Abstract
TNF-related apoptosis-inducing ligand (TRAIL), a member of the TNF superfamily, was suggested to contribute to HIV-1 pathogenesis by inducing CD4+ T cell death characteristic of AIDS. We previously reported HIV-1-mediated, TRAIL-induced apoptosis in primary CD4+ T cells in vitro and observed elevated levels of plasma TRAIL in HIV-1-infected patients. The present study elucidates the unresolved mechanism by which HIV-1 induces TRAIL expression on primary CD4+ T cells. We demonstrate that the expression of TRAIL by primary CD4+ T cells is regulated by IFN-α that is produced by HIV-1-stimulated plasmacytoid dendritic cells (pDCs). We also found that IFN-induced TRAIL is mediated by signal transducers and activators of transcription 1 and 2. We show that IFN-α production by HIV-1-activated pDCs is blocked by an early viral entry inhibitor of CD4-gp120 binding, but not by inhibitors of viral coreceptor binding. Our in vitro data are supported by the demonstration that anti-IFN-α and -β Abs inhibit apoptosis and TRAIL expression in CD4+ T cells from HIV-1-infected patients. Our findings suggest a potential unique role of pDCs in the immunopathogenesis of HIV-1 infection by inducing the death molecule TRAIL.
TNF-related apoptosis-inducing ligand (TRAIL), a TNF superfamily member (1), has two death receptors (DRs) that induce apoptosis (2) (DR4 and DR5) and three other receptors that lack the death domain and engage ligand without initiating apoptosis (3). TRAIL protein is expressed on the cell membrane (mTRAIL) or is secreted (soluble TRAIL) (1), and both the soluble and membrane-bound forms induce apoptosis of cells expressing functional DRs (4). TRAIL may contribute to HIV-1 immunopathogenesis because CD4+ and CD8+ T cells from HIV-1-infected patients are more susceptible to TRAIL-induced apoptosis in vitro than T cells from healthy donors (5-7). TRAIL induced the selective apoptosis of uninfected CD4+ T cells in HIV-1-infected hu-PBL-NOD-SCID mice (8). TRAIL, produced by monocytes exposed to the HIV-1 Tat protein, induced apoptosis of uninfected CD4+ T cells (9). We recently reported that plasma TRAIL levels in HIV-1-infected patients directly correlate with viral load (10). We also reported that CD4+ T cells exposed to HIV-1 underwent apoptosis by a TRAIL-DR5-dependent mechanism, which was inhibited by anti-type I IFN Abs (11). These latter results suggest that type I IFN plays a fundamental role in HIV-1-induced TRAIL expression by CD4+ T cells.
Type I IFN (IFN-α/β) has antiviral activity, including activity against HIV-1 (12). IFN-α/β are produced mainly by plasmacytoid dendritic cells (pDCs) (13). pDCs are located in blood and lymphoid tissue (13-15) and participate in innate immune responses against viruses by producing large amounts of IFN-α, -β, and -ω (16, 17). Interestingly, pDCs secrete type I IFN when stimulated by infectious HIV-1 or chemically inactivated HIV-1 virions (18). The majority of plasma HIV-1 is not infectious (19, 20), raising the possibility that such virions contribute to HIV-1 pathogenesis and CD4+ T cell loss, even without mediating productive infection.
The IFN receptor has two subunits, one common with the type II IFN receptor (IFNAR1) and one specific for type I IFN (IFNAR2). IFNAR2 binds type I IFN with high affinity (21). IFN-α/β binding triggers receptor dimerization and activation, leading to phosphorylation of a tyrosine residue on IFNAR1, creating a docking site for the signal transducers and activators of transcription (STAT) 2 (22). STAT2 and STAT1 are subsequently phosphorylated and form heterodimeric complexes (23). The STAT heterodimer associates with IFN regulatory factor-9, translocates to the nucleus, and binds to the IFN-stimulated gene response element, located upstream from several IFN-regulated genes (24), including TRAIL (25).
HIV-1-induced TRAIL expression by primary CD4+ T cells, its dependence on type I IFN, and the cellular source of type I IFN have not been clarified. We demonstrate here that both inactivated and infectious HIV-1 stimulate pDCs to produce IFN-α, which induces TRAIL expression by CD4+ T cells. These findings provide a unique role for pDCs in TRAIL-mediated apoptosis of CD4+ T cells in HIV-1 infection and progression to AIDS.
Materials and Methods
HIV-1-Infected Patients. Blood was collected from 13 HIV-1-infected patients enrolled in a U.S. Air Force natural history protocol at Wilford Hall Medical Center. Plasma HIV-1 RNA was measured by quantitative RT-PCR; data were expressed as copies per milliliter (Amplicor Monitor, Roche Diagnostics; detection limit 50 copies per milliliter). Patient and control blood were studied under institutional review board-approved protocols and consent forms from the National Cancer Institute and Wilford Hall Medical Center. Ficoll-purified peripheral blood mononuclear cells (PBMC) from patients' blood were cultured 2 d in medium supplemented with 50% autologous plasma in the absence or presence of anti-IFN Abs at 5 μg/ml (Biosource, Camarillo, CA). Cells were tested for apoptosis and TRAIL expression.
Preparation of Noninfectious Aldrithiol-2 (AT-2) HIV-1. HIV-1MN (X4) and HIV-1Ada (R5) were inactivated with 1 mM AT-2 for 18 h at 4°C (AT-2 HIV-1), as described (26). Elimination of infectivity of HIV-1 by AT-2 was verified for the viral preparations used in this article. Microvesicles, isolated from uninfected cell cultures, were used as a negative control (27).
