Abstract
Eukaryotic cells have evolved complex signaling networks to sense environmental stress and to repair stress-induced damage. IFN regulatory factor-3 (IRF-3) is a transcription factor that plays a central role in the host response to viral infection. Although the main activity of IRF-3 characterized to date has been its role in the induction of IFN-α and -β after virus infection, recent evidence indicates additional roles for IRF-3 in the response to DNA damage and in virus-induced apoptosis. Here we identify IRF-3 as the first in vivo target for DNA-dependent protein kinase (DNA-PK). Phosphorylation of IRF-3 by DNA-PK after virus infection results in its nuclear retention and delayed proteolysis. These results expand the known roles of DNA-PK and provide a functional link between the cellular machineries that regulate the innate immune response and that sense and respond to DNA damage. As such this study contributes to a more integrated view of the cellular responses to various cellular stress signals.
Cells have evolved intricate networks of stress detection, decision making, and DNA repair to respond to the variety of endogenous and environmental stresses to which they are exposed. Virus infection represents a rather specific type of stress to which animal cells must respond for the survival of the entire organism. Although adaptive immunity contributes significantly to the elimination of an infection, the initial rapid innate immune response to a virus occurs at the level of infected cells to prevent the spread of the virus to neighboring cells. The transcriptional induction of IFN-α and -β is central to the cellular antiviral response and occurs rapidly after infection (reviewed in ref. 1). These cytokines have pleiotropic effects including inhibition of cell proliferation and induction of cell death on both the infected and the uninfected neighboring cells. In addition, type I IFNs trigger a strong proinflammatory response at the site of infection. Although the potency of the IFNs is critical for the effective elimination of virus infection, if uncontrolled these cytokines could have pathogenic consequences to the host. In recent years, complex regulatory mechanisms have been uncovered that govern the production of these cytokines. One of the proteins involved in sensing a viral infection and triggering the production of IFNs is IFN regulatory factor-3 (IRF-3; refs. 2–8).
IRF-3 belongs to a family of the IRF transcription factors characterized by a conserved DNA binding domain with a signature tryptophan pentad. The nine cellular factors identified thus far have been shown to participate in a large number of biological processes including the regulation of cell proliferation, hematopoietic development, antiviral defense, and response to DNA damage (reviewed in refs. 9 and 10). As mentioned above, the main known role of IRF-3 is the induction of type I IFNs in response to virus infection. The activity of IRF-3 is tightly regulated at multiple levels to ensure rapid but brief response. In the absence of stimuli, IRF-3 exists in the cytoplasm in a latent inactive form incapable of binding to DNA. It is believed that an intramolecular interaction between the regulatory C terminus and sequences in the N-terminal portion of the protein keeps IRF-3 in this inactive conformation (11, 12). After virus infection, IRF-3 is phosphorylated rapidly and translocates into the nucleus, where it activates the transcription of its target genes. Furthermore, whereas inactive IRF-3 is very stable, it is degraded rapidly after activation via the ubiquitin-proteasome pathway (2–5, 7, 13). The degradation of active IRF-3 is clearly an important mechanism contributing to the control of the level of the IFN response. To date, however, little is known about the regulation of this proteolytic pathway.
Recent evidence also has suggested roles for IRF-3 in DNA damage response and in virus-induced apoptosis (14, 15). However, although the nuclear translocation and transactivating functions of IRF-3 after DNA damage and virus infection are similar, the signaling pathways leading to its activation after these two stress stimuli clearly are different (16). The unexpected involvement of IRF-3 in the DNA damage response and recent characterizations of the DNA damage-sensing machineries as more general stress sensors suggest the possibility of cross talk between the DNA damage and IFN response pathways. The initial events in response to DNA damage are known to include activation of the phosphatidylinositol 3-kinase (PI3-kinase)-like proteins ATM, ATR, and DNA-dependent protein kinase (DNA-PK). These kinases, in turn, regulate the activity of a number of target proteins ultimately controlling cell cycle checkpoints (e.g., Chk1, Chk2, and p53), DNA repair (e.g., BRCA1, RPA-p34, and Nibrin), and apoptosis (p53 and IKK; for review see refs. 17–19).
In this study, we have identified IRF-3 as an in vivo target for DNA-PK and provide evidence for a function for DNA-PK in regulating the cellular antiviral response. We demonstrate that DNA-PK is activated in response to virus infection and phosphorylates IRF-3 on Thr-135. The phosphorylation of Thr-135 down-regulates the proteolysis of active IRF-3 by causing its nuclear retention. These results establish an important in vivo role for DNA-PK in regulating IRF-3 activity.
Materials and Methods
Cell Lines, Expression Vectors, and Antibodies.
