Abstract
Cancer-associated fibroblasts induce malignant behavior in genetically initiated but nontumorigenic human prostatic epithelium. The genetic basis for such transformation is still unknown. By using Affymetrix GeneChip technology, we profiled genomewide gene expression of transformed [tumorigenic benign prostatic hyperplasia (BPH1)CAFTD] and parental (nontumorigenic BPH1) cells. We identified differentially expressed genes, which are associated with tumorigenesis or tumor progression. One striking finding is that a significant portion of the down-regulated genes belongs to interferon (IFN)-inducible molecules. We show that IFN inhibited the tumorigenic BPH1CAFTD cell proliferation and colony formation in vitro and inhibited tumor growth in xenografts in vivo. Expression of the IFN-inducible molecules correlates with the growth-inhibiting effects of IFN. In addition, these genes are reported to be mapped mainly to two chromosomal regions, 10q23–26 and 17q21, which are frequently deleted in human prostate cancers. Furthermore, in silico data-mining with the GeneLogic database revealed that expression of the IFN-inducible genes was down-regulated in approximately 30% of the 49 clinically characterized samples of prostatic adenocarcinomas. Collectively, we show that there seems to be a direct link between IFN-inducible molecules and prostatic tumor progression. These findings suggest IFN-inducible molecules as potential therapeutic targets for the treatment of prostate cancer.
Prostate cancer is the most frequent malignancy in aged men and the second leading cause of male cancer death in the United States. Because of histological heterogeneity and the difficulty of conventional genetic analysis, our knowledge about the genes specifically involved in prostate carcinogenesis is still very limited. Gene expression profiling, using a powerful DNA microarray technique, has recently proven to be an effective way to globally analyze genes involved in carcinogenesis of many types of tissues (1, 2) including the prostate (3–5).
Our previous studies have demonstrated that the nontumorigenic human benign prostatic hyperplasia (BPH)1 cell line and its tumorigenic sublines provide a convenient model system for studying prostate tumorigenesis (6–8). The BPH1 line, immortalized by means of a large T antigen oncogene, fails to form tumors even though it survives for several months after being grafted into immunodeficient mice (6). However, these cells become tumorigenic if they are recombined with carcinoma-associated fibroblasts (CAFs) derived from human prostatic carcinomas before grafting (7). The conversion of the nontumorigenic cells to tumorigenic cells elicited by prostate CAF seems to be irreversible, because when epithelial cells isolated from these primary tumors are grafted to new hosts in the absence of CAF, tumors form (8). Advantages of the BPH1 cells include their clonal derivation and relative consistency. Primary prostate tumor tissue, in contrast, is often heterogeneous with mixed cell types and various tumor grades.
In the present experiments, we have used RNAs extracted from the nontumorigenic parental BPH1 line and from two tumorigenic sublines, BPH1CAFTD-01 and -06. We examined the expression profiles of approximately 12,000 probe sets by using Affymetrix (Santa Clara, CA) GeneChip U95A. We found 179 up-regulated and 95 down-regulated probe sets in tumorigenic BPH1CAFTD cells compared with the parental nontumorigenic BPH1 cells. It is particularly interesting that 18 of the 95 down-regulated hits were IFN-inducible molecules. DNA synthesis analysis revealed that IFNs inhibited proliferation of both types of BPH cells. Soft agar assays indicated that colony formation of BPH1CAFTD cells was inhibited when IFN-γ was included in the cultures. In addition, xenograft experiments in nude mice showed that treatment with IFN-γ greatly reduced tumor volumes. Reverse transcription (RT)-PCR analysis revealed that a cyclin-dependent kinase inhibitor, p21, is significantly down-regulated in the tumorigenic BPH1CAFTD cells, and treatment of these cells with IFN-γ reversed its expression back to the basal levels of nontumorigenic BPH1 cells. Most importantly, the deceased expression of the IFN-inducible genes was confirmed in approximately 30% of the 49 clinical samples of prostatic adenocarcinomas.
Materials and Methods
Cell Culture and RNA Preparation.
Parental BPH1 and BPH1CAFTD cells were maintained in RPMI medium 1640 plus 5% FBS (GIBCO/BRL) in 5% CO2-humidified tissue culture incubator. Total RNAs were isolated by using RNeasy columns (Qiagen, Chatsworth, CA), treated with DNase I (Roche Molecular Biochemicals) for 40 min at room temperature, and cleaned with RNeasy columns.
Affymetrix GeneChip Probe Array Analyses.
Oligo microarray experiments were performed and analyzed as described (9). Briefly, after quality determination on test arrays, the samples were hybridized for 16 h at 45°C to Affymetrix Human Genome arrays (U95A). The arrays were washed and then stained with streptavidin-phycoerythrin (genome arrays were amplified with an anti-streptavidin Ab). The arrays were scanned with the GeneArray scanner (Agilent Technologies, Palo Alto, CA). Raw data were collected and analyzed by using Affymetrix microarray suite and data mining tools software. Experiments were done in six replicates for two BPH1 samples. Mann–Whitney pairwise comparison (9) was performed to identify genes that were differentially expressed. Each of the six parental BPH1 samples was compared with BPH1CAFTD samples resulting in 36 pairwise comparisons for both samples. Genes with concordance exceeding 80.6% were considered statistically significant (P < 0.05). Gene lists from both comparisons were then loaded into genespring software (Silicon Genetics, Redwood City, CA) to look for common Affymetrix probe sets in both lists. This resulted in 179 probe sets increased in common to both lists and 95 in common that decreased expression relative to BPH1CAFTD.
GeneLogic (Gaithersburg, MD) BioExpress Database.
A commercially available database, BioExpress, was used to confirm our experimental data. At the time of this work, the database included 49 human prostate tumors and 26 benign prostate hyperplasia samples that had been analyzed on Affymetrix GeneChips (U95 series). Average difference expression values as reported by the database were used to identify a subset of 14 prostate tumors, which demonstrated a down-regulation of the 18 IFN-inducible genes, when compared with prostate hyperplasia samples.