Isolation and Culture of T Cells and pDCs. All in vitro experiments were performed by using PBMC isolated by density centrifugation from peripheral blood of HIV-1-seronegative blood bank volunteers. Cells were cultured in RPMI medium 1640 (Invitrogen) containing 10% FBS (Sigma) and 1% Pen-Strep-Glut (Invitrogen). CD4+ cells were purified from PBMC by positive selection with anti-CD4 (Miltenyi Biotec, Auburn, CA). The purity of CD4+ cells after purification was >98%. The pDCs (CD4+, CD123+, and BDCA-2+) were isolated from PBMC by using the BDCA-2 isolation kit (Miltenyi Biotec). pDCs were removed from positively isolated CD4+ cells by using the same blood dendritic cell antigen-2 kit.
Incubation of Leukocytes with HIV-1, gp120, and Tat. Isolated CD4+ and CD8+ cells were cultured with AT-2 HIV-1MN or AT-2 HIV-1Ada at 50 ng/ml p24CA equivalent. Infectious HIV-1MN [tissue culture 50% infective dose (TCID50) = 106] and HIV-1Ada (TCID50 = 1,000) were used at the same concentrations. We verified HIV-1 infection by measuring p24 levels in culture supernatants by ELISA (Beckman Coulter). pDCs were cultured with infectious and noninfectious HIV-1MN or HIV-1Ada, gp120MN, gp120SF162, gp160MN, or Tat (AIDS Research and Reference Reagent Program, Bethesda) at different concentrations for 24 h, and the supernatants were collected and tested in cultures of CD4+ T cells. To eliminate effects of alloreactivity, T cells and pDCs used in each individual experiment were isolated from PBMC of the same donor.
Blocking Assay. pDCs were cultured for 24 h with AT-2 HIV-1 in the presence of soluble CD4-IgG (sCD4), which blocks the interaction between envelope gp120 and the CD4 receptor molecule, or in the presence of AMD-3100 (AMD; AIDS Research and Reference Reagent Program) or regulated activation, normal T expressed and secreted (RANTES) (Pepro-Tech, Rocky Hill, NJ), which blocks gp120 binding to the CXCR4 and CCR5, respectively. sCD4, AMD, and RANTES were used at 1 μg/ml. Supernatants of those cultures were tested for IFN-α levels, and pDCs were stained by using Abs against CXCR4 or CCR5 (Pharmingen).
Detection of STAT1 and Membrane TRAIL by Flow Cytometry. Isolated CD4+ cells were cultured for 24 h in the absence or presence of AT-2 HIV-1MN or AT-2 HIV-1Ada. After two washes, cells were incubated for 20 min at room temperature with phycoerythrin-conjugated mouse IgG1 anti-human TRAIL mAb (RIK-2) (eBioscience, San Diego, CA), PercP-conjugated anti-CD4 mAb, APC-conjugated anti-CD3 mAb (BD Bioscience, San Jose, CA), or isotype-matched control Abs (at 5 μg/ml each) in PBS containing 2% mouse serum (Sigma). Cells were washed twice in ice-cold PBS and analyzed by flow cytometry. Intracellular staining was performed for STAT1 detection by using the Fix and Perm kit (Caltag, Burlingame, CA) and STAT1 phosphorylated mAb (BD Bioscience), according to the manufacturer's instructions. FACS analysis was performed on CD3+CD4+-gated cells.
Introduction of Abs into Cells. To introduce Abs into intact cells without permeabilization, we used a Chariot kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. The procedure was used to introduce blocking Abs to STAT1, STAT2, and/or isotype control Abs (Upstate Biotechnology, Lake Placid, NY) into primary CD4+ T cells. After a 4-h incubation with chariot reagent complexed to Ab, HIV-1 was added to the cell cultures, and mTRAIL was monitored on CD4+ T cells after 24 h by FACS. We cultured CD4+ T cells with an empty chariot (no Abs, mock) and without HIV-1 as internal negative control.
Type I IFN Detection. Type I IFN levels were detected by ELISA (R & D Systems).
Western Blot. Isolated CD4+ cells were cultured with infectious or noninfectious HIV-1MN or HIV-1Ada in the presence or absence of anti-IFN-α and -β Abs (R & D Systems), recombinant IFN-α (rIFN-α) (50 ng/ml), or rIFN-β (50 ng/ml). After 24 h, cells were pelleted and lysed [1% Nonidet P-40/200 mM NaCl/50 mM Tris, pH 7.5/protease inhibitor mixture (Roche Diagnostics)]. Total protein was quantified by using bicinchoninic acid (Pierce). Protein samples (10 μg) were run on a 15% SDS/PAGE gel (Bio-Rad). After transfer to a nitrocellulose membrane, blots were blocked in 5% milk/Tris-buffered saline with triton and then incubated with anti-STAT1 or -STAT2 (Upstate Biotechnology), followed by anti-mouse horseradish peroxidase-conjugated secondary Ab (Jackson ImmunoResearch). Enhanced chemiluminescence was performed, and bands were visualized on Hyperfilm (Amersham Pharmacia). β-Actin mAb was used as a loading control (Sigma).
Statistical Analysis. Experiments were repeated at least four times, and P values were determined by using a two-tailed Student's t test. P < 0.05 was considered statistically significant.
