, the total glutamate concentration, as estimated by NMR areas, did not increase with increasing glucose concentration, in opposition to one recent report (25), but in agreement with another (26). Interestingly, the spectra show that total tissue aspartate decreased with increasing glucose in the incubation media.
Figure 2.
13C NMR spectra of extracts of 832/13 cells after a 4-h incubation with [U-13C6]glucose. The robustly glucose-responsive INS-1-derived cell line 832/13 (21) was incubated with 3, 6, or 12 mM [U-13C6]glucose, followed by extraction of cellular glutamate for NMR analysis. GluC2, GluC3, and GluC4 are the resonances of carbons 2, 3, and 4 of glutamate, respectively. AspC3 is the resonance for the C3 carbon of tissue aspartate. An expanded Glu-C4 resonance from cells incubated with 12 mM glucose is shown the top of the figure. The arrows designate three small resonances (a singlet and a doublet) that could have been derived only by entry of a TCA cycle intermediate into the pyruvate pool feeding acetyl-CoA (see text). The two small arrows over the GluC2 resonance identify a doublet (D23) whose area is particularly sensitive to pyruvate cycling.
Next, we compared NMR spectra from two robustly glucose responsive cell lines, 832/13 and 834/40, with those generated from two lines with lesser glucose responsiveness, 832/1 and 832/2 (spectra not shown). Comparisons of spectra of cell extracts after a 4-h incubation period with 12 mM [U-13C6]glucose revealed two distinct and consistent differences. First, the singlet and doublet components evident in C4 (see the expanded resonance of Fig. 2) were larger in spectra of the highly responsive cells vs. the poorly responsive cells. These peaks can arise only from labeled TCA cycle intermediates that leave the cycle and reenter a pyruvate pool destined for conversion to acetyl-CoA. Their presence proves that a portion of pyruvate is derived from a TCA cycle intermediate in these cells. A second difference between these spectra was a consistently larger D23 component (marked by the arrows over the C2 resonance of Fig. 2) in spectra of the robustly glucose responsive cells. We have shown that the D23 component of the C2 resonance is a sensitive marker of pyruvate cycling in 13C NMR spectra of glutamate derived from rat liver (27).
Modeling.
The differences in NMR spectra between robustly and poorly glucose-responsive cell lines described in the previous paragraph indicate that these cells are fundamentally different with regard to mitochondrial metabolism of glucose-derived intermediates. To gain a more comprehensive and quantitative measure of these differences, the relative areas of all multiplets contributing to glutamates C2, C3, and C4 in these spectra were measured by using standard deconvolution methods. These data were used along with the C4/C3 peak area ratio as input for an isotopomer analysis by using the program tcacalc (http://www2.swmed.edu/rogersmr/tcacalc1.htm). The fittings were performed systematically by starting with the simplest metabolic model and adding pathways as needed to achieve the best agreement between calculated and experimental spectra. If addition of a new pathway did not improve the fit, this pathway was considered unnecessary and removed from further models.
A simple, direct analysis of the spectrum (the C4Q multiplet area multiplied by the C4/C3 ratio; see ref. 20) indicated that the fraction of acetyl-CoA derived from [U-13C6]glucose increased with increasing glucose concentration over an identical range in all four cell lines, constituting approximately ≈40, ≈70, and ≈85% at 3, 6, and 12 mM, respectively, in each line (Table 1). Thus, the fraction of glucose that enters the TCA cycle via PDH-catalyzed conversion to acetyl-CoA is not a determinant of the degree of glucose responsiveness in these cell lines.
Table 1.