HEC1B and NIH 3T3 cells were maintained in DMEM + 10% FBS. M059J cells and M059J cells reconstituted with a chromosomal copy of DNA-PK (kindly provided by Cordula U. Kirchgessner (20) were maintained in a 1:1 ratio of F12/DMEM supplemented with 10% FBS. The reconstituted cell line was kept under selection in 250 μg/ml G418. HT-29 cells were grown in McCoy + 10% FBS. Point mutants of IRF-3 were generated by using the GENEEDITOR system from Promega. To generate His-6-tagged proteins, wild type and point mutants of IRF-3 were cloned into pET-32a vector from Novus (Littleton, CO). Proteins were purified on nickel-nitrilotriacetic acid agarose from Qiagen (Chatsworth, CA).
For immunoprecipitation experiments, monoclonal SL-12 α-IRF-3 antibody (5) was covalently crosslinked to protein A/G beads by using a standard protocol. To generate the polyclonal antibody specific for IRF-3 phosphorylated at Thr-135, a phosphopeptide spanning the site was synthesized, coupled to keyhole limpet hemocyanin, and used to immunize rabbits. The antibody was affinity-purified by using the phosphopeptide coupled to CNBr-activated Sepharose and depleted of any reactivity for the unphosphorylated protein by incubating with full-length IRF-3 coupled to the beads. Monoclonal α-DNA-PK antibody (Ab-2) was purchased from NeoMarkers (Fremont, CA).
Treatments.
Virus infection of cells with Sendai virus (SV) has been described (5). For UV treatment, cells were rinsed in PBS once and exposed to 30 J/m2 of the 254-nm light in a Stratalinker (Stratagene). For leptomycin B treatment, the drug was added to the final concentration of 100 ng/ml at 3 h post virus infection.
Lysis Buffers, Immunoprecipitations, and Western Blotting.
For coimmunoprecipitation experiments, lysates were prepared by KCl extraction as described (13). The phosphate at Thr-135 is very labile, and its detection is impaired greatly if cell pellets are frozen, lysates are sonicated, or blots are stripped. The following protocol was used for immunoprecipitation-Western blots to detect the phosphorylation. Cells were lysed in a modified RIPA buffer (20 mM sodium phosphate, pH 7.2/300 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS/2 mM EDTA/2 mM EGTA/50 mM NaF/10 mM β-glycerophosphate/1 mM Na3VO4). A mixture of protease inhibitors from PharMingen was added at the suggested dilution. Lysates were homogenized by passing through a 21-gauge needle, rotated at 4°C for 30 min, and spun for 10 min at 18,000 × g. Lysate (300–500 μg) was used for immunoprecipitation with the crosslinked SL-12 antibody. β-Glycerophosphate (10 mM) was included in the blocking solution for Western blotting.
In Vitro Phosphorylation and Kinase Assay.
The partially purified DNA-PK was purchased from Promega. The phosphorylation assay was performed according to the manufacturer's instructions. In the case of large-scale phosphorylation, a large excess of the kinase was used, and the reactions were incubated at 30°C for 1–1.5 h. The phosphorylated protein was repurified away from the kinase by nickel-affinity chromatography. For the detection of DNA-PK kinase activity in SV-infected or UV-treated cells, nuclear extracts were prepared from treated HT-29 cells according to a modified method originally described by Dignam et al. (21). Kinase reactions were performed by using SignaTECT DNA-PK assay following the manufacturer's protocol (Promega) except that the extracts were not depleted of DNA, and no activation buffer was added to the reaction. Briefly, 10 μg of nuclear extract were used to phosphorylate a biotinylated p53-derived peptide substrate (peptide 15; ref. 22) for 5 min at 30°C in the presence of [γ-32P]ATP. After termination of the reaction, the peptide was captured and washed on a streptavidin-coated membrane. The amount of 32P-labeled substrate was determined by phosphorimaging analysis.
Immunofluorescence.
For detection of nuclear IRF-3 staining, cells were fixed with 2% paraformaldehyde in PBS for 15 min at room temperature and permeabilized with −20°C methanol for 10 min. Staining then was performed with the SL-12 antibody by using a standard protocol. The secondary antibody was Cy3-labeled goat anti-mouse from Jackson ImmunoResearch. For the detection of phosphorylated IRF-3, cells were fixed with 3% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 4 min at room temperature. The phospho-specific antiserum that had been affinity-purified by using peptide beads and depleted of nonphospho-specific reactivity with IRF-3 beads was further selected on a polyvinylidene difluoride membrane with immobilized full-length IRF-3 that had been phosphorylated in vitro by DNA-PK. After extensive washing, membrane strips were incubated with the coverslips to allow passive transfer of the antibody onto the cells. After incubation with the phospho-specific primary antibody, light crosslinking with 1.5% paraformaldehyde for 2 min was performed to prevent dissociation of this low-affinity antibody. The secondary antibody was Cy3-labeled goat anti-rabbit from Jackson ImmunoResearch. In both cases cells were counterstained with 4′,6-diamidino-2-phenylindole and analyzed by confocal microscopy.
Confocal Microscopy.