TaqMan (Perkin–Elmer/Applied Biosystems) Real-Time Quantitative RT-PCR Analysis.
TaqMan RT-PCR analysis was performed with the 5′-exonuclease assay by using fluorescent nonextendible oligonucleotide probes (TaqMan PCR detector 7700) as described (10, 11). Cells were grown in Falcon 96-well plates. High-quality RNA was prepared by using Applied Biosystems PRISM 6700 Automated Nucleic Acid Workstation and subjected to TaqMan RT-PCR. Initial RT-PCR amplifications were also examined by agarose gel electrophoresis to ensure that bands were visible only at the expected molecular weights. We used specific probes and primers for human IFN-inducible 56-kD protein, iip56 (probe, TCT CAC AGA GTT CTC AAA GTC AGC AGC CA; forward primer, TGA GCG GGC CCT GAG A; reverse primer, ATA TCT GGG TGC CTA AGG ACC TT), human IFN-stimulated 54-kD protein, isp54 (probe, TCC ATT CTT GCC AGC CTC CAT GCT; forward primer, CCA ATG ATA ATC TCT TCC GTG TCT G; reverse primer, TCT GCG TCT TCA TAC TGA TCT GCT), human ifp35 (probe, TGC CCA TAT AGG AGG TCT GTA TGT TCA CCA AC; forward primer, CAC TGG CCT GGG CTT GG; reverse primer, ATG TGT GAC CCC TCC GCA), human cig49 (probe, TGA CTG CGC CCT GGC CCA; forward primer TGT CAG CAT CTG AGC TTG AGG A; reverse primer, AGG AGC TCT CTG GGA CTG GAG), human aprf (probe, CGG CCA GAG AGC CAG GAG CAT C; forward primer, GGA GGC ATT CGG GAA GTA TTG; reverse primer, CGC TAC CTG GGT CAG CTT CA), human p21 (probe, TGG CCT GGA CTG TTT TCT CTC GGC; forward primer, AAC ACC TTC CAG CTC CTG TAA CAT A; reverse primer, GGG AAC CAG GAC ACA TGG G), and human gapdh (probe, CTGGCATTGCCCTCAACGACCAC; forward primer, CTCCTCCACCTTTGACGCTG; reverse primer, CATACCAGGAAATGAGCTTGACAA).
BrdUrd Immunohistochemistry, Microscopy, and Image Acquisition.
BPH1 and BPH1CAFTD-01 cells were grown either in the presence or absence of IFN-γ for 24 h. BrdUrd (1:5000, Amersham Pharmacia Biosciences) was added to the medium for the last 5 h. Cells were then fixed and processed for anti-BrdUrd immunocytochemistry as described.
Tritiated-Thymidine Incorporation Assays.
To measure DNA synthesis, an identical number of BPH1 and BPH1CAFTD-01cells (4 × 103 per well) were plated in 96-well plates. [3H]thymidine (1 μCi per well) was added for 16 h at 24 h of culture. Cells were harvested by using a Tomtec (Orange, CT) cell harvester as described (11, 12). Data were collected from 8 culture wells of each experimental group and expressed as mean ± SE. Two-tailed unpaired t test was used for statistical analysis.
Colony Formation Assay and Xenograft Experiment.
Cell cutter and colony formation assay were performed as described (8). Briefly, cells were plated in duplicates in 6-well plates at the density of 105, 104, 103 cells per well in 0.3% agar in 1× culture medium over the bottom layer containing 0.5% agar in culture medium. Cells were grown for 3 weeks and fed gently with 1 ml of medium 3 times a week. Cultures were then stained overnight with 10 μg/ml of tetrazolium violet in medium. Colonies larger than 200 μm in diameter were counted. Male athymic nude mice were injected s.c. with 5 × 106 BPH1CAFTD-01 cells in 0.2 ml of culture medium. After grafting, mice were injected s.c. daily with 5 × 106 units/kg of body weight (high dose) or 5 × 105 units/kg of body weight (low dose) IFN-γ. Tumor sizes were measured 3 times a week over a 27-day period.
Results
Gene Expression Profiling in BPH1 Cells.
To identify genetic elements involved in prostate tumor progression, we performed global gene expression analysis of the BPH1-based cell models by using Affymetrix GeneChip technology. Six independent RNA samples were prepared respectively from parental (BPH1) and tumorigenic (BPH1CAFTD) cell lines. To minimize the chance of getting false positives, two independent tumorigenic cell lines (BPH1CAFTD-01 and BPH1CAFTD-06) were used in the study. In addition we focused only on genes that showed the same patterns of expression in both cell lines. Isolated RNAs were labeled and hybridized to the Affymetrix oligo probe arrays. Experiments and data analyses were performed by using the U95A full-length arrays as described (9) and in Materials and Methods.
Of a total of about 12,000 probe sets, we discovered 179 showing significantly increased expression in both tumorigenic BPHCAFTD cell strains. To prioritize the increased calls, the gene list generated in this experiment was crossreferenced to the data sets generated from a variety of different human tumor specimens that were analyzed for gene expression by using Affymetrix GeneChip technologies. Genes that showed increased expression in tumorigenic BPH1CAFTD cells and that were also present in primary human prostate tumors, but absent in pooled normal human epithelial tissues, are listed in Table 1. Among this selected population of genes several potential contributors to the tumorigenesis were found to be up-regulated in BPH1CAFTD cells (Table 1). For example, matrilysin (MMP7), which has elevated expression levels in prostate (13) and breast (14) tumors. Expression of matrilysin is reported to be regulated by mitogenic growth factors such as fibroblast growth factor (FGF; ref. 15) and epidermal growth factor (EGF; ref. 16). Matrilysin has been shown to be involved in the cleavage of integrin (17) and implicated in cancer invasion and metastasis (18, 19). Human cox-2 is an inducer of angiogenesis. Its expression is up-regulated in prostate cancers (20). Inhibitors of cox-2 suppress angiogenesis and inhibit cancer cell growth (21). Our data also revealed a possible involvement of glypican-4 in prostate tumorigenesis. Glypicans are one of the heparin-binding proteoglycans, which affect the binding of heparin-binding growth factors, such as FGF, heregulin, EGF, etc., to their receptors. Recently, there has been rising interest in the roles of glypicans in tumorigenesis (22, 23). Expression of glypican-1 and -4 is up-regulated in breast cancer cells, and decreased expression of glypican-1 is found to be associated with attenuated response of the breast cancer cells to a variety of growth factors (24). Increased expression of these genes with tumor progression of BPH1 to BPH1CAFTD cells suggests that the BPH1 cell models may be useful to dissect out the molecular events underlying prostate tumorigenesis.