Results
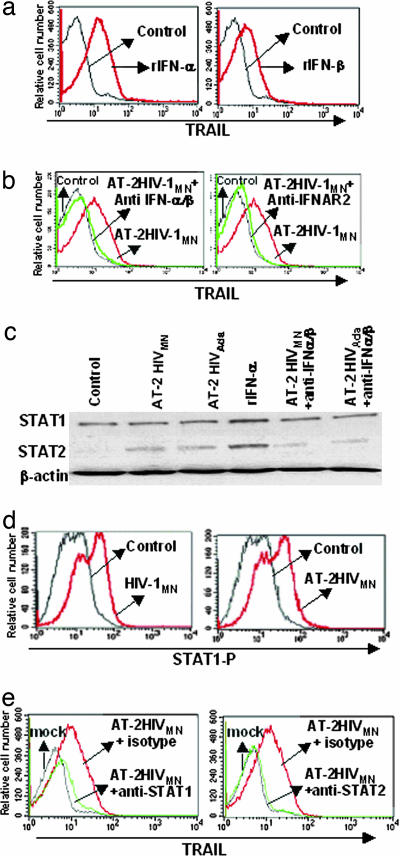
Induction of TRAIL, STAT1, and STAT2 Expression in CD4+ T Cells by HIV. We tested the effect of recombinant type I IFN on mTRAIL expression by primary isolated CD4+ cells. Both rIFN-α and -β induced mTRAIL expression on CD4+ T cells (distinguished by gating on CD3+CD4+ T cells by flow cytometry) (Fig. 1a). Cells cultured with media alone did not express mTRAIL (control). Exposure of cells to AT-2 HIV-1MN resulted in expression of mTRAIL on a CD4+ T cell, which was blocked by Abs against IFN-α/β or IFNAR2 (Fig. 1b). Similar results were obtained by using infectious HIV-1 particles (data not shown). Because STAT1 and STAT2 are type I IFN-signaling molecules, we studied their expression in CD3+CD4+ T cells after culture with HIV-1 (Fig. 1c). HIV-1 exposure did not significantly increase STAT1 expression compared with microvesicles or untreated cells (controls). However, STAT2 levels were increased by exposure of cells to noninfectious HIV-1MN or HIV-1Ada. The same results were obtained by using infectious HIV-1MN or HIV-1Ada (data not shown). We verified that rIFN-α also up-regulated STAT1 and STAT2 expression (positive controls). Anti-IFN-α/β Abs inhibited STAT2 expression in cells cultured with HIV-1, demonstrating that HIV-1-mediated STAT2 expression was due to type I IFN. Because we did not see increased expression of STAT1 in cells cultured with HIV-1, we used flow cytometry to measure expression of the active form of STAT1, phosphorylated STAT1. Infectious and AT-2 HIV-1 induced high levels of phosphorylated STAT1 on CD4+ T cells compared with controls (P = 0.0001) (Fig. 1d). To further demonstrate that STAT1 and STAT2 were responsible for TRAIL expression in CD4+ T cells, we used the Chariot system to deliver blocking anti-STAT1, anti-STAT2, or isotype control Abs into the cytoplasm. Both anti-STAT1- and -STAT2-blocking Abs greatly reduced HIV-1-mediated TRAIL expression on CD4+ T cells compared with isotype control Abs (Fig. 1e). These results demonstrate that HIV-1 induced increased expression of STAT2 and phosphorylation of STAT1, resulting in TRAIL expression by CD4+ T cells.
Fig. 1.
Regulation of TRAIL expression on CD4+ T cells. (a) Detection of mTRAIL by flow cytometry on CD4+ T cells after a 24-h incubation with IFN-α or rIFN-β.(b) HIV-1MN-induced mTRAIL expression on CD4+ T cells is inhibited by anti-IFN-α/β- or anti-IFNAR2-blocking Abs. (c) STAT1 and STAT2 expression determined by Western blot in CD4+ cells after a 24-h culture with AT-2 HIV-1MN, AT-2 HIV-1Ada, rIFN-α, AT-2 HIV-1MN plus anti-IFN-α/β Abs, or AT-2 HIV-1Ada plus anti-IFN-α/β Abs. β-Actin was used as a loading control. (d) Intracellular staining of phosphorylated STAT1 (STAT1-P) in CD4+ T cells after a 24-h culture with HIV-1MN or AT-2 HIV-1MN. Control is CD4+ T cells cultured with microvesicles or media. (e) Inhibition of STAT1 and STAT2 protein in CD4+ T cells. Anti-STAT1, anti-STAT2, and isotype Abs were delivered into CD4+ T cells by using a Chariot kit. Cells were cultured for 24 h with AT-2 HIVMN plus isotype Ab, AT-2 HIVMN plus anti-STAT1, or -STAT2 Abs. Chariot was also used without Abs as negative control (mock). Cells were stained for mTRAIL and acquired and analyzed by flow cytometry. Data are representative of four experiments