Samples were mounted in antifade and imaged through a 63 × 1.2-numerical aperture water immersion (Fig. 2B and 3A) or 10 × 0.25-numerical aperture dry (Fig. 2B) lens on a Zeiss LSM510 confocal microscope using 364 (4′,6-diamidino-2-phenylindole) and 543-nm (Cy3) laser illumination and emission filters appropriate for each dye. The pinhole was opened to a diameter such that most of the intensity from a cell was contained within a single optical section. Photomultiplier gain and offset were adjusted to give no underflow in the background and no overflow with the brightest sample in a set and then held at those values with a fixed pinhole diameter for the entire experiment such that the pixel intensities would be directly comparable between samples. Transmitted light differential interference contrast microscopy or phase-contrast images (data not shown) were collected simultaneously with the epifluorescence images to define cell and nuclear boundaries.
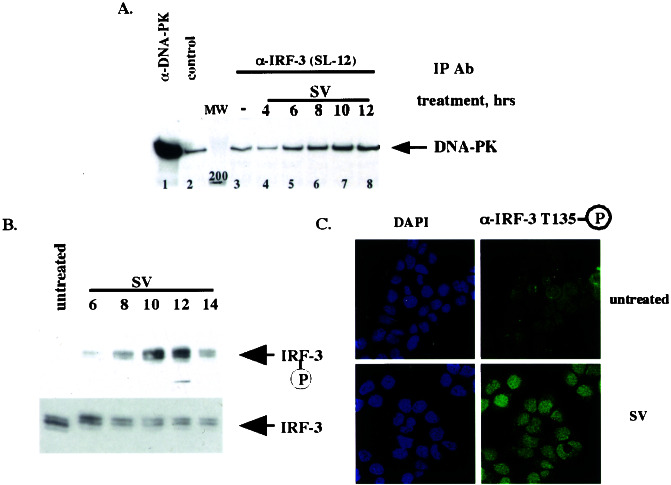
Figure 2.
DNA-PK binds and phosphorylates IRF-3 in vivo after virus infection. (A) IRF-3 and DNA-PK interact in vivo after virus infection. HEC1B cells were treated with virus (SV) for an indicated number of hours. Immunoprecipitations (IP) then were performed with control, α-DNA-PK, or α-IRF-3 (SL-12) antibody. Immunocomplexes were resolved by SDS/PAGE, and Western blotting was performed with the α-DNA-PK antibody. Fifty percent of the input is shown in the α-DNA-PK immunoprecipitation. (B) Thr-135 is phosphorylated in vivo with the kinetics similar to that of IRF-3–DNA-PK complex formation. After the treatment in A, immunoprecipitations were performed with the α-IRF-3 antibody crosslinked to protein A/G beads followed by Western blotting using affinity-purified phospho-specific antibody 2115 (Upper) or the α-IRF-3 antibody (Lower). (C) Phosphorylated IRF-3 is localized to the nucleus. HEC1B cells were treated with virus and stained with the phospho-Thr-135-specific antibody (see Materials and Methods).
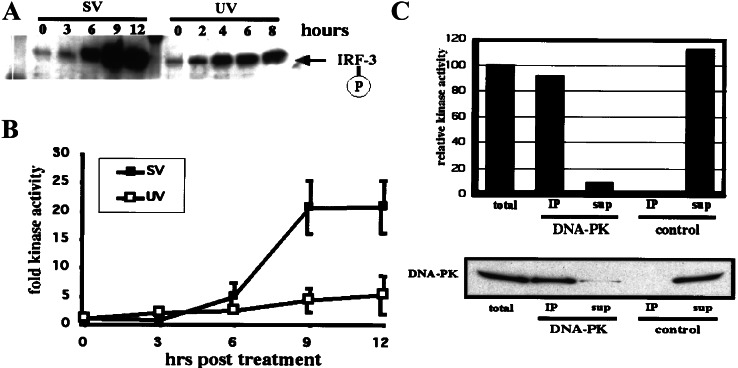
Figure 3.
Virus infection induces DNA-PK kinase activity. (A) Virus-induced phosphorylation of Thr-135 on IRF-3 occurs with faster kinetics in HT-29 cells than in HEC1B cells (compare with Fig. 2B). HT-29 cells were infected with SV for up to 12 h and harvested, and immunoprecipitations (IP) were performed as described for Fig. 2B. Phospho-Thr-135 was detected by Western blotting using affinity-purified phospho-specific antibody. Also shown is the UV-induced phosphorylation of Thr-135 on IRF-3 in HT-29 cells assayed at 2-h intervals up to 8 h. (B) HT-29 cells were infected with SV or treated with UV light and assayed at 3-h time points. Kinase activity was tested in nuclear extracts by phosphorylation of a mutant p53 peptide. Values representing three independent experiments were plotted relative to the activity in untreated extracts. (C) Virus infection activates predominantly DNA-PK. Immunoprecipitations using control antibody (normal mouse serum) or DNA-PK-specific antibody were performed with nuclear extract of HT-29 cells that had been infected with SV for 12 h. Kinase activity was determined as described above. Values were plotted relative to the control precipitation. In addition, precipitated and depleted protein was separated by 3–8% Tris-acetate PAGE. Immunoblotting was performed by using the DNA-PK-specific antibody. sup, supernatant.
Microinjections.