Contribution of [U-13C6]glucose to acetyl-CoA in two highly responsive (832/13 and 834/40) and two poorly responsive (832/2 and 832/1) INS-1-derived β cells
| Experimental condition | Fraction of acetyl-CoA derived from
[U-13C6]glucose
|
|||
|---|---|---|---|---|
| 832/13 (n = 4) | 834/40 (n = 3) | 832/1 (n = 2) | 832/2 (n = 4) | |
| 3 mM glucose | 45 ± 3% | 41 ± 3% | 47 ± 1% | 39 ± 2% |
| 6 mM glucose | 73 ± 1% | — | — | 68 ± 1% |
| 12 mM glucose | 84 ± 1% | 86 ± 3% | 80 ± 4% | 84 ± 4% |
A fit of all multiplet data (C2, C3, and C4) to a model that excludes anaplerosis (Fig. 1 Top) did not produce spectra resembling those generated with living cells. Furthermore, the appearance of C4S and C4D34 components in the C4 resonance (Fig. 2) shows that some fraction of pyruvate is derived from TCA cycle intermediates. This observation further supports the inadequacy of a model that does not include the operation of a combined carboxylation/decarboxylation pathway. However, a fit of all multiplet data to a metabolic model involving a single pool of pyruvate feeding both PDH and PC (Fig. 3 Top) resulted in severe underestimation of C4/C3 and the C3 triplet contribution, and a divergence of carboxylation and decarboxylation fluxes. This divergence was especially problematical for NMR data collected on cells incubated with 12 mM [U-13C6]glucose. Thus, this simple metabolic model is also not an acceptable representation of glucose metabolism in INS-1-derived cells.
Figure 3.
Two metabolic models used for fitting of 13C multiplet data of Fig. 2. (A) A single pyruvate pool feeds both PDH and PC. Any pyruvate derived from the TCA cycle via a catabolic decarboxylation pathway also randomly mixes with [U-13C3]pyruvate coming from [U-13C6]glucose. (B) The pyruvate pool involved in cycling with TCA cycle intermediates is sequestered in a compartment that does not allow full equilibration with [U-13C3]pyruvate coming from [U-13C6]glucose.
A relatively minor change in the metabolic model resulted in dramatic improvement in the fit to the experimental spectra. This metabolic model, shown schematically at the bottom of Fig. 3, includes two pools of pyruvate, one derived from [U-13C6]glucose that is destined for PDH-catalyzed conversion to acetyl-CoA and a second pool that engages in direct exchange with the TCA cycle. This latter exchange is hereafter referred to as pyruvate cycling. The fitting also revealed that the two pools of pyruvate do not mix randomly but rather that there is modest “leakage” of pyruvate isotopomers derived from the TCA cycle-exchangeable pool into the pyruvate pool destined for PDH-catalyzed metabolism to acetyl-CoA.
NMR Analysis Reveals a Strong Correlation Between Pyruvate Cycling and GSIS.
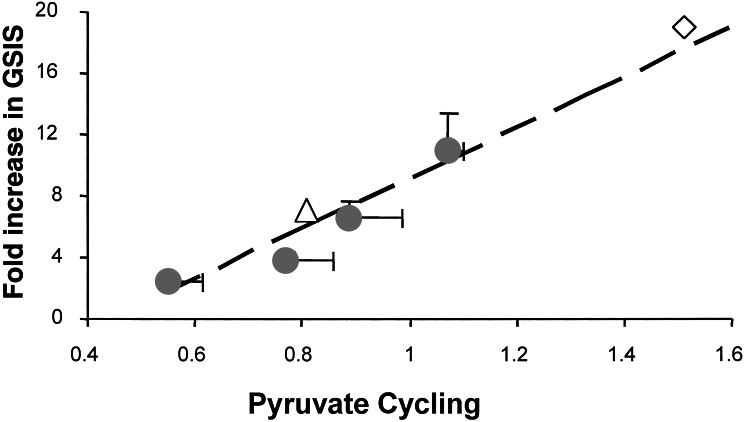
All NMR spectra generated from the INS-1 cell lines were fitted to the preferred “two-pyruvate pool” model of Fig. 3. As shown in Table 1, glucose oxidation (as estimated by the fractional contribution of [U-13C6]glucose to acetyl-CoA formation) increased with increasing glucose concentration to a similar degree in each of the four clonal cell lines studied. Thus, this integrated analysis resulted in the same conclusion about the lack of a role of PDH-catalyzed conversion of glucose to acetyl-CoA as deduced from our earlier direct analysis of the C4 resonance. In contrast, the isotopomer analysis revealed that pyruvate cycling was increased to a significantly larger extent as glucose was raised from 3 to 12 mM in the two lines with robust GSIS vs. the two less-responsive lines (Table 2). In relating pyruvate cycling to insulin secretion, we did not rely on historical data for insulin secretion from our INS-1 cell lines. Instead, we measured insulin secretion in the media collected from the same tissue culture plates used for the NMR analysis. This action resulted in the plot of GSIS vs. pyruvate cycling flux shown in Fig. 4. Remarkably, the capacity for GSIS was found to be linearly related to pyruvate cycling in our four differentially glucose-responsive cell lines (solid points in the graph of Fig. 4), with a correlation coefficient of r2 = 0.92. Thus, pyruvate cycling rather than PDH-catalyzed pyruvate conversion to acetyl-CoA correlates with the capacity for GSIS in the INS-1-derived cell lines.