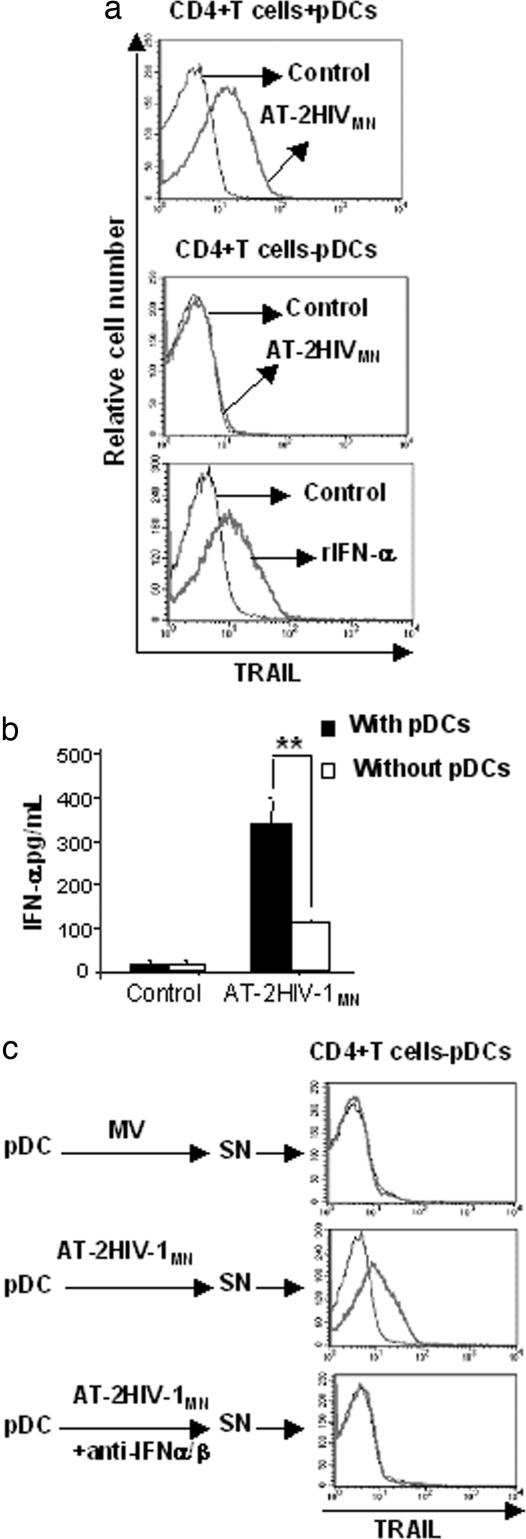
Role of pDCs in Type I IFN-Mediated TRAIL Expression by CD4+ T Cells. In Fig. 1, we showed that TRAIL expression by HIV-1-exposed CD4+ T cells depended on type I IFN. Because we used positive selection to isolate CD4+ cells, it is possible that some pDCs, which express CD4, were present in the cultures. Therefore, we studied isolated CD3+CD4+ T cells and pDCs to distinguish the contribution of these two CD4+ subpopulations to IFN production in response to HIV-1. We found that 0.5-1% of positively selected CD4+ cells in our isolated population were pDCs (CD123+ BDCA-2+) (data not shown). To test whether these pDCs were responsible for IFN-α production, we first depleted PBMC of pDCs and then performed CD4-positive selection. We reduced the percentage of pDCs in CD4+ cells to 0.03%. The two resulting populations (undepleted or depleted of pDCs) were cultured with AT-2 HIV-1MN for 24 h. CD4+ T cells expressed mTRAIL only when cultured with AT-2 HIV-1 in the presence of pDCs (Fig. 2a Top). The depletion of pDCs resulted in the loss of mTRAIL expression on HIV-1-exposed CD4+ T cells (Fig. 2a Middle). Addition of recombinant IFN-α to CD4+ T cells depleted of pDCs (Fig. 2a Bottom) restored mTRAIL expression. Culture supernatants of CD4+ cells undepleted or depleted of pDCs were tested for their IFN-α content. The supernatants of cultures that contained pDCs produced much more IFN-α than the pDC-depleted cultures (CD4+ T cells-pDCs) (Fig. 2b). It is possible that the IFN-α we detected in supernatants of cells depleted of pDCs were due to residual pDCs. To further demonstrate that IFN-α/β were produced by pDCs and induced mTRAIL expression by CD4+ T cells, we cultured isolated pDCs with AT-2 HIV-1MN or microvesicles (control) for 24 h. Supernatants from these cultures were added to CD4+ cells depleted of pDCs for an additional 24 h. Supernatants from pDCs cultured with AT-2 HIV-1MN induced mTRAIL expression by CD4+ T cells, in contrast to supernatants from pDCs cultured with microvesicles (Fig. 2c). Finally, to demonstrate that type I IFN was required for inducing mTRAIL expression by CD4+ T cells, we cultured isolated pDCs with AT-2 HIV-1MN and IFN-α/β blocking Abs. The supernatants of these cell cultures did not induce mTRAIL expression on CD4+ T cells. Thus, the induction of mTRAIL on CD4+ T cells requires IFN-α/β production by pDCs (Fig. 2c Bottom). Similar results were obtained by using infectious HIV-1MN, AT-2 HIV-1Ada, or infectious HIV-1Ada (data not shown).
Fig. 2.