Microinjections were performed by using the CompiC Inject automated system (Luigs and Neumann, Ratingen, Germany) at the European Molecular Biology Laboratory (Heidelberg, Germany). Subsequent images were taken by using a Leica fluorescent microscope equipped with an ×63 objective.
Results
IRF-3 Is Phosphorylated by DNA-PK in Vitro.
In our analysis of mechanisms possibly involved in IRF-3 regulation, we examined the primary sequence of the protein for potential phosphorylation sites by using the SCANSITE database (23). This analysis, based on peptide library screens to determine the substrate specificity of various protein kinases, led to the identification of Thr-135 as a potential target for phosphorylation by DNA-PK (Fig. 1A; ref. 24). We therefore sought to determine whether DNA-PK was involved in regulating IRF-3 activity after virus infection, the main known activating signal for IRF-3.
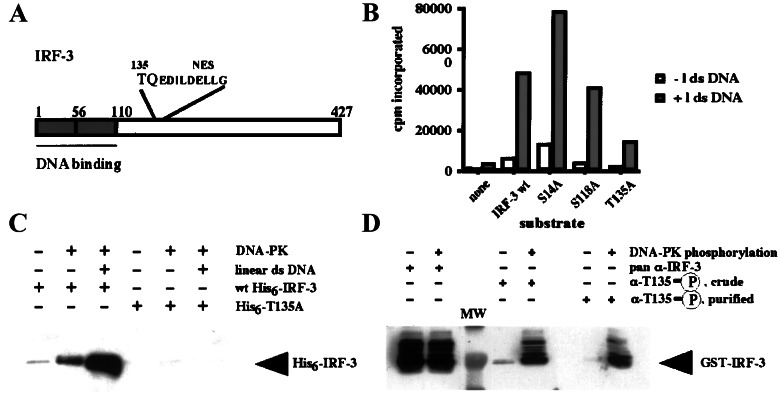
Figure 1.
DNA-PK phosphorylates IRF-3 on Thr-135 in vitro. (A) Schematic of the primary structure of IRF-3 protein showing the relative location of Thr-135. (B) A representative in vitro kinase assay using partially purified DNA-PK fraction and His-tagged IRF-3. One microgram of bacterially produced wild-type (wt) or mutant His6-IRF-3 was incubated with 25 units of DNA-PK for 5 min at 30°C in the presence of [γ-32P]ATP. The kinase reactions were stopped, and incorporation of the radiolabel was quantitated by binding to the P81 paper (Whatman). (C) Phospho-Thr-135-specific antiserum detects DNA-PK-induced phosphorylation of wild-type but not T135A mutant IRF-3. Equal amounts of bacterially produced wild-type or mutant His6-IRF-3 were incubated with DNA-PK in either the absence or presence of the linearized double-stranded DNA (dsDNA). Small aliquots of the reactions were subjected to Western blotting with the crude phopsho-Thr-135-specific serum. (D) Affinity purification of the phospho-Thr-135-specific antiserum removes the remaining reactivity for nonphosphorylated IRF-3. Equal amounts of unphosphorylated and phosphorylated glutathione S-transferase (GST)-IRF-3 were subjected to Western blotting with the pan-α-IRF-3 (SL-12) antibody, crude phospho-specific antiserum, or affinity-purified antiserum.
As an initial step, bacterially produced IRF-3 was incubated with partially purified DNA-PK to examine whether DNA-PK could phosphorylate IRF-3 in vitro. Fig. 1B shows the results of a representative in vitro kinase assay using wild-type IRF-3, a T135A mutant, as well as substitution mutants (S14A and S118A) in the two other potential sites identified as targets for PI3-kinase-like proteins. Although the Ser-14 and Ser-118 sites conform to a more widely known and loosely defined consensus of S/TQ, neither site scored highly in the SCANSITE database search for DNA-PK. Although the specific activity of DNA-PK varied with different preparations of the kinase and linear dsDNA, the overall pattern shown in Fig. 1B was highly reproducible. The results of the assays indicated that the dsDNA-dependent IRF-3-directed kinase activity in the DNA-PK fraction was specific for Thr-135.
To examine in vivo phosphorylation of IRF-3 on Thr-135, an antibody was raised against a phosphopeptide spanning Thr-135. The antibody was characterized for the dependence of its reactivity using IRF-3 phosphorylated by DNA-PK. Fig. 1C shows that incubation of the wild-type but not the T135A mutant form of His-6–IRF-3 with the DNA-PK fraction led to the appearance of phospho-specific antibody reactivity. The addition of dsDNA, a known activator of DNA-PK, to the kinase reaction enhanced this reactivity further. Even the crude serum was strongly selective for the DNA-PK-phosphorylated form of IRF-3, and any remaining reactivity for unphosphorylated protein was removed through affinity purification (Fig. 1D). The affinity-purified phospho-specific antibody subsequently was used for the Western blotting and immunofluorescence experiments described below.
DNA-PK Binds and Phosphorylates IRF-3 in Vivo After Virus Infection.