Table 2.
Pyruvate cycling in two highly responsive (832/13 and 834/40) and two poorly responsive (832/2 and 832/1) INS-1-derived β cells
| Experimental condition | Pyruvate cycling (relative to TCA cycle
flux = 1)
|
|||
|---|---|---|---|---|
| 832/13 (n = 4) | 834/40 (n = 3) | 832/1 (n = 2) | 832/2 (n = 4) | |
| 3 mM glucose | 0.30 ± 0.06 | 0.36 ± 0.08 | 0.50 ± 0.08 | 0.38 ± 0.07 |
| 6 mM glucose | 0.65 ± 0.03 | — | — | 0.50 ± 0.05 |
| 12 mM glucose | 1.07 ± 0.03 | 0.89 ± 0.08 | 0.55 ± 0.06 | 0.77 ± 0.09 |
Figure 4.
Linear relationship between GSIS and pyruvate cycling. The filled circles are data from INS-1-derived cell lines 832/1, 832/2, 834/40, and 832/13 INS-1 cells (ref. 21; in order of increasing capacity for GSIS). The open diamond represents 832/13 cells incubated with 12 mM glucose plus 10 mM DMM, a stimulatory metabolite for pyruvate cycling. The open triangle represents 832/13 cells incubated with 12 mM glucose plus 5 mM PAA, an inhibitor of PC and pyruvate cycling.
Stimulation or Inhibition of Pyruvate Cycling Has Proportional Effects on GSIS.
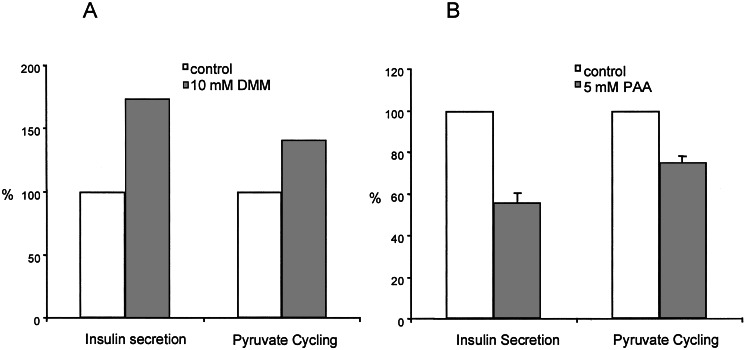
To investigate further the relationship between pyruvate cycling and GSIS, we measured the effects of stimulators and inhibitors of this pathway. First, we tested the cell permeant ester of malate, DMM, reasoning that this agent should increase anaplerosis and hence stimulate pyruvate cycling. Addition of DMM to 832/13 cells had little effect on insulin secretion at 3 mM glucose but nearly doubled insulin secretion at 12 mM glucose (Fig. 5). An isotopomer analysis of 13C spectra collected on cell extracts after incubation with 12 mM [U-13C6]glucose plus DMM indicated that DMM stimulated pyruvate cycling by 40%. This result is shown as the open diamond (⋄) point on the plot of Fig. 4. Thus, pyruvate cycling is proportional to GSIS when anaplerosis is stimulated by addition of DMM. As a second test, PAA, a well known inhibitor of PC (12, 28), was added to 832/13 cells under stimulatory conditions (12 mM glucose) and, in all cases, insulin secretion was inhibited by PAA (Fig. 5). This observation demonstrates that PC-catalyzed carboxylation of pyruvate plays an important role in GSIS in these cells. Furthermore, a 13C isotopomer analysis of 832/13 cells after incubation with 12 mM [U-13C6]glucose and 5 mM PAA confirmed that pyruvate cycling was lower by 25%. This result is shown as the open triangle (▵) on the plot of Fig. 4. Thus, under both stimulatory (DMM) and inhibitory (PAA) conditions, there is a linear correspondence between GSIS and pyruvate cycling as measured by NMR.
Figure 5.