Role of pDCs in mTRAIL expression and IFN-α production by CD4+ cells. (a) Isolated CD4+ cells were depleted of pDCs and cultured with HIV-1MN or rIFN-α. After 24 h, mTRAIL expression was detected on CD4+ T cell by flow cytometry. Undepleted CD4+ cells cultured with HIV-1MN were used as positive control for mTRAIL expression. (b) Isolated CD4+ cells depleted or not of pDCs were cultured for 24 h with AT-2 HIVMN and supernatants were tested for their IFN-α content. (c) Isolated pDCs were cultured with AT-2 HIV-1MN or AT-2 HIVAda in presence or absence of anti-IFN-α/β Abs for 24 h and culture supernatants (SN) were used to stimulate CD4+ cells depleted of pDCs. After 24 h mTRAIL expression on CD4+ T cells was determined by flow cytometry. Data in a and c are representative of four independent experiments. Data in b are mean values with SEs of four independent experiments for each condition tested. **, P < 0.01, determined by a two-tailed Student t test
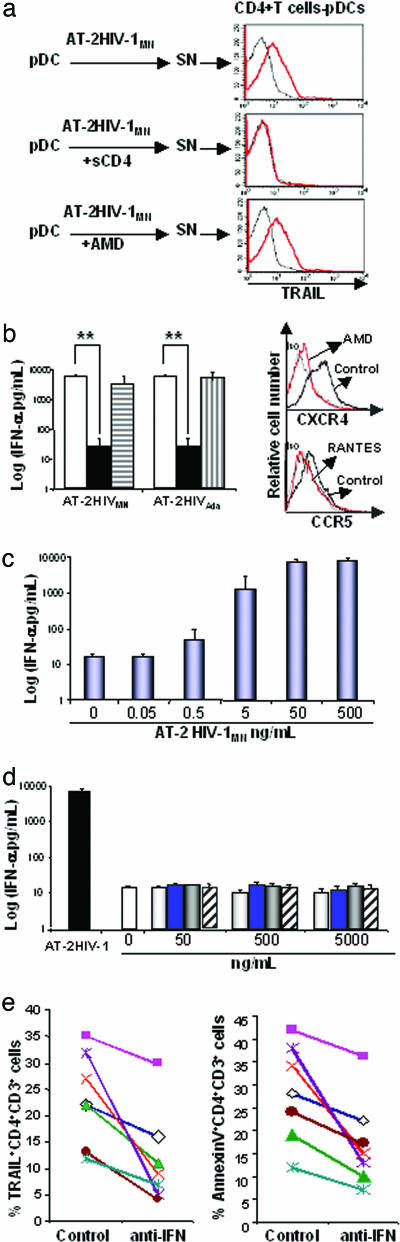
Role of Virus-Cell Interaction in TRAIL Expression by CD4+ T Cells and Type I IFN Production by pDCs. We tested whether TRAIL expression induced by AT-2 HIV-1 would be inhibited by: (i) sCD4, which blocks the binding of viral gp120 to cellular CD4; or (ii) AMD or RANTES, which blocks gp120 binding to the CXCR4 or CCR5 coreceptor, respectively. Supernatants from cultures of pDCs with AT-2 HIV-1MN in the presence of sCD4 did not induce mTRAIL on CD4+ T cells (Fig. 3a). Thus, the binding of gp120 to CD4 is an essential step in this mechanism. In contrast, supernatants from cultures of pDCs incubated with AT-2 HIV-1MN plus AMD still induced mTRAIL on CD4+ T cells. Furthermore, IFN-α production by pDCs exposed to AT-2 HIV-1 was inhibited by sCD4 but not by AMD or RANTES (Fig. 3b Left). These results are consistent with a triggering mechanism that depends on early CD4/gp120-binding events, but which does not require postbinding events such as coreceptor engagement. We verified that AMD and RANTES bound CXCR4 and CCR5, in our experiment, by staining pDCs with anti-CXCR4 or -CCR5 Abs. AMD and RANTES bound CXCR4 and CCR5, respectively, preventing the binding of coreceptor staining Abs (Fig. 3b Right). We titrated AT-2 HIV-1MN and showed that IFN-α production by pDCs was HIV-1 concentration-dependent (Fig. 3c). To further test whether binding of viral gp120 or gp160 to cellular CD4 on pDCs was sufficient to induce type I IFN production, we cultured pDCs with gp120MN, gp120SF (M tropic), and gp160MN. None of these preparations stimulated pDCs to produce type I IFN (Fig. 3d). Because Tat was shown to induce TRAIL production by monocytes (9), we tested the effect of Tat on pDCs. Tat did not induce IFN-α production by pDCs even at high concentration (5 μg/ml) (Fig. 3d).
Fig. 3.
Role of CD4-gp120 interaction in IFN-α/β production by pDCs. (a) Isolated pDCs were culture 24 h with AT-2 HIV-1MN in presence of sCD4 or AMD at 5 μg/ml. Culture supernatants (SN) were used to culture CD4+ cells depleted of pDCs for an additional 24 h. mTRAIL expression on CD4+ T cells was determined by flow cytometry. (b) (Left) IFN-α levels in SN of pDCs cultured with AT-2 HIV-1 and viral entry inhibitors were quantified by ELISA. Control (open bar); sCD4 (solid bar); AMD (horizontal lined bar); RANTES (vertical lined bar). (Right) pDCs cultured with or without AMD or RANTES and were stained with anti-CXCR4 or anti-CCR5 Abs and analyzed by flow cytometry. Isotype control (Iso) was used as negative control. (c) Titration of AT-2 HIV-1 for inducing INF-α production by isolated pDCs. (d) pDCs were culture for 24 h with gp120SF (open bar), gp120MN (blue bar), gp160MN (gray bar), or Tat (diagonal bar) at several concentrations (0-5,000 ng/ml). IFN-α levels were quantified by ELISA. (e) PBMC from 13 patients' blood were cultured for 2 d in medium supplemented with 50% autologous plasma in the absence or presence of anti-IFN-α/β Abs at 5 μg/ml. Cells were stained for CD3, CD4, TRAIL, and Annexin V and analyzed by flow cytometry. Data from seven samples are shown. Data in a are representative of four experiments for each condition tested. Data in b-d are mean values with SEs of six experiments for each condition tested. **, P < 0.01 determined by a two-tailed Student t test
Finally, we tested whether anti-type I IFN Abs inhibited TRAIL expression on CD4+ T cells from HIV-1-infected patients. PBMC from patients were cultured for 2 d in the presence or absence of anti-IFN-α/β Abs. Cells were then stained for CD4, CD3, and TRAIL and analyzed by flow cytometry. Anti-IFN-α/β Abs reduced TRAIL expression on CD4+CD3+ cells in 13/13 patients tested (10-1,000% reduction, P = 0.001) (Fig. 3e Left). Abs against IFN-α/β reduced apoptosis (Annexin V+ cells) of CD4+ T cells in 13/13 patients (10-250% reduction, P = 0.0008) (Fig. 3e Right). We show seven representative examples in Fig. 3e. Three of the patients illustrated were on highly active antiretroviral therapy (HAART), and four were not. We did not observe any effect of HAART on these parameters. Taken together, these results demonstrate that infectious or noninfectious HIV-1 induce IFN-α, leading to TRAIL expression on CD4+ T cells in vitro and in an ex vivo model.