To address whether DNA-PK phosphorylates IRF-3 in vivo after viral infection, we examined whether the two proteins interact. Coimmunoprecipitation experiments in HEC1B cells demonstrated virus-induced formation of a specific complex between IRF-3 and DNA-PK (Fig. 2A). Although some background DNA-PK binding was observed with control antibody (lane 2) or beads alone (data not shown), increasing levels of DNA-PK were coimmunoprecipitated with IRF-3 by using the anti-IRF-3 antibody SL-12 after virus infection. In HEC1B cells, maximum binding occurred 8–12 h after virus infection; however, the kinetics of the interaction varied in different cell lines (see below). The phosphorylation of Thr-135 in vivo after virus infection was examined next (Fig. 2B). IRF-3 was immunoprecipitated with the SL-12 anti-IRF-3 antibody, and phosphorylation of Thr-135 was examined by Western blot analysis using the phospho-specific antiserum. Fig. 2B shows that strong phosphorylation of Thr-135 was observed 10–12 h after viral infection, with kinetics similar to that observed for complex formation between DNA-PK and IRF-3 in these cells (Fig. 2A). Immunofluorescence experiments using affinity-purified Thr-135 phospho-specific antiserum that had been selected further on a polyvinylidene difluoride membrane with immobilized full-length IRF-3 phosphorylated by DNA-PK in vitro as described in Materials and Methods showed that Thr-135-phosphorylated IRF-3 localized predominantly to the nucleus, suggesting that it is the transcriptionally active form of IRF-3 that is phosphorylated (Fig. 2C).
Virus Infection Induces DNA-PK Activity.
Phosphorylation of IRF-3 at Thr-135 after virus infection is not restricted to HEC1B cells, and also occurs in other cell lines examined. In HT-29 cells, the phosphorylation of Thr-135 after virus infection occurred with a faster kinetics (Fig. 3A) than was seen in HEC1B cells (Fig. 2B). In addition, the kinetics and magnitude of IRF-3 Thr-135 phosphorylation after virus infection or UV treatment differed as shown in HT-29 cells (Fig. 3A) and as observed in other cell lines such as HEC1B cells (data not shown). To examine whether the in vivo phosphorylation of IRF-3 on Thr-135 was caused by DNA-PK, nuclear extracts were prepared at various times after treatment from HT-29 cells that had either been infected with SV or exposed to UV radiation. These extracts were subjected to kinase assays by using a p53-derived biotinylated peptide featuring Ser-15 as substrate. This residue is known to be a target for the PI3-kinase family members ATM, ATR, and DNA-PK (22, 25, 26). The peptide-specific kinase activity could be detected by 6 h after virus infection and reached a maximum of 21-fold activity after 9 h (Fig. 3B). In comparison, UV treatment resulted in only a 5-fold induction by 12 h. To examine whether DNA-PK contributed to this in vivo kinase activity, the nuclear extract of the SV-infected cells was subjected to immunoprecipitation by using an antiserum to DNA-PK or a control serum. Anti-DNA-PK antibodies pulled down 92% of the total kinase activity, with 8% remaining in the supernatant, whereas a control antibody brought down none of the activity (Fig. 3C Upper). Western blot analysis of the same immunoprecipitates revealed the presence of corresponding amounts of DNA-PK in the pellet (94%) and supernatant (6%; Fig. 3C Lower). The correlation between the protein amount and the kinase activity verified that DNA-PK constitutes the main member of the PI3-kinase family activated after virus infection.
DNA-PK Affects the Kinetics of IRF-3 Degradation.
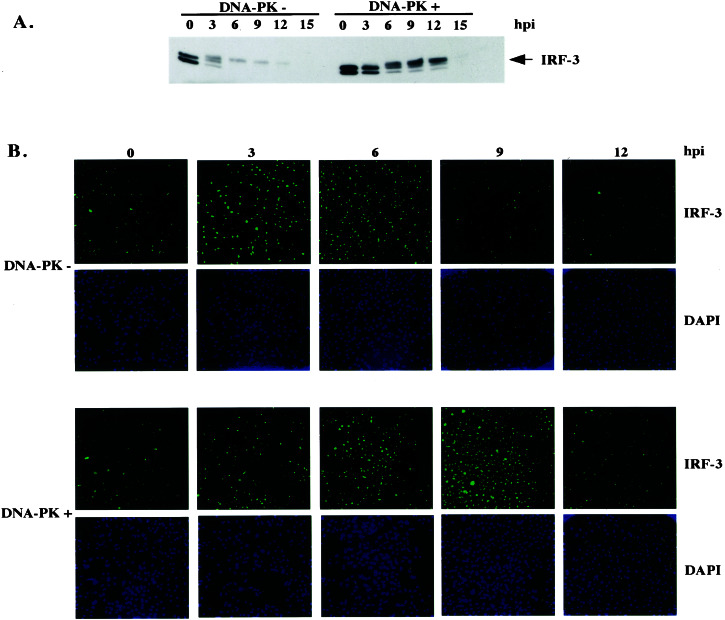
It has been well established that virus infection-induced activation of IRF-3 depends on the phosphorylation of a cluster of Ser and Thr residues in the C terminus of the protein (3, 11), although the specific kinase(s) responsible for this activation has not been defined yet. The initial activation of IRF-3 in response to virus infection therefore would not be expected to depend on DNA-PK, which phosphorylates IRF-3 at a distinct site. That DNA-PK is not responsible for the virus activation of IRF-3 was verified by analyzing the electrophoretic mobility of IRF-3 in both DNA-PK-positive and -negative cell lines after SV infection. As shown in Fig. 4A, the characteristic infection-induced forms of IRF-3 that migrated more slowly than the two isoforms of IRF-3 seen in untreated cells were detected in both DNA-PK-positive and -negative cells. Strikingly, however, the kinetics of virus-induced activation and degradation of IRF-3 were markedly different between the DNA-PK-negative and -positive cell lines, with a slightly slower activation and a significantly slower degradation seen in the DNA-PK-positive cells (Fig. 4A). In agreement with these data, immunofluorescence experiments performed under fixation and permeabilization conditions that lead to the retention of only the nuclear form of IRF-3 (see Materials and Methods) revealed a more rapid disappearance of virus-induced nuclear IRF-3 in DNA-PK-negative cells (Fig. 4B).