A stimulatory substrate (DMM) and an inhibitor (PAA) of pyruvate cycling have proportional effects on insulin secretion. Insulin secretion was measured from 832/13 cells during incubation with 12 mM [U-13C6]glucose. A shows insulin secretion and pyruvate cycling in the presence and absence of 10 mM DMM. B shows insulin secretion and pyruvate cycling in the presence and absence of 5 mM PAA, an inhibitor of PC. Data represent the mean ± SE for 4 independent determinations.
One potential concern with regard to our conclusions about the link between pyruvate cycling and insulin secretion is that pyruvate cycling flux, as estimated by the NMR isotopomer method, is expressed as a ratio relative to TCA cycle flux. If absolute flux of pyruvate through PDH was different in glucose-responsive vs. unresponsive cells, this could mean that no real change in PC-catalyzed pyruvate cycling flux had actually occurred. If PDH-catalyzed glucose oxidation differed in the two cell lines, this should be reflected by a change in O2 consumption. However, direct O2 consumption measurements on actively respiring cells showed no difference between highly or poorly responsive cells (6.82 ± 0.54 nmol/min/mg). This result supports our conclusion of a significant difference in pyruvate cycling activity in robustly and poorly glucose-responsive cell lines.
Discussion
Regulation of insulin secretion by glucose requires metabolism of the hexose in islet β cells. Decades of research on the specific metabolic pathways that link glucose metabolism to insulin secretion have resulted in confusing and often contradictory findings. Thus, key signaling functions have been ascribed to such diverse events as cytosolic generation of reducing equivalents (29, 30), reducing equivalent shuttles such as the glycerol phosphate and malate/aspartate shuttles that are very active in β cells (31), PDH-catalyzed fuel oxidation (3), and anaplerosis (10, 12). Approaches that have implicated these pathways have included the use of pharmacologic agents, genetic engineering, and measurement of the concentrations of individual metabolic intermediates such as malate or citrate during glucose stimulation. Although informative, each of these methods can be criticized. Thus, use of pharmacologic agents such as iodoacetate for inhibition of glycolytic flux through glyceraldehyde-3-phosphate dehydrogenase may be subject to other interpretations because of nonspecific effects of the drug. In another study, reducing equivalent shuttles were blocked by genetic knock out of mitochondrial glycerol phosphate dehydrogenase and coapplication of an inhibitor of malate/aspartate shuttle activity (31). Although the authors convincingly demonstrated ablation of GSIS in islets treated in this fashion, the importance of reducing equivalent shuttles for regulation of GSIS in normal islets relative to other coupling factors cannot be accurately determined from this approach. Finally, static measurement of specific metabolic intermediates has been used as an indicator of anaplerosis. Thus, there have been reports of increased levels of citrate (12) or malate (11) during glucose stimulation of islets or insulinoma cells. However, a transient rise in a specific metabolic intermediate does not demonstrate a sustained flux of intermediates through a particular metabolic pathway.
In the current study, we applied 13C NMR to the study of a set of robustly and poorly glucose-responsive insulinoma cell lines. This approach has allowed us to develop a “metabolic fingerprint” of glucose metabolism that can be derived in the absence of pharmacologic or genetic manipulation of the system. Furthermore, because this analysis is conducted on cells in metabolic steady-state, the results provide a history of glucose metabolism in the mitochondria over the entire period of glucose stimulation, rather than relying on static, transient assays of random intermediates. The combination of novel cellular models and the application of a comprehensive tool for metabolic analysis yielded insights into aspects of glucose metabolism that may be important for stimulation of insulin secretion.
In β cells, pyruvate derived from glucose has two primary fates, one to generate reducing equivalents via PDH and the TCA cycle and another to provide a three-carbon anaplerotic substrate that can enter the TCA cycle pool of intermediates via PC. Others have demonstrated that pyruvate enters these two competing pathways to a similar extent in islet β cells (7–10), although absolute estimates of the relative contribution of PC-catalyzed pyruvate metabolism have varied from 40–67%. This high anaplerotic input of carbon into the cycle of course requires an equivalent cataplerotic output at metabolic steady-state. Pyruvate cycling in β cells has been proposed to occur via a pyruvate-malate cycle (11) or a pyruvate-citrate cycle (12), both of which culminate with malic enzyme-catalyzed conversion of malate into pyruvate. The feasibility of such cycling was supported by the demonstration of significant levels of malic enzyme in pancreatic islets (7). In this study, we have used the term “pyruvate cycling” to refer to flux through either the pyruvate-malate or pyruvate-citrate cycles.