Discussion
We show in this article that pDCs produce IFN-α after culture with infectious or inactivated HIV-1, and that IFN-α production depends on the interaction of viral gp120 with cellular CD4. The resulting IFN-α induces STAT1 and STAT2 activation in primary CD4+ T cells, leading to TRAIL expression. Taken together, these results suggest a potential role for pDCs in the immunopathogenesis of HIV-1 infection by inducing expression of the death molecule TRAIL.
A large proportion of plasma HIV-1 particles are noninfectious (19, 20). Therefore, our findings that noninfectious HIV-1 induce TRAIL expression on uninfected CD4+ T cells underscore the potential role of replication-deficient HIV-1 particles in contributing to HIV-1 immunopathogenesis. Here, we show that noninfectious particles are not inert and can induce type I IFN and TRAIL expression on CD4+ T cells. Furthermore, another report (18) indicated that noninfectious HIV-1 induced pDCs activation and maturation in vitro. Thus, we raise the possibility that CD4+ T cells and pDCs could be potentially activated in patients by HIV-1 particles without resulting in productive infection. Type I IFN signal transduction involves different molecules, including those of the STAT pathway. We demonstrate that after HIV-1 exposure, primary CD4+ T cells expressed phosphorylated STAT1 and overexpressed STAT2, resulting in mTRAIL expression. Abs against IFN-α/β greatly reduced mTRAIL and STAT2 expression. We showed that both STAT1 and STAT2 are required for TRAIL regulation, because the blockade of either inhibited TRAIL expression on CD4+ T cells. This result indicates that the STAT pathway is involved in TRAIL regulation in primary CD4+ T cells by HIV-1.
Type I IFNs produced by pDCs have been suggested to protect against AIDS progression (28, 29). However, as with many aspects of the immune response to HIV-1 infection, induction of type I IFN may be a double-edged sword. Our data suggest that pDCs may contribute to CD4+ T cell depletion by producing type I IFN after exposure to HIV-1, leading to TRAIL expression on uninfected CD4+ T cells. We also showed that infectious and noninfectious X4- and R5-tropic HIV-1 virions induce type I IFN production by pDCs, underscoring the potentially broad importance of these cells in HIV-1 immunopathogenesis. We found that HIV-1 induced TRAIL and IFN-α production by monocytes, which may contribute to the high levels of both cytokines in patients (10), despite the reported reduction of blood pDCs in progressive HIV-1 disease (17, 30). This reduction of circulating pDCs may be due to their redistribution to lymphoid tissue (31), which could result in IFN production, leading to TRAIL expression by CD4+ T cells and apoptosis of DR5+ CD4+ T cells (11). This hypothesis is consistent with our finding that HIV-1 induced TRAIL production in ex vivo tonsil cultures (10). It is possible that the concentration of HIV-1 used in our in vitro experiments is higher than would be detected in blood. However, it is likely that much pathogenesis occurs in the lymphoid tissue where pDCs and HIV-1 are more concentrated than in the circulation.
sCD4 blocked IFN-α production by pDCs in response to HIV-1 exposure. Interestingly, our finding that AMD did not significantly inhibit IFN-α production suggests that coreceptor binding is not required for HIV-1-induced IFN-α production by pDCs. Thus, only the earliest interaction between HIV-1 and CD4 on pDCs is essential for TRAIL production. Our results show that free gp120 or gp160 are not sufficient to induce type I IFN, raising the possibility that the conformation of gp120 on HIV-1 particles or its combination with other envelope molecules may be important (32). Previous studies suggest that native virion-associated gp120 may be more potent than soluble monomeric recombinant gp120 in inducing effects dependent on authentic interactions with cell surface receptors (33).
The role of type I IFN in HIV-1 disease is complex and controversial. IFN levels have been reported in blood during acute infection and correlates directly with HIV-1 viremia (34-36). Levels of IFN decrease coincident with the development of neutralizing Abs and reduction of viral load. Paradoxically, IFN-α reappears in blood during late-stage disease, as an indicator of poor clinical prognosis (34, 35). IFN-α was reported to inhibit HIV-1 replication in vitro (12) and was used for therapy in AIDS patients with Kaposi's sarcoma (37). However, type I IFN induced in lymph nodes of macaques infected with simian immunodeficiency virus (SIV) did not control viral replication (38). Our findings indicate that, in addition to beneficial antiviral activity, type I IFN may exert pathogenic effects by inducing TRAIL expression on CD4+ T cells, leading to their apoptosis. Our observation that IFN-α/β mediate HIV-1-induced mTRAIL expression on CD4+ T cells raises the possibility of considering inhibition of type I IFNs as a potential therapeutic approach. This strategy is consistent with reports that loss of uninfected T cells is induced by IFN-α and can be blocked by anti-IFN-α Abs (39). Finally, we showed that Abs against type I IFN inhibited apoptosis and TRAIL expression in CD4+ T cells from HIV-1-infected patients. This result strongly suggests that IFN-α/β contribute to CD4+ T cell depletion in vivo.