Table 1.
List of genes that are increased in tumorigenic BPH cells, which are also present in primary human prostate tumors, but not in normal epithelial tissue pools
| Affymetrix IDs | Accession no. | Description | Fold increase
|
|
|---|---|---|---|---|
| BPH1CAFTD-01 | BPH1CAFTD-06 | |||
| 546_at | S76965 | Protein kinase inhibitor | 1.84 ± 0.17 | 3.11 ± 0.3 |
| 668_s_at | L22524 | Human matrilysin | 53.14 ± 25.21 | 113.55 ± 55.23 |
| 1069_at | U04636 | Human cyclooxygenase-2 (hCox-2) | 2.47 ± 0.39 | 2.98 ± 0.44 |
| 1736_at | M62402 | Human insulin-like growth factor binding protein 6 | 2.28 ± 0.28 | 3.32 ± 0.37 |
| 1788_s_at | U48807 | Human MAP kinase phosphatase (MKP-2) | 3.99 ± 1.24 | 2.98 ± 0.91 |
| 33933_at | X63187 | HE4 extracellular proteinase inhibitor homologue | 2.22 ± 0.35 | 2.04 ± 0.28 |
| 35330_at | AJ012737 | Muscle isoform filamin | 8.87 ± 2.39 | 26.55 ± 4.44 |
| 35732_at | AL031427 | Novel protein | 2.23 ± 0.52 | 2.39 ± 0.55 |
| 36491_at | D82345 | NB thymosin beta | 4.63 ± 1.88 | 13.06 ± 5.45 |
| 36508_at | AF030186 | Glypican-4 (GPC4) | 4.90 ± 1.26 | 7.53 ± 1.55 |
| 36671_at | M27396 | Human asparagine synthetase | 2.16 ± 0.31 | 2.19 ± 0.27 |
| 38827_at | AF038451 | Secreted cement gland protein XAG-2 homolog | 1.93 ± 0.53 | 2.70 ± 0.63 |
IFN-Inducible Genes Comprise a Major Group of Down-Regulated Genes in Tumorigenic BPH1CAFTD Cells.
We also discovered a total of 122 and 209 probe sets that passed the Mann–Whitney U test to show significantly decreased expression in BPH1CAFTD-01 and BPH1CAFTD-06 cells, respectively, with 95 hits showing decreased expression in both cell lines. Among this group of 95 we identified a major set of 18, nearly 20% of the total 95 hits, which are IFN-inducible molecules. Consistently, none of these genes appeared in the increased gene calls, indicating a dramatic inhibition of IFN pathway in the tumorigenic BPH1CAFTD cells. These 18 hits actually represented 11 distinct genes, which are listed in Table 2. A quantitation of the relative decrease of expression of each of these genes is also listed in Table 2. These genes represent downstream effects in a variety of biological events thought to be regulated by IFN. Because the genes in Table 2 can be stimulated by type I and/or type II IFN (25), the inhibition of the IFN pathway is not restricted to either type of IFN.
Table 2.
IFN-inducible genes that are down-regulated in tumorigenic BPH cells
| Affymetrix ID | Accession no. | Description | Fold decrease
|
|
|---|---|---|---|---|
| BPH1CAFTD-01 | BPH1CAFTD-06 | |||
| 909_g_at | M14660 | IFN-stimulated 54-kD protein | 6.98 ± 1.24 | 20.03 ± 7.28 |
| 908_at | 4.76 ± 0.89 | 9.61 ± 1.72 | ||
| 38389_at | X04371 | 2-5A synthetase | 6.91 ± 1.2 | 21.32 ± 5.03 |
| 37014_at | M33882 | p78 protein | 5.11 ± 0.82 | 7.29 ± 1.16 |
| 1358_s_at | U22970 | IFN-inducible peptide (6–16) | 4.89 ± 1.07 | 7.31 ± 1.7 |
| 38584_at | AF026939 | CIG49 | 3.69 ± 0.85 | 7.92 ± 1.41 |
| 464_s_at | U72882 | Leucine zipper protein (IFP35) | 3.65 ± 0.46 | 4.16 ± 0.68 |
| M97935_5_at | M97935 | Transcription factor ISGF-3 | 3.46 ± 1.01 | 7.85 ± 4.57 |
| M97935_MB_at | 2.28 ± 0.36 | 3.22 ± 0.38 | ||
| 32859_at | 2.75 ± 0.42 | 4.36 ± 0.72 | ||
| 33339_g_at | M97936 | 2.44 ± 0.31 | 3.66 ± 0.7 | |
| 33338_at | 2.07 ± 0.3 | 2.63 ± 0.33 | ||
| 289_at | L29277 | DNA-binding protein (APRF) | 1.92 ± 0.2 | 1.92 ± 0.17 |
| 39708_at | 1.88 ± 0.17 | 1.99 ± 0.12 | ||
| 35735_at | M55542 | Guanylate binding protein isoform I (GBP-2) | 4.53 ± 1.42 | 45.85 ± 24.14 |
| 1107_s_at | M13755 | IFN-induced 17-kD/15-kD protein | 2.41 ± 0.54 | 3.27 ± 0.76 |
| 915_at | M24594 | IFN-inducible 56-kD protein | 2.91 ± 0.41 | 5.69 ± 0.68 |
| 32814_at | 1.70 ± 0.24 | 3.18 ± 0.54 | ||
IFN Signaling Machinery Is Intact in BPH1CAFTD Cells.