Figure 4.
DNA-PK affects kinetics of IRF-3 degradation. (A) Immunoprecipitation-Western blot showing total levels of IRF-3 in M059J (DNA-PK−) and M059J with a chromosomal copy of DNA-PK (DNA-PK+) cells that had been infected with virus for indicated amounts of time. Immunoprecipitations were carried out by using the α-IRF-3 (SL-12) antibody covalently crosslinked to protein A/G beads. Immunoblotting was performed with the same antibody. (B) Immunofluorescence experiment showing the time course of nuclear IRF-3 staining in the same cell lines. After infection, cells were fixed in 2% paraformaldehyde and permeabilized with −20°C methanol to reveal almost exclusively nuclear IRF-3 staining. Cells were stained with the SL-12 antibody and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Subsequently, low-magnification confocal images were generated.
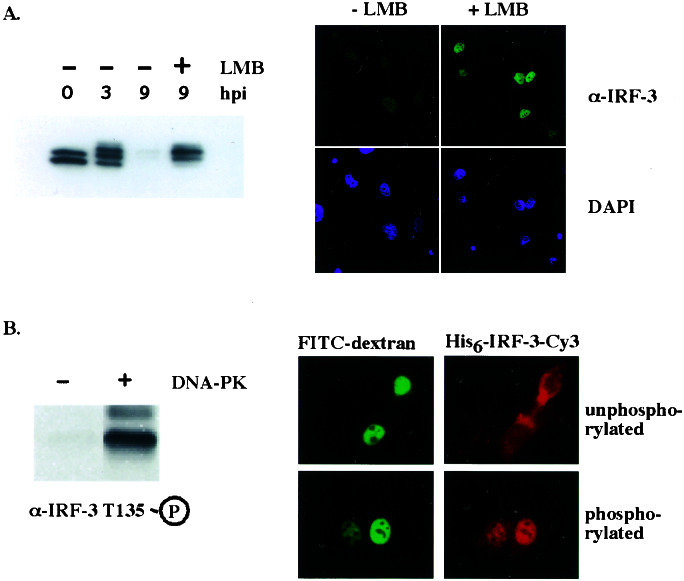
Thr-135 Phosphorylation Prevents IRF-3 Degradation by Causing Its Nuclear Retention.
The observed correlation between the kinetics of degradation of active IRF-3 and the disappearance of its nuclear staining prompted us to examine whether IRF-3 export into the cytoplasm was necessary for its degradation. IRF-3 possesses a classic nuclear export sequence, and its nuclear export depends on the activity of Crm-1 (3, 12). Because Thr-135 is immediately proximal to the rather hydrophobic nuclear export sequence (Fig. 1A), its phosphorylation potentially could affect the ability of the export machinery to bind IRF-3 and delay its export from the nucleus for degradation. To test whether the reexport of activated IRF-3 into the cytoplasm was required for its degradation, the specific Crm-1 inhibitor, leptomycin B, was added to cells after virus infection. For this experiment, we used DNA-PK-negative human glioblastoma M059J cells, in which activation of IRF-3 was observed by 3 h and almost complete degradation was observed by 9 h postinfection (Fig. 4A). Leptomycin B treatment of these cells at 3 h postinfection prevented the degradation of IRF-3 (Fig. 5A Upper) and resulted in nuclear retention at 9 h postinfection (Fig. 5A Lower). These results suggested that the slower kinetics of IRF-3 degradation in the presence of DNA-PK activity could be caused by a decreased export of Thr-135-phosphorylated IRF-3. To test the hypothesis that Thr-135 phosphorylation specifically inhibited IRF-3 export, we determined the localization of microinjected IRF-3 that had been phosphorylated in vitro by DNA-PK (Fig. 5B). When fluorescently labeled unphosphorylated IRF-3 was microinjected into the nuclei of NIH 3T3 cells, efficient export was observed after a 1-h incubation at 37°C. In contrast, in vitro phosphorylation of IRF-3 by DNA-PK (Fig. 5B Upper) resulted in nuclear retention of the protein (Fig. 5B Lower). Taken together, these data indicate that DNA-PK-dependent Thr-135 phosphorylation of IRF-3 inhibits nuclear export of IRF-3 for degradation and thereby prolongs the half-life of the activated form of this transcription factor.