Sooty mangabey monkeys do not exhibit apoptosis, CD4+ T cell depletion, or an AIDS-like syndrome when infected by SIV (40). In marked contrast to rhesus macaques, which develop AIDS, pDCs from Sooty mangabey monkeys do not produce IFN-α when exposed to SIV. Nevertheless, the pDCs of Sooty mangabey monkeys produce IFN-α in response to other stimuli, suggesting that activation of an IFN-related inflammatory cascade in response to SIV may contribute to the overall immuopathogenesis of SIV infection and, by implication, HIV infection.∥ The above findings in this natural host species are consistent with our data and hypothesis that type I IFN contributes to HIV-1 immunopathogenesis by inducing TRAIL-mediated apoptosis of CD4+ T cells.
Acknowledgments
We thank J. D. Lifson (Science Applications International Corporation-National Cancer Institute, Frederick, MD) for generously providing the AT-2 HIV-1 particles and reviewing this manuscript; and the Fondation pour la Recherche Médicale for financial support. This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, and the National Institutes of Health Intramural AIDS Targeted Antiviral Program.
Author contributions: J.-P.H. and G.M.S. designed research; J.-P.H. and A.W.H. performed research; S.A.A., M.J.D., and M.D. contributed new reagents/analytical tools; J.-P.H., A.B., and M.D. analyzed data; and J.-P.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TRAIL, TNF-related apoptosis-inducing ligand; mTRAIL, membrane TRAIL; pDC, plasmacytoid dendritic cell; DR, death receptor; STAT, signal transducers and activators of transcription; AT-2, Aldrithiol-2; SIV, simian immunodeficiency virus; sCD4, soluble CD4-IgG; PBMC, peripheral blood mononuclear cells; rIFN, recombinant IFN; AMD, AMD-3100; RANTES, regulated activation, normal T expressed and secreted.
Footnotes
Feinberg, M. B., HIV Pathogenesis Keystone Symposia, April 9-15, 2005, Banff, Canada, abstr. no. 22.
References
- 1.Wiley, S. R., Schooley, K., Smolak, P. J., Din, W. S., Huang, C. P., Nicholl, J. K., Sutherland, G. R., Smith, T. D., Rauch, C., Smith, C. A., et al. (1995) Immunity 3, 673-682. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhary, P. M., Eby, M., Jasmin, A., Bookwalter, A., Murray, J. & Hood, L. (1997) Immunity 7, 821-830. [DOI] [PubMed] [Google Scholar]
- 3.Pan, G., O'Rourke, K., Chinnaiyan, A. M., Gentz, R., Ebner, R., Ni, J. & Dixit, V. M. (1997) Science 276, 111-113. [DOI] [PubMed] [Google Scholar]
- 4.Herbeuval, J. P., Lambert, C., Sabido, O., Cottier, M., Fournel, P., Dy, M. & Genin, C. (2003) J. Natl. Cancer Inst. 95, 611-621. [DOI] [PubMed] [Google Scholar]
- 5.Katsikis, P. D., Garcia-Ojeda, M. E., Torres-Roca, J. F., Tijoe, I. M., Smith, C. A. & Herzenberg, L. A. (1997) J. Exp. Med. 186, 1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lum, J. J., Pilon, A. A., Sanchez-Dardon, J., Phenix, B. N., Kim, J. E., Mihowich, J., Jamison, K., Hawley-Foss, N., Lynch, D. H. & Badley, A. D. (2001) J. Virol. 75, 11128-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeremias, I., Herr, I., Boehler, T. & Debatin, K. M. (1998) Eur. J. Immunol. 28, 143-152. [DOI] [PubMed] [Google Scholar]
- 8.Miura, Y., Misawa, N., Maeda, N., Inagaki, Y., Tanaka, Y., Ito, M., Kayagaki, N., Yamamoto, N., Yagita, H., Mizusawa, H., et al. (2001) J. Exp. Med. 193, 651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang, Y., Tikhonov, I., Ruckwardt, T. J., Djavani, M., Zapata, J. C., Pauza, C. D. & Salvato, M. S. (2003) J. Virol. 77, 6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbeuval, J. P., Boasso, A., Grivel, J. C., Hardy, A. W., Anderson, S. A., Dolan, M. J., Chougnet, C., Lifson, J. D. & Shearer, G. M. (2005) Blood 105, 2458-2464. [DOI] [PubMed] [Google Scholar]
- 11.Herbeuval, J. P., Grivel, J. C., Boasso, A., Hardy, A. W., Chougnet, C., Dolan, M. J., Yagita, H., Lifson, J. D. & Shearer, G. M. (2005) Blood, in press. [DOI] [PMC free article] [PubMed]
- 12.Yamamoto, J. K., Barre-Sinoussi, F., Bolton, V., Pedersen, N. C. & Gardner, M. B. (1986) J. Interferon Res. 6, 143-152. [DOI] [PubMed] [Google Scholar]
- 13.Siegal, F. P., Kadowaki, N., Shodell, M., Fitzgerald-Bocarsly, P. A., Shah, K., Ho, S., Antonenko, S. & Liu, Y. J. (1999) Science 284, 1835-1837. [DOI] [PubMed] [Google Scholar]