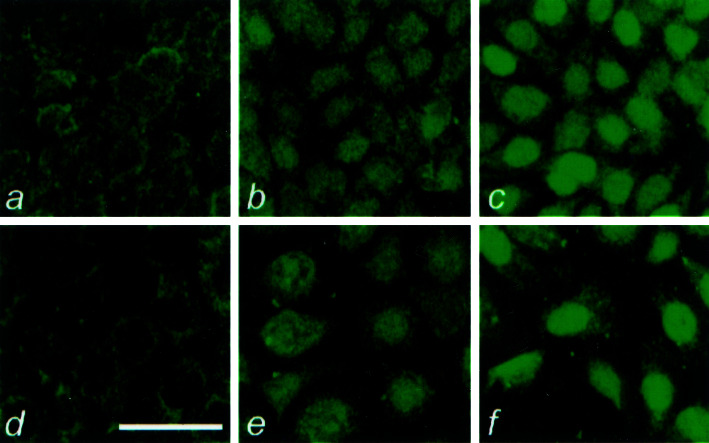
To examine whether the down-regulation of IFN-inducible genes is caused by any defects of IFN signaling machinery, we performed immunocytochemistry in cultured BPH cells with an Ab specifically recognizing phosphorylated signal transducers and activator-1 (STAT1). Previous experiments have shown that STAT1 phosphorylation is a critical event of the IFN signaling pathway. Activation of STAT1 leads to a nuclear translocation of the phosphorylated STAT1 protein (25). As shown in Fig. 1, only weak immunostaining reactivity was seen in the cytoplasm of the cultured BPH1 and BPH1CAFTD-01 cells, and the nuclear staining was not detected (Fig. 1 a and d). However, 15 min after exposure to IFN-γ, intense nuclear staining was observed (Fig. 1 c and f). Similar results were also observed in cultures treated with IFN-α (Fig. 1 b and e). These observations suggest that IFN signaling machinery for both types of IFNs is intact in tumorigenic BPH1CAFTD cells.
Figure 1.
Induction of STAT1 phosphorylation and nuclear translocation by IFNs in BPH1 and BPH1CAFTD cells. Immunocytochemical labeling of antiphosphorylated STAT1 in cultured cells. BPH1CAFTD-01 (a–c) and BPH1 (d–f) cells were cultured in normal medium or 15 min after treatment with IFN-α (b and e) and IFN-γ (c and f), receptively. Note that although there is only a weak diffused cytoplasmic signal in the untreated cells, IFN induces a rapid nuclear translocation of phosphorylated STAT1 protein. [Bar = 50 μm.]
IFN Directly Inhibits Tumorigenic BPH1CAFTD Cell Growth Both in Vitro and in Vivo.
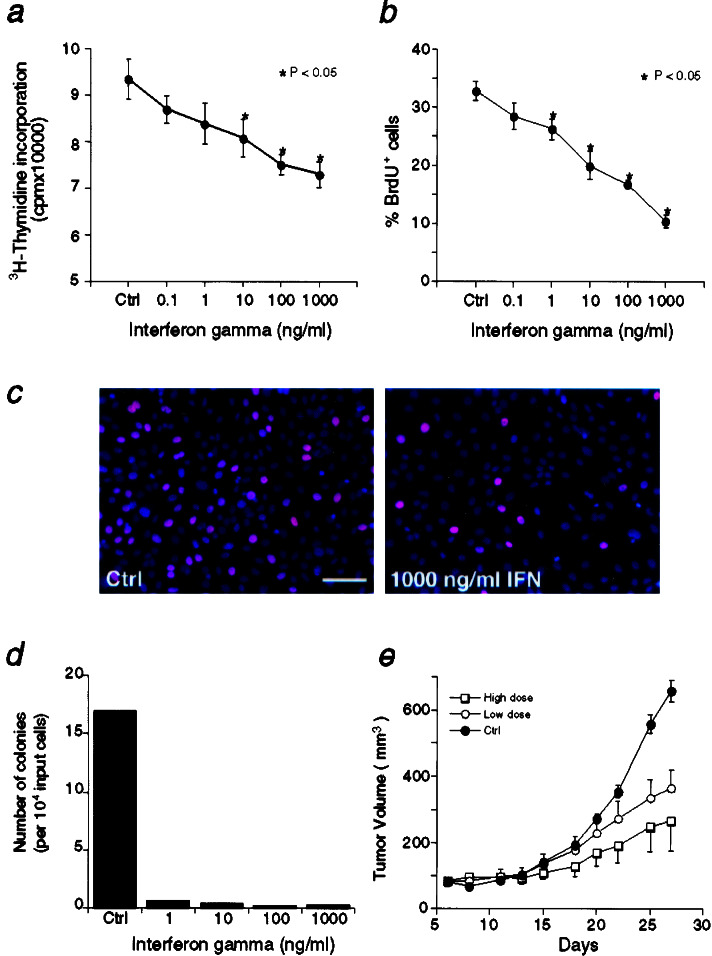
To provide further supporting evidence that IFN is directly involved in prostate tumorigenesis, we performed cell proliferation analysis measuring [3H]thymidine and BrdUrd incorporation. Treatment with IFN-γ significantly inhibited DNA synthesis by tumorigenic BPH1CAFTD-01 cells in a dose-dependent manner (Fig. 2a). Similar growth-inhibiting effects were also observed for IFN-α (data not shown). Cell counts of mitotic (BrdUrd-positive) cells in the BPH1CAFTD-01 cell cultures confirmed that inclusion of IFN-γ significantly reduced the number of proliferating cells (Fig. 2 b and c).
Figure 2.