Figure 5.
Phosphorylation of Thr-135 prevents IRF-3 degradation by causing its nuclear retention. (A) Crm-1-dependent export of active IRF-3 is required for its degradation. M059J cells were infected with virus for 9 h. Three hours after the infection, when IRF-3 had been activated and accumulated in the nucleus, cells were treated with leptomycin B (LMB) or left untreated for the rest of the infection. IRF-3 protein levels (Upper) and nuclear staining (Lower) were compared as described for Fig. 4. hpi, hours postinfection. (B) Phosphorylation of IRF-3 by DNA-PK prevents its nuclear export. Bacterially produced His-tagged IRF-3 was phosphorylated with DNA-PK in vitro and fluorescently labeled with Cy3. The phosphorylated protein was microinjected into the nuclei of NIH 3T3 cells, and its export was compared with that of unphosphorylated IRF-3. FITC-conjugated dextran served as a coinjection marker.
Discussion
There are a variety of host defenses that have evolved to counter viral infections. In addition to adaptive immunity, several mechanisms synergize at the cellular level to establish an antiviral state. Secretion of IFN-α and -β is central to this response and induces an antiviral state in neighboring cells, inhibits cell proliferation, and can induce apoptosis. The potent cellular activities of the type I IFNs together with their ability to trigger a strong inflammatory response at the site of infection require tight regulation. Therefore, although the effective elimination of the viral infection needs to be achieved, extensive tissue damage has to be avoided.
In this study we found that DNA-PK is an important regulator of one of the main factors in the establishment of the antiviral state, IRF-3. We have found that viral infection activates DNA-PK, allowing it to bind and phosphorylate IRF-3. IRF-3 is a potent substrate for DNA-PK in vitro, and the activity of DNA-PK in vivo correlates with the extent of phosphorylation of IRF-3 Thr-135. Our data suggest that DNA-PK is the principle kinase responsible for the in vivo phosphorylation of IRF-3 on Thr-135. Furthermore, we found that IRF-3 phosphorylated on Thr-135 localized almost exclusively to the nucleus, and that this posttranslational modification down-regulated the export of IRF-3 from the nucleus to the cytoplasm for degradation by the ubiquitin-proteasome pathway.
Because DNA-PK is located primarily within the nucleus, it seems likely that IRF-3 first must be activated and translocated into the nucleus to be a substrate for DNA-PK phosphorylation. Physiologically, DNA-PK phosphorylation of Thr-135 on IRF-3 extends the half-life of the transcriptionally active IRF-3, thereby contributing to the regulation of its activity in the antiviral response.
Viruses have evolved numerous strategies to evade the critical components of the IFN response (reviewed in ref. 27). The virus used in this study, SV, belongs to the family of paramyxoviruses and produces no dsDNA in the course of its life cycle, which could activate DNA-PK by conventional means. Therefore, the mechanism of DNA-PK activation after viral infection remains to be determined, but several possible scenarios can be envisioned. First, it has been recognized for some time that certain specific protein–protein interactions can activate DNA-PK independent of the presence of dsDNA. One example involves activation of the kinase and stimulation of its DNA binding ability by the high mobility group (HMG) proteins HMGI and HMGY (28, 29). Interestingly, the HMG proteins form an integral part of the virus-inducible IFN-β enhancesome and thus are well positioned to activate DNA-PK in close proximity to IRF-3. A second mechanism could involve the virus- and IFN-stimulated dsRNA-dependent protein kinase, PKR. Although not involved in virus-induced activation of IRF-3, PKR is itself activated rapidly in the course of virus infection and exhibits antiviral and antiproliferative activities (reviewed in ref. 30). More recent data have characterized PKR as a general stress sensor (reviewed in ref. 31). Interestingly, a recent report placed PKR upstream of a PI3-kinase-like activity in the DNA damage-induced phosphorylation of p53 (32), which suggests that PKR may have the potential for regulating the activity of the DNA-PK/ATM family members.
The importance of DNA-PK activity in cellular response to virus infection is underscored further by the finding that the kinase is targeted for proteasomal degradation by herpes simplex virus type I (33). In addition, IRF-3 is targeted by the HPV16 E6 oncoprotein, and the transcriptional activity of IRF-3 toward the IFNβ promoter is reduced in the presence of E6 (13). The effect of E6 expression on Thr-135 phosphorylation of IRF-3 and virus-induced apoptosis remains to be investigated.