- 14.Galibert, L., Maliszewski, C. R. & Vandenabeele, S. (2001) Semin. Immunol. 13, 283-289. [DOI] [PubMed] [Google Scholar]
- 15.Colonna, M., Trinchieri, G. & Liu, Y. J. (2004) Nat. Immunol. 5, 1219-1226. [DOI] [PubMed] [Google Scholar]
- 16.Kadowaki, N., Antonenko, S., Lau, J. Y. & Liu, Y. J. (2000) J. Exp. Med. 192, 219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soumelis, V., Scott, I., Liu, Y. J. & Levy, J. (2002) Hum. Immunol. 63, 1206-1212. [DOI] [PubMed] [Google Scholar]
- 18.Fonteneau, J. F., Larsson, M., Beignon, A. S., McKenna, K., Dasilva, I., Amara, A., Liu, Y. J., Lifson, J. D., Littman, D. R. & Bhardwaj, N. (2004) J. Virol. 78, 5223-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrov, D. S., Willey, R. L., Sato, H., Chang, L. J., Blumenthal, R. & Martin, M. A. (1993) J. Virol. 67, 2182-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piatak, M., Jr., Saag, M. S., Yang, L. C., Clark, S. J., Kappes, J. C., Luk, K. C., Hahn, B. H., Shaw, G. M. & Lifson, J. D. (1993) Science 259, 1749-1754. [DOI] [PubMed] [Google Scholar]
- 21.Domanski, P. & Colamonici, O. R. (1996) Cytokine Growth Factor Rev. 7, 143-151. [DOI] [PubMed] [Google Scholar]
- 22.Yan, H., Krishnan, K., Greenlund, A. C., Gupta, S., Lim, J. T., Schreiber, R. D., Schindler, C. W. & Krolewski, J. J. (1996) EMBO J. 15, 1064-1074. [PMC free article] [PubMed] [Google Scholar]
- 23.Darnell, J. E., Jr., Kerr, I. M. & Stark, G. R. (1994) Science 264, 1415-1421. [DOI] [PubMed] [Google Scholar]
- 24.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651-662. [DOI] [PubMed] [Google Scholar]
- 25.Sato, K., Hida, S., Takayanagi, H., Yokochi, T., Kayagaki, N., Takeda, K., Yagita, H., Okumura, K., Tanaka, N., Taniguchi, T., et al. (2001) Eur. J. Immunol. 31, 3138-3146. [DOI] [PubMed] [Google Scholar]
- 26.Rossio, J. L., Esser, M. T., Suryanarayana, K., Schneider, D. K., Bess, J. W., Jr., Vasquez, G. M., Wiltrout, T. A., Chertova, E., Grimes, M. K., Sattentau, Q., et al. (1998) J. Virol. 72, 7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bess, J. W., Jr., Gorelick, R. J., Bosche, W. J., Henderson, L. E. & Arthur, L. O. (1997) Virology 230, 134-144. [DOI] [PubMed] [Google Scholar]
- 28.Siegal, F. P., Fitzgerald-Bocarsly, P., Holland, B. K. & Shodell, M. (2001) AIDS 15, 1603-1612. [DOI] [PubMed] [Google Scholar]
- 29.Soumelis, V., Scott, I., Gheyas, F., Bouhour, D., Cozon, G., Cotte, L., Huang, L., Levy, J. A. & Liu, Y. J. (2001) Blood 98, 906-912. [DOI] [PubMed] [Google Scholar]
- 30.Chehimi, J., Campbell, D. E., Azzoni, L., Bacheller, D., Papasavvas, E., Jerandi, G., Mounzer, K., Kostman, J., Trinchieri, G. & Montaner, L. J. (2002) J. Immunol. 168, 4796-4801. [DOI] [PubMed] [Google Scholar]
- 31.Servet, C., Zitvogel, L. & Hosmalin, A. (2002) Curr. Mol. Med. 2, 739-756. [DOI] [PubMed] [Google Scholar]
- 32.Graham, D. R., Chertova, E., Hilburn, J. M., Arthur, L. O. & Hildreth, J. E. (2003) J. Virol. 77, 8237-8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esser, M. T., Bess, J. W., Jr., Suryanarayana, K., Chertova, E., Marti, D., Carrington, M., Arthur, L. O. & Lifson, J. D. (2001) J. Virol. 75, 1152-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minagawa, T., Mizuno, K., Hirano, S., Asano, M., Numata, A., Kohanawa, M., Nakane, A., Hachimori, K., Tamagawa, S., Negishi, M., et al. (1989) Life Sci. 45, iii-vii. [DOI] [PubMed] [Google Scholar]
- 35.von Sydow, M., Sonnerborg, A., Gaines, H. & Strannegard, O. (1991) AIDS Res. Hum. Retroviruses 7, 375-380. [DOI] [PubMed] [Google Scholar]
- 36.Abb, J., Zachoval, R., Zachoval, V. & Deinhardt, F. (1987) Infection 15, 425-426. [DOI] [PubMed] [Google Scholar]
- 37.Frissen, P. H., de Wolf, F., Reiss, P., Bakker, P. J., Veenhof, C. H., Danner, S. A., Goudsmit, J. & Lange, J. M. (1997) J. Infect. Dis. 176, 811-814. [DOI] [PubMed] [Google Scholar]
- 38.Abel, K., Alegria-Hartman, M. J., Rothaeusler, K., Marthas, M. & Miller, C. J. (2002) J. Virol. 76, 8433-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zagury, D., Lachgar, A., Chams, V., Fall, L. S., Bernard, J., Zagury, J. F., Bizzini, B., Gringeri, A., Santagostino, E., Rappaport, J., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silvestri, G., Fedanov, A., Germon, S., Kozyr, N., Kaiser, W. J., Garber, D. A., McClure, H., Feinberg, M. B. & Staprans, S. I. (2005) J. Virol. 79, 4043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]