IFN-γ inhibits cell proliferation and tumorigenesis of BPH1CAFTD cells. (a) Tritiated thymidine incorporation assay. (b) Labeling index of BrdUrd-positive cells. (c) Dual labeling of BrdUrd immunocytochemistry (visualized with Texas red-conjugated secondary Ab) and 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining (blue) of BPH1CAFTD-01cells in control culture (c Left) and in a culture treated with IFN-γ (c Right). Note that the number of proliferating cells was greatly reduced in the presence of IFN-γ (asterisks indicating P < 0.05 in a and b). (d) Colony formation assays of BPH1CAFTD-01 cells in soft agar assays. Note that although BPH1CAFTD cells readily form colonies in control cultures, very few colonies are formed in the cultures treated with IFN-γ. (e) IFN inhibits tumor growth after BPH1CAFTD-01 cells are grafted into nude mice. Note that significant reduction in tumor volume was observed in mice treated with either low or high doses of IFN. For ease of viewing, error bars are depicted unidirectionally in treatment groups. [Bar = 100 μm (c).]
To determine whether IFN-γ would directly influence tumorigenesis of BPH cells, we performed colony-formation assays in soft agar cultures. As shown in Fig. 2d, addition of IFN-γ inhibited colony formation, whereas in control cultures many colonies were formed.
We then extended our in vitro experiments to an in vivo setting. We carried out xenograft experiments in nude mice by s.c. injection of the tumorigenic BPH1CAFTD-01 cells. Although tumors were formed in the mice of all three groups, including the saline control, low dose (5 × 105 units/kg), and high dose (5 × 106 units/kg) of IFN-γ, there was a dramatic reduction in tumor growth in IFN-γ treated mice. After 27 days, 8 of 10 mice in the control group, but only 3 of 10 in high-dose group and 4 of 10 in the low-dose group, had tumors larger than 400 mm3. Although tumors grew aggressively as a function of time in the mice receiving saline injection, tumor growth was significantly inhibited in the mice receiving IFN-γ injection (P < 0.05, Fig. 2e). No significant difference was observed between the two groups treated with different dosages of IFN (P = 0.538), although tumors in the high-dose groups showed a tendency of smaller volume (Fig. 2e).
Induction of IFN-Inducible Molecules Correlates to Growth-Inhibitory Effects.
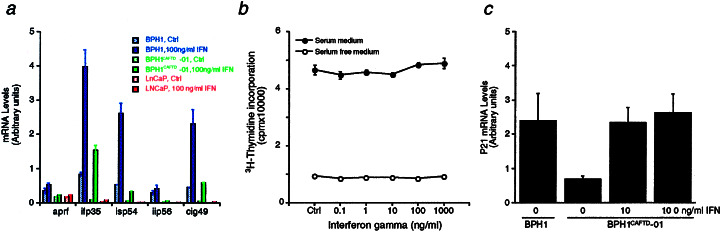
Thus, in tumorigenic BPH1CAFTD cells, the expression of IFN-inducible molecules was suppressed. In addition, IFNs inhibited BPH1CAFTD cell growth in vitro and in vivo. These observations prompted us to determine the effects of IFN on the expression of IFN-inducible molecules during growth inhibition of BPH1CAFTD cells. We performed TaqMan RT-PCR analysis with probes and primers specifically designed for 5 of the 11 genes listed in Table 2. We found that all of the 5 genes examined were indeed up-regulated after IFN (100 ng/ml) treatment (Fig. 3a). In sharp contrast, lymph node-derived prostate cancer (LNCaP) cells [shown to have defects in IFN signaling (26)] did not show up-regulated expression of the 5 genes tested on IFN (100 ng/ml) treatment (Fig. 3a). Furthermore, challenging LNCaP cells with IFN-γ had no significant effect on cell proliferation as measured by [3H]thymidine incorporation (Fig. 3b). To ensure that LNCaP cells do not respond to IFN, the proliferation assay was also performed in defined serum-free medium, so as to eliminate possible factors that might obscure the effects of IFN (Fig. 3b, lower lines). Thus, we demonstrated that there is a direct correlation between the ability of IFN to induce the expression of IFN-inducible molecules and its ability to suppress prostate cancer cell growth.
Figure 3.
Correlation of IFN-inducible molecule expression with growth inhibition. (a) Quantitative RT-PCR analysis of expression levels of five IFN-inducible genes in BPH1, BPH1CAFTD-01, and LNCaP cells in the absence or presence of 100 ng/ml of IFN-γ. The five IFN-inducible genes include aprf, ifp35, isp54, iip56, and cig49. Note that expression levels of all five genes are elevated after treatment of IFN-γ in both BPH1 and BPH1CAFTD-01 cells. In contrast, no elevation was seen in the LNCaP cells. (b) Tritiated thymidine incorporation assay in LNCaP cells indicates that IFN-γ has no effect on the proliferation of LNCaP cells in either serum-containing medium or serum-free medium. (c) Expression levels of p21 in cultured tumorigenic and nontumorigenic BPH1 cells. Quantitative RT-PCR analysis of BPH1 and BPH1CAFTD-01 cells in the absence or presence of IFN-γ.
IFN Treatment Elevates Expression Levels of p21 in Tumorigenic BPH Cells.
As an attempt to understand the possible mechanism by which IFN-γ regulates proliferation of BPH1CAFTD cells, we performed quantitative RT-PCR analysis of gene expression of cell cycle-related genes including p21 and p27, cyclin-dependent kinase inhibitors, in the tumorigenic BPH1CAFTD cells after IFN-γ treatment. We found that although there was no change in the expression levels of p27 (data not shown), the expression of p21 was significantly lower in the tumorigenic BPH1CAFTD cells, compared with the parental nontumorigenic cells (Fig. 3c). After IFN-γ treatment, however, there was an up-regulation of p21 in tumorigenic BPH1CAFTD cells to levels comparable to the nontumorigenic BPH1 cells (Fig. 3c).
Down-Regulation of IFN-Inducible Genes Is Validated in Clinical Samples of Prostatic Tumors.