Application of 13C NMR and isotopomer analysis of glutamate allowed us to obtain a quantitative measure of pyruvate cycling in four INS-1-derived clonal cell lines with varying capacities for GSIS. This analysis was made possible by our finding that sufficient 13C-enriched glutamate can be detected in as few as 1.5 × 108 cells after a 4-h incubation with [U-13C6]glucose. By NMR-based isotopomer analysis of glutamate and fitting of the data to models of mitochondrial metabolism of pyruvate, we have found that pyruvate cycling activity is closely correlated with the degree of glucose responsiveness in our INS-1-derived cell lines. In sharp contrast, the fraction of [1,2-13C2]acetyl-CoA entering the TCA cycle in these cells is proportional to the amount of [U-13C6]glucose in the incubation medium and does not vary between poorly and highly responsive cells. It is interesting to note, however, that the fractional contribution of glucose to acetyl-CoA increases much more dramatically between 3 and 6 mM glucose (43–70%) than it does between 6 and 12 mM glucose (70–85%). This observation suggests that the glucose contribution to acetyl-CoA is saturable and may not reach 100% even at very high glucose concentrations. The unlabeled substrate that contributes the remaining acetyl-CoA was not identified in these experiments.
One interesting feature of the 13C isotopomer NMR analysis performed here on cell extracts was the clear identification of two metabolically distinct pools of pyruvate. Pyruvate compartmentation has been identified in other mammalian tissues (32–35) but, to our knowledge, has not been detected in β cells before this study. Peuhkurinen et al. (35) proposed that one pool of pyruvate in the myocyte is associated more closely with glycolysis and tissue lactate, whereas a second “peripheral” pool is in close communication with extracellular pyruvate and mitochondrial pyruvate. Our 13C analysis indicated that substantial pyruvate cycling occurs in a β cell compartment that is functionally separate from glycolytically produced pyruvate destined for PDH. The subcellular location of this process was not identified here—it could reflect a pool of pyruvate undergoing active carboxylation/decarboxylation within the mitochondrial matrix or it could involve transport of a TCA cycle intermediate from the mitochondria to the cytosol whereupon it forms pyruvate that is later recarboxylated via PC. Our methods cannot differentiate between these two processes, but the observation that the pyruvate cycling pool does not fully mix with glycolytic pyruvate destined for PDH hints at a cycling pathway that may be confined to the mitochondrial matrix. The exact amount of mixing between these pools of pyruvate is difficult to quantify but is estimated by our metabolic model to be 10% or less.
Although this study demonstrates a direct proportionality between GSIS and pyruvate cycling, it leaves open the question of the identity of relevant coupling factors. Our results do not seem to point to ATP as the most likely factor, because neither the pyruvate-malate nor pyruvate-citrate cycling pathways are energy-yielding. We also found no difference in total tissue glutamate as measured by NMR at either low or high glucose in cells with different glucose responsiveness. Thus, glutamate does not seem to be implicated as a relevant coupling factor in our studies, in agreement with one published report, but not with another (25, 26). One byproduct of the pyruvate-citrate cycling pathway that has received extensive consideration is malonyl-CoA (13–16). However, our own work indicates that blockade of glucose-induced increases in malonyl-CoA levels by adenovirus-mediated expression of malonyl-CoA decarboxylase (MCD) in native and derived INS-1 cell lines does not interfere with GSIS (15, 16). However, our results with MCD expression can be reconciled with the current study if one notes that degradation of malonyl CoA would not be expected to prevent recycling via the citrate-pyruvate pathway. One common product of the pyruvate-citrate and pyruvate-malate cycling pathways is NADPH, produced as a product of the reaction catalyzed by the malic enzyme. Two-photon excitation microscopy has been used to show that total NADH/NADPH increases by ≈30 μM in pancreatic islets after maximal glucose stimulation, and that most of this increase can be traced to mitochondrial NADH/NADPH (36). Others have shown that abolishing all shuttle systems that transport cytosolic NADH into mitochondria also disrupts GSIS (31). Both studies suggest a connection between mitochondrial reducing equivalents and GSIS, consistent with the NMR results reported here. Currently, no NAPDH-regulated molecule that would be analogous to ATP-regulated K+ channels in control of insulin secretion is known but further investigation of potential signaling partners for NADPH in β cells may be warranted. Although the metabolic model described here in insulinoma cells is intriguing, one should not necessarily assume that pyruvate cycling also correlates with GSIS in islet β cells. Unfortunately, the amount of glutamate that one can derive from even a large number of pancreatic islets currently precludes the use of NMR to perform this assay in the β cell. Nevertheless, the observations that mitochondrial NADH/NADPH increases after maximal glucose stimulation (36) whereas pyruvate is partitioned nearly equally between PDH and PC in pancreatic islets (7–10) are certainly consistent with an important role of pyruvate cycling in primary cells as well.