The identification of DNA-PK as a regulator of IRF-3 pathways provides for the rapid, efficient, and tightly controlled induction of antiviral defense. The fast and specific engagement of DNA-PK in the antiviral response is remarkable. With its proven ability to phosphorylate and activate proteins such as p53, DNA-PK is uniquely positioned to rapidly engage the cellular defense machinery in response to viral infection. Such phosphorylation-dependent regulation of nuclear–cytoplasmic shuttling by a transcription factor is not unprecedented. Recently, for example, a very similar regulatory mechanism was described for p53 (34). In that report, the authors demonstrated that the DNA damage-induced phosphorylation of Ser-15 in the vicinity of a newly identified N-terminal nuclear export sequence inhibits nuclear export of p53. Interestingly, this residue is the known target for the DNA-PK family of kinases. Thus, such stress-dependent retention of transcription factors in the nucleus through phosphorylation in the vicinity of nuclear export sequence by stress-responsive kinases may emerge as a general theme.
Acknowledgments
We thank Dr. B. Price (Dana–Farber Cancer Institute) for the M059J cells and Dr. C. Kirchgessner (Stanford University) for the M059J cells reconstituted with a chromosomal copy of DNA-PK (34). We are grateful to Dr. M. Yoshida (University of Tokyo) for providing leptomycin B. Our strong appreciation goes to Drs. R. Pepperkok, R. Saffrich, and P. Bastiaens (European Molecular Biology Laboratory, Heidelberg, Germany) for hosting A.Y.K. at the Advanced Light Microscopy Facility and helping with microinjection and imaging experiments. This work was supported by National Institutes of Health Grants GM60216 (to J.M.M.), GM56203 (to L.C.C.), and P01AI42257 (to P.M.H.). A.Y.K. was a Howard Hughes predoctoral fellow, and M.T. was supported by the Deutsche Forschungsgemeinschaft.
Abbreviations
- IRF-3
IFN regulatory factor
- PI3-kinase
phosphatidylinositol 3-kinase
- DNA-PK
DNA-dependent protein kinase
- SV-Sendai virus
dsDNA, double-stranded DNA
References
- 1.Biron C A, Sen G C. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 1. Philadelphia: Lippincott; 2001. pp. 321–352. [Google Scholar]
- 2.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 5.Wathelet M, Lin C H, Parekh B, Ronco L V, Howley P M, Maniatis T. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 6.Juang Y, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver B K, Kumar K P, Reich N C. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 9.Harada H, Taniguchi T, Tanaka N. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 10.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant M J, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 11.Lin R, Mamane Y, Hiscott J. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar K P, McBride K M, Weaver B K, Dingwall C, Reich N C. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronco L V, Karpova A Y, Vidal M, Howley P M. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim T, Kim T Y, Song Y H, Min I M, Yim J, Kim T K. J Biol Chem. 1999;274:30686–30689. doi: 10.1074/jbc.274.43.30686. [DOI] [PubMed] [Google Scholar]
- 15.Heylbroeck C, Balachandran S, Servant M J, DeLuca C, Barber G N, Lin R, Hiscott J. J Virol. 2000;74:3781–3792. doi: 10.1128/jvi.74.8.3781-3792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Servant M J, ten Oever B, LePage C, Conti L, Gessani S, Julkunen I, Lin R, Hiscott J. J Biol Chem. 2001;276:355–363. doi: 10.1074/jbc.M007790200. [DOI] [PubMed] [Google Scholar]
- 17.Smith G C, Jackson S P. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 18.Dasika G K, Lin S C, Zhao S, Sung P, Tomkinson A, Lee E Y. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- 19.Zhou B B, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 20.Hoppe B S, Jensen R B, Kirchgessner C U. Radiat Res. 2000;153:125–130. doi: 10.1667/0033-7587(2000)153[0125:cotrmc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 21.Dignam J D, Martin P L, Shastry B S, Roeder R G. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 22.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaffe M B, Leparc G G, Lai J, Obata T, Volinia S, Cantley L C. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 24.O'Neill T, Dwyer A J, Ziv Y, Chan D W, Lees-Miller S P, Abraham R H, Lai J H, Hill D, Shiloh Y, Cantley L C, Rathbun G A. J Biol Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- 25.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 26.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roulston A, Marcellus R C, Branton P E. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe F, Shirakawa H, Yoshida M, Tsukada K, Teraoka H. Biochem Biophys Res Commun. 1994;202:736–742. doi: 10.1006/bbrc.1994.1992. [DOI] [PubMed] [Google Scholar]
- 29.Yumoto Y, Shirakawa H, Yoshida M, Suwa A, Watanabe F, Teraoka H. J Biochem (Tokyo) 1998;124:519–527. doi: 10.1093/oxfordjournals.jbchem.a022143. [DOI] [PubMed] [Google Scholar]
- 30.Katze M G. Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 31.Williams B R. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 32.Cuddihy A R, Li S, Tam N W, Wong A H, Taya Y, Abraham N, Bell J C, Koromilas A E. Mol Cell Biol. 1999;19:2475–2484. doi: 10.1128/mcb.19.4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkinson J, Lees-Miller S P, Everett R D. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Xiong Y. Science. 2001;292:1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]