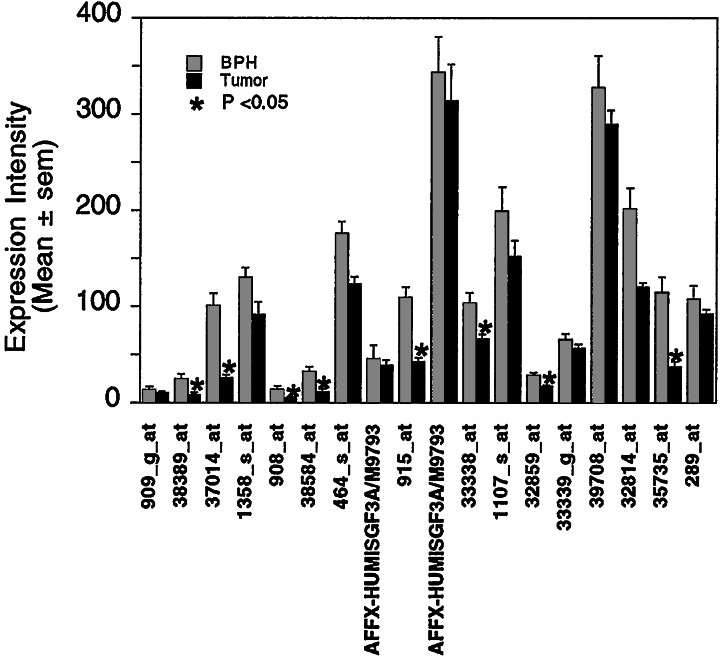
To extend our findings in more complex clinical human prostate adenocarcinomas, the expression of the 18 IFN-inducible hits identified above was examined by querying the GeneLogic database (see Materials and Methods). Among a total of 49 pathologically characterized prostatic adenocarcinoma samples, we identified a subgroup of 14 malignant tumors (approximately 30% of the total samples) in which the expression of the IFN-inducible molecules were down-regulated as compared with the nonmalignant human BPH samples, consistent with what we discovered from our BPH1 cell model studies. All 18 probe sets showed a trend of decreased expression in malignant samples, and the differences in 8 of the 18 hits were statistically significant (Welch t test, P < 0.05, as marked by asterisks in Fig. 4).
Figure 4.
Comparison of tumor vs. BPH expression values for 18 IFN-inducible probe sets. Fourteen prostate tumors were identified in the GeneLogic BioExpress Database that demonstrated low-level expression of the 18 IFN-inducible probe sets. Average expression value (i.e., average of the average differences) is depicted for the 14 prostate tumors compared with the average of the 26 BPH samples. Asterisks depict those probe sets that demonstrate statistical significance (P < 0.05, using a Welch t test).
Discussion
By using an Affymetrix oligo microarray technique, we compared gene expression profiles of tumorigenic vs. nontumorigenic BPH1 cells. Of a total of 12,000 probe sets we identified 274 that are differentially expressed in tumorigenic compared with nontumorigenic BPH1 cell strains. Many of the genes, such as matrilysin, cox-2, and glypican, identified in this study have been previously demonstrated to be associated with carcinogenesis. However, to our knowledge, the present work for the first time links glypican-4 with prostate tumor progression. Moreover, the genes listed in Table 1 are also present in primary human prostate tumor samples but absent in pooled human epithelial tissues, indicating a similar trend toward expression in malignant tissue vs. nonmalignant samples. These findings suggest that the BPH1 cell model, coupled with DNA microarray technique and in silico data mining, can be a valid way to identify genes associated with prostate tumor progression. To uncover novel molecules involved in prostate tumor progression, we used Affymetrix human expressed sequence tag chips (U95B-E).
Previous studies have suggested therapeutic potential of IFNs for the treatment of cancer (27). However, the anti-tumor mechanisms of IFN have not been completely understood. It has been shown that IFN can cause cell cycle arrest, down-regulation of her2 expression, and inhibits metastatic potential in a few metastatic prostate cancer cell lines (28, 29), but the direct link between down-regulation of the IFN pathway and prostate tumorigenesis was still lacking. On the other hand, mice deficient in the IFN-γ receptor or stat1 develop spontaneous or chemically induced tumors more rapidly than wild-type mice (30). IFN-γ and lymphocytes act together to prevent various tumor development and shape tumor immunogenicity (31). Therefore, IFNs are mainly believed to play an indirect immunosurveillance role that is not specific for prostate cancer. In the present study, using genomewide gene expression-profiling approaches, we demonstrated that the IFN signaling pathway is indeed suppressed when BPH1 cells become tumorigenic. The suppression of IFN signaling resulted in a decreased expression of a large group of IFN-inducible molecules in tumorigenic BPH1CAFTD cells. Our in vitro cell proliferation and colony formation assays coupled with in vivo xenograft studies provided strong support for the notion that IFNs can directly act on BPH1CAFTD cells to elicit growth-inhibiting effects. Consistent with the antiproliferative effects of IFN, we showed that the expression level of p21 was down-regulated in the tumorigenic BPH1CAFTD cells and that treatment with IFN-γ brought p21 expression levels back to the basal levels of parental nontumorigenic BPH1 cells.
There are currently several models available to study prostate carcinogenesis. For example, the TRAMP model is generated by targeted expression of a transgene carrying the rat probasin gene fused to the SV40 T antigen (32). This model has been used in various prostate cancer studies, including tumor initiation, progression, metastasis, and prevention (32, 33).By using tissue recombination technology, the functional consequences of the retinoblastoma (Rb) tumor suppressor in development and progression of prostate cancer and the susceptibility to hormonal carcinogenesis have been addressed (34). Our studies using BPH1 cell lines are an example of another model. These cells grown in culture permit a very controllable sample. The advantage of such a sample is the reduction of complexity found in tumors in situ, which will have a variety of other cell types and potential necrotic sites contained in the harvest. We readily admit that a cell culture model does have its limitations. One such limitation is neither the parental BPH1 nor its derivatives express androgen receptor (AR). The lack of AR expression in BPH1 cells prevents them from being an ideal model to study hormonal responses in prostate cancer. Results obtained from any given model system are subject to further validation by using clinical samples. In this light, the expression of the 18 IFN-inducible hits was screened in clinical samples by using in silico biology approaches. By mining the GeneLogic database, which is generated by profiling gene expression in a large sets of human clinical samples, we identified a subgroup of prostate adenocarcinomas in which the expression of these hits was lower than in nonmalignant human BPHs. Because of the high heterogeneity of human tumor samples, we think that this portion is a significant one, given the fact that even Her2, which is the drug target for Herceptin, a highly effective mAb to treat breast cancer, is overexpressed only in about 25% of the breast cancer patients (35). The complexity in human tumor samples makes it difficult to identify genes that may be associated with carcinogenesis in just a subset of tumors. The down-regulation of IFN-inducible genes in the subset of tumors would not have been identified if only real clinical samples were analyzed, demonstrating the value of using the cell model system to study complex disease.