Acknowledgments
The authors thank Teresa Eversole for outstanding technical assistance. This work was supported by National Institutes of Health Grants HL-34557 (to A.D.S.), DK42582 (to C.B.N.), and RR-02584 for support of the Rogers Magnetic Resonance Center.
Abbreviations
- GSIS
glucose-stimulated insulin secretion
- TCA
tricarboxylic acid
- PC
pyruvate carboxylase
- PDH
pyruvate dehydrogenase
- PAA
phenylacetic acid
- DMM
dimethylmalate
- OAA
oxaloacetate
- INS
rat insulinoma cell
References
- 1.Newgard C B, McGarry J D. Annu Rev Biochem. 1995;64:689–719. doi: 10.1146/annurev.bi.64.070195.003353. [DOI] [PubMed] [Google Scholar]
- 2.Newgard C B, Matschinsky F M. In: Handbook of Physiology. Jefferson J, Cherrington A, editors. Vol. 2. London: Oxford Univ. Press; 2001. pp. 125–152. [Google Scholar]
- 3.Heimberg H, DeVos A, Vandercammen A, Van Schaftingen E, Pipeleers D, Schuit F. EMBO J. 1993;12:2873–2879. doi: 10.1002/j.1460-2075.1993.tb05949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald M J. Arch Biochem Biophys. 1995;319:128–132. doi: 10.1006/abbi.1995.1274. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald M J, McKenzie D I, Walker T M, Kaysen J H. Horm Metab Res. 1992;24:158–160. doi: 10.1055/s-2007-1003284. [DOI] [PubMed] [Google Scholar]
- 6.Brun T, Roche E, Assimacopoulos-Jeannet F, Corkey B E, Kim K H, Prentki M. Diabetes. 1996;45:190–198. doi: 10.2337/diab.45.2.190. [DOI] [PubMed] [Google Scholar]
- 7.McDonald J. J Biol Chem. 1995;270:20051–20058. [PubMed] [Google Scholar]
- 8.Malaisse W J, Best L, Kawazu S, Malaisse-Lagae F, Sener A. Arch Biochem Biophys. 1983;224:102–110. doi: 10.1016/0003-9861(83)90193-5. [DOI] [PubMed] [Google Scholar]
- 9.Khan A, Ling C Z, Landau B R. J Biol Chem. 1996;271:2539–2542. doi: 10.1074/jbc.271.5.2539. [DOI] [PubMed] [Google Scholar]
- 10.Schuit F, DeVos A, Farfari S, Moens K, Pipeleers D, Brun T, Prentki M. J Biol Chem. 1997;272:18572–18579. doi: 10.1074/jbc.272.30.18572. [DOI] [PubMed] [Google Scholar]
- 11.MacDonald M J. Arch Biochem Biophys. 1982;213:643–649. doi: 10.1016/0003-9861(82)90594-x. [DOI] [PubMed] [Google Scholar]
- 12.Farfari S, Schulz V, Corkey B, Prentki M. Diabetes. 2000;49:718–726. doi: 10.2337/diabetes.49.5.718. [DOI] [PubMed] [Google Scholar]
- 13.Corkey B E, Glennon M C, Chen K S, Deeney J T, Matschinsky F M, Prentki M. J Biol Chem. 1989;264:21608–21612. [PubMed] [Google Scholar]
- 14.Prentki M, Vischer S, Glennon M, Regazzi R, Deeney J, Corkey B E. J Biol Chem. 1992;267:5802–5810. [PubMed] [Google Scholar]
- 15.Antinozzi P, Segall L, Prentki M, McGarry J D, Newgard C B. J Biol Chem. 1998;273:16146–16154. doi: 10.1074/jbc.273.26.16146. [DOI] [PubMed] [Google Scholar]
- 16.Mulder H, Lu D, Finley J, An J, Cohen J, Antinozzi P, McGarry J D, Newgard C B. J Biol Chem. 2001;276:6479–6484. doi: 10.1074/jbc.M010364200. [DOI] [PubMed] [Google Scholar]