Coincidentally, cytogenetic mapping of the IFN-inducible molecules, using the tools embedded in genespring, a gene expressing analysis software used in this study, revealed two chromosomal regions, 10q and 17q, which harbor three and two of the IFN-inducible molecules, respectively. aprf and ifp35 are located in the region of 17q21, whereas isp54 is mapped to 10q23–25, iip56 is mapped to 10q25–26, and cig49 is mapped to 10q24. Furthermore, previous genetic studies have indicated that loss of 17q21–23 or 10q23–25 is a frequent event in prostate cancer progression. These regions are thought to harbor putative tumor suppressors and metastasis-suppressor genes (36, 37). Although one major tumor suppressor that has been characterized in 10q region is pten (38), these findings together suggest that in addition to the deletion of pten, there could be loss or down-regulation of the IFN-inducible molecules in prostate cancers.
It is well demonstrated that ligand-dependent activation of IFN pathway involves a cascade of protein phosphorylation, which requires the specific type I and type II receptors, the Janus kinases (JAKs), and STATs [for review, see Stark et al. (25)]. Phosphorylated STATs translocate to the nucleus where target gene transcription is activated. One simple interpretation for our finding that expression of 11 distinct IFN-inducible genes is suppressed in the tumorigenic BPH1CAFTD cells could be that the receptor-STAT pathway is impaired in BPH1CAFTD cells. However, our observations that STAT phosphorylation is intact and that IFNs are able to induce gene expression in these cells are against this model. The second explanation for the suppression of IFN pathway in BPH1CAFTD cells might be that parental BPH1 cells produce more IFNs than the BPH1CAFTD cells. Although no evidence was obtained at mRNA levels (data not shown), such a possibility cannot be completely ruled out because there may be other subtypes of IFNs that have not been identified yet. On the other hand, IFN signaling has been reported to cross paths with others. For example, ras/AP-1 signaling antagonizes the JAK/STAT pathway of IFN signaling in the regulation of expression of the macrophage scavenger receptor gene, via a mechanism of competition for limited amount of CBP and p300 (39). By analogy, it is conceivable that induction of IFN-inducible molecules in the BPH1CAFTD cells may be suppressed by a convergence with an antagonizing signaling pathway. It is possible that in the BPH1CAFTD cells a crossinhibition pathway somehow suppresses, but does not eliminate, the activation of IFN pathway. In this way, the BPH1CAFTD cells remain responsive to IFN treatment. In the nontumorigenic parental BPH1 cells, however, such suppression is not activated, resulting in a higher expression of IFN-inducible genes.
The question, then, is whether suppression of IFN signaling is important in tumorigenesis. To address this issue, we performed experiments to examine the correlation of growth-inhibitory effects of IFN and the induction of the expression of IFN-inducible molecules. To appreciate the importance of the 10q23–25 and 17q21–23 regions, five genes located in these regions were tested (Fig. 3a). Our data showed that treatment of the BPH1CAFTD cells with either IFN-α or -γ significantly induced the expression of all five genes. In contrast, in LNCaP cells, which are known to have defects in IFN signaling (26), there was no induction of these genes. Coincidentally, IFNs did not inhibit LNCaP cell growth. Such a correlation indicates the importance of a direct IFN effect on the suppression of tumor progression. It is known that in IFN-mediated immunotherapies some patients fail to respond to IFN (27). Our study provides a possible explanation for this phenomenon, that is, the unresponsive patients might have lost the IFN-inducible genes. Such a loss would eliminate the direct effects of IFNs. In this light, our work not only stresses the importance of pharmacogenomics but also suggests an alternative direction for prostate cancer treatment, that is, to target the IFN-inducible molecules, in particular, for those patients who are resistant to IFN therapy.
Acknowledgments
We thank V. Smith, P. Polakis, and K. Hillan for giving us the access to their Affymetrix experiment database; T. Wu, K. Jung, and Z. Zhang for assistance in bioinformatics; M. Ostland for assistance in biostatistics; and J. Zavala-Solorio for assistance with xenograft experiments.
Abbreviations
- BPH
benign prostatic hyperplasia
- LNCaP
lymph node-derived prostate cancer
- RT
reverse transcription
- CAF
carcinoma-associated fibroblasts
- STAT
signal transducers and activator-1
References
- 1.Perou C, Sorlie T, Eisen M, van de Rijn M, Jeffrey S, Rees C, Pollack J, Ross D, Johnsen H, Akslen L, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Clark E, Golub T, Lander E, Ro H. Nature (London) 2001;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 3.Dhanasekaran S, Barrette T, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta K, Rubin M, Chinnaiyan A. Nature (London) 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 4.Luo J, Duggan D, Chen Y, Sauvageot J, Ewing C, Bittner M, Trent J, Isaacs W. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 5.Welsh J, Sapinoso L, Su A, Kern S, Wang-Rodriguez J, Moskaluk C, Frierson H J, Hampton G. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 6.Hayward S W, Dahiya R, Cunha G R, Bartek J, Deshpande N, Narayan P. In Vitro Cell Dev Biol Anim. 1995;31:14–24. doi: 10.1007/BF02631333. [DOI] [PubMed] [Google Scholar]
- 7.Olumi A F, Grossfeld G D, Hayward S W, Carroll P R, Tlsty T D, Cunha G R. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward S W, Wang Y Z, Cao M, Hom Y K, Zhang B, Grossfeld G D, Sudilovsky D, Cunha G R. Cancer Res. 2001;61:8135–8142. [PubMed] [Google Scholar]
- 9.Jin H, Yang R, Awad T, Wang F, Li W, Williams S, Ogasawara A, Shimada B, Williams P, de Feo G, Paoni N. Circulation. 2001;103:736–742. doi: 10.1161/01.cir.103.5.736. [DOI] [PubMed] [Google Scholar]
- 10.Zheng J, Gao W-Q. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 11.Shou J, Ross S, Koeppen H, de Sauvage F, Gao W-Q. Cancer Res. 2001;61:7291–7297. [PubMed] [Google Scholar]
- 12.Zheng J L, Helbig C, Gao W-Q. J Neurosci. 1997;17:216–226. doi: 10.1523/JNEUROSCI.17-01-00216.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto K, Kihira Y, Matuo Y, Usui T. J Urol. 1998;160:1872–1876. [PubMed] [Google Scholar]
- 14.Heppner K J, Matrisian L M, Jensen R A, Rodgers W H. Am J Pathol. 1996;149:273–282. [PMC free article] [PubMed] [Google Scholar]
- 15.Klein R D, Maliner-Jongewaard M S, Udayakumar T S, Boyd J L, Nagle R B, Bowden G T. Prostate. 1999;41:215–223. doi: 10.1002/(sici)1097-0045(19991201)41:4<215::aid-pros1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 16.Sundareshan P, Nagle R B, Bowden G T. Prostate. 1999;40:159–166. doi: 10.1002/(sici)1097-0045(19990801)40:3<159::aid-pros3>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 17.von Bredow D C, Nagle R B, Bowden G T, Cress A E. Exp Cell Res. 1997;236:341–345. doi: 10.1006/excr.1997.3711. [DOI] [PubMed] [Google Scholar]
- 18.Agnihotri R, Crawford H C, Haro H, Matrisian L M, Havrda M C, Liaw L. J Biol Chem. 2001;276:28261–28267. doi: 10.1074/jbc.M103608200. [DOI] [PubMed] [Google Scholar]
- 19.Abdel-Ghany M, Cheng H C, Elble R C, Pauli B U. J Biol Chem. 2001;276:25438–25446. doi: 10.1074/jbc.M100478200. [DOI] [PubMed] [Google Scholar]
- 20.Kirschenbaum A, Liu X, Yao S, Levine A C. Urology. 2001;58:127–131. doi: 10.1016/s0090-4295(01)01255-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu X H, Kirschenbaum A, Yao S, Lee R, Holland J F, Levine A C. J Urol. 2000;164:820–825. doi: 10.1097/00005392-200009010-00056. [DOI] [PubMed] [Google Scholar]
- 22.Kleeff J, Ishiwata T, Kumbasar A, Friess H, Buchler M W, Lander A D, Korc M. J Clin Invest. 1998;102:1662–1673. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filmus J. Glycobiology. 2001;11:19R–23R. doi: 10.1093/glycob/11.3.19r. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda K, Maruyama H, Guo F, Kleeff J, Itakura J, Matsumoto Y, Lander A D, Korc M. Cancer Res. 2001;61:5562–5569. [PubMed] [Google Scholar]
- 25.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein D, O'Leary M, Mitchen J, Borden E C, Wilding G. J Urol. 1991;146:1173–1177. doi: 10.1016/s0022-5347(17)38034-5. [DOI] [PubMed] [Google Scholar]
- 27.Kuratsukuri K, Nishisaka N, Jones R, Wang C, Haas G. Urol Oncol. 2000;5:265–273. doi: 10.1016/s1078-1439(00)00086-7. [DOI] [PubMed] [Google Scholar]
- 28.Hobeika A, Etienne W, Cruz P, Subramaniam P, Johnson H. Int J Cancer. 1998;77:138–145. doi: 10.1002/(sici)1097-0215(19980703)77:1<138::aid-ijc21>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Kominsky S L, Hobeika A C, Lake F A, Torres B A, Johnson H M. Cancer Res. 2000;60:3904–3908. [PubMed] [Google Scholar]
- 30.Kaplan D, Shankaran V, Dighe A, Stockert E, Aguet M, Old L, Schreiber R. Proc Natl Acad Sci USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shankaran V, Ikeda H, Bruce A T, White J M, Swanson P E, Old L J, Schreiber R D. Nature (London) 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 32.Greenberg N M, DeMayo F, Finegold M J, Medina D, Tilley W D, Aspinall J O, Cunha G R, Donjacour A A, Matusik R J, Rosen J M. Proc Natl Acad Sci USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gingrich J, Barrios R, Morton R, Boyce B, DeMayo F, Finegold M, Angelopoulou R, Rosen J, Greenberg N. Cancer Res. 1996;56:4096–4102. [PubMed] [Google Scholar]
- 34.Wang Y, Hayward S, Donjacour A, Young P, Jacks T, Sage J, Dahiya R, Cardiff R, Day M, Cunha G. Cancer Res. 2000;60:6008–6017. [PubMed] [Google Scholar]
- 35.Slamon D, Pegram M. Semin Oncol. 2001;28:13–19. doi: 10.1016/s0093-7754(01)90188-5. [DOI] [PubMed] [Google Scholar]
- 36.Gray I C, Phillips S M, Lee S J, Neoptolemos J P, Weissenbach J, Spurr N K. Cancer Res. 1995;55:4800–4803. [PubMed] [Google Scholar]
- 37.Chekmareva M A, Hollowell C M, Smith R C, Davis E M, LeBeau M M, Rinker-Schaeffer C W. Prostate. 1997;33:271–280. doi: 10.1002/(sici)1097-0045(19971201)33:4<271::aid-pros8>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Ernsting B R, Wishart M J, Lohse D L, Dixon J E. J Biol Chem. 1997;272:29403–29406. doi: 10.1074/jbc.272.47.29403. [DOI] [PubMed] [Google Scholar]
- 39.Horvai A, Xu L, Korzus E, Brard G, Kalafus D, Mullen T, Rose D, Rosenfeld M, Glass C. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]