- 17.Noel R J, Antinozzi P, McGarry J D, Newgard C B. J Biol Chem. 1997;272:18621–18627. doi: 10.1074/jbc.272.30.18621. [DOI] [PubMed] [Google Scholar]
- 18.Sherry A D, Malloy C R. In: NMR in Physiology and Biomedicine. Gillies R J, editor. San Diego: Academic; 1994. pp. 439–449. [Google Scholar]
- 19.Sherry A D, Malloy C R. In: In Vivo Carbon-13 NMR, Biological Magnetic Resonance. Berliner L J, Robitaille P-ML, editors. Vol. 15. New York: Kluwer Academic/Plenum; 1998. pp. 59–97. [Google Scholar]
- 20.Malloy C R, Sherry A D, Jeffrey F M H. Am J Physiol. 1990;259:H987–H995. doi: 10.1152/ajpheart.1990.259.3.H987. [DOI] [PubMed] [Google Scholar]
- 21.Hohmeier H, Mulder H, Chen G, Henkel-Reiger R, Prentki M, Newgard C B. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Hohmeier H E, Gasa R, Tran V, Newgard C B. Diabetes. 2000;49:562–570. doi: 10.2337/diabetes.49.4.562. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg H L. Essays Biochem. 1966;2:1–31. [Google Scholar]
- 24.Carvalho R A, Babcock E E, Jeffrey F M H, Sherry A D, Malloy C R. Magn Reson Med. 1999;42:197–200. doi: 10.1002/(sici)1522-2594(199907)42:1<197::aid-mrm26>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Maechler P, Wollheim C B. Nature (London) 1999;402:685–689. doi: 10.1038/45280. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald M J, Fahien L A. J Biol Chem. 2000;275:34025–34027. doi: 10.1074/jbc.C000411200. [DOI] [PubMed] [Google Scholar]
- 27.Jones J G, Naidoo R, Sherry A D, Jeffrey F M H, Cottam G L, Malloy C R. FEBS Lett. 1997;412:131–137. doi: 10.1016/s0014-5793(97)00764-3. [DOI] [PubMed] [Google Scholar]
- 28.Bahl J J, Matsuda M, DeFronzo R A, Bressler R. Biochem Pharmacol. 1997;53:67–74. doi: 10.1016/s0006-2952(96)00660-0. [DOI] [PubMed] [Google Scholar]
- 29.Dukes I D, McIntyre M S, Mertz R J, Philipson L H, Roe M W, Spencer B, Worley J F. J Biol Chem. 1994;269:10979–10982. [PubMed] [Google Scholar]
- 30.Mertz R J, Worley J F, Spencer B, Johnson J H, Dukes I D. J Biol Chem. 1996;271:4838–4845. doi: 10.1074/jbc.271.9.4838. [DOI] [PubMed] [Google Scholar]
- 31.Eto K, Tsubamoto Y, Terauchi Y, Sugiyama T, Kishimoto T, Takahashi N, Yamauchi N, Kubota N, Murayama S, Aizawa T, et al. Science. 1999;283:981–931. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- 32.Mowbray J, Ottaway J H. Eur J Biochem. 1973;36:362–368. doi: 10.1111/j.1432-1033.1973.tb02920.x. [DOI] [PubMed] [Google Scholar]
- 33.Mowbray J, Ottaway J H. Eur J Biochem. 1973;36:369–379. doi: 10.1111/j.1432-1033.1973.tb02921.x. [DOI] [PubMed] [Google Scholar]
- 34.Peuhkurinen K J, Hassinen I E. Biochem J. 1982;202:67–76. doi: 10.1042/bj2020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peuhkurinen K J, Hiltunen J K, Hassinen I E. Biochem J. 1983;210:193–198. doi: 10.1042/bj2100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson G H, Knobel S M, Arkhammar P, Thastrup O, Piston D W. Proc Natl Acad Sci USA. 2000;97:5203–5207. doi: 10.1073/pnas.090098797. [DOI] [PMC free article] [PubMed] [Google Scholar]