Abstract
Despite numerous recent advances in our understanding of the molecular mechanisms underlying receptor tyrosine kinase down-regulation and degradation in response to growth factor binding, relatively little is known about ligand-independent receptor tyrosine kinase degradation mechanisms. In a screen for proteins that might regulate the trafficking or localization of the ErbB3 receptor, we have identified a tripartite or RBCC (RING, B-box, coiled–coil) protein that interacts with the cytoplasmic tail of the receptor in an activation-independent manner. We have named this protein Nrdp1 for neuregulin receptor degradation protein-1. Northern blotting reveals ubiquitous distribution of Nrdp1 in human adult tissues, but message is particularly prominent in heart, brain, and skeletal muscle. Nrdp1 interacts specifically with the neuregulin receptors ErbB3 and ErbB4 and not with epidermal growth factor receptor or ErbB2. When coexpressed in COS7 cells, Nrdp1 mediates the redistribution of ErbB3 from the cell surface to intracellular compartments and induces the suppression of ErbB3 and ErbB4 receptor levels but not epidermal growth factor receptor or ErbB2 levels. A putative dominant-negative form of Nrdp1 potentiates neuregulin-stimulated Erk1/2 activity in transfected MCF7 breast tumor cells. Our observations suggest that Nrdp1 may act to regulate steady-state cell surface neuregulin receptor levels, thereby influencing the efficiency of neuregulin signaling.
The neuregulins comprise a subfamily of at least four epidermal growth factor (EGF)-like growth factors that influence a variety of cellular events, including proliferation, differentiation, migration, survival, and fate. The most thoroughly examined neuregulin, neuregulin-1 (NRG1; also called heregulin, neu differentiation factor, glial growth factor, or acetylcholine receptor-inducing activity), has been shown to play essential roles in the development of cardiac and neural tissues and also has been shown to play a role in the postsynaptic development of the neuromuscular junction (1). The neuregulins bind to ErbB3 or ErbB4, members of the EGF receptor family of receptor tyrosine kinases, and can activate the ErbB2 receptor through a heterodimerization mechanism (2–5).
Recent studies suggest that the fidelity of signaling through ErbB and other growth factor receptors is ensured by the maintenance of a relatively narrow range of receptors at the cell surface site of signaling (6). For example, in the nematode Caenerhabditis elegans mutations in the proteins lin-2 or lin-7 mislocalize the worm ErbB receptor let-23 and abrogate its function in vulval cell fate determination (7). Overexpression of the let-23 receptor can compensate for the mislocalization defect, suggesting that the accumulation of threshold levels of ErbB receptors at a specific cell surface location is necessary for biological activity. However, receptor overexpression also can lead to disease states such as the genesis or progression of tumors. Hence, mechanisms must exist for localizing and maintaining a precise concentration of growth factor receptors at the site of signal reception. Interestingly, it has been reported that ErbB2, ErbB3, and ErbB4 are somewhat unique among examined receptor tyrosine kinases in that they exhibit impaired ligand-induced internalization, down-regulation, and degradation (8, 9), underscoring the need for very tight regulation of cell surface receptor concentration before ligand stimulation.
Conceptually, steady-state levels of cell surface proteins are maintained as a result of a balance between synthesis, delivery, retention, and degradation. Although synthesis, delivery, and retention play crucial roles in establishing the pattern of cell surface protein expression, degradation is necessary to remove overproduced or mislocalized proteins and to maintain the dynamic equilibrium of expressed protein. Anchoring or trafficking proteins involved in these processes are predicted to interact with receptors in an activation-independent manner. In the present study we used the yeast two-hybrid system to identify proteins that are capable of constitutively associating with the intracellular domain of ErbB3. One candidate that emerged from this screen appears to be involved in the suppression of cell surface neuregulin receptor levels and could thereby contribute to the maintenance of the threshold levels necessary for proper signaling.
Materials and Methods
Yeast Two-Hybrid Screens, Cloning, Constructs, and Antibody Preparation.
The region of cDNA encoding the entire intracellular portion of bovine ErbB3 (residues W665 to I1335) was amplified and subcloned into the pAS1 bait plasmid. This construct was cotransformed with a plasmid library encoding human brain cDNAs fused to the activation domain of GAL4 into the Y190 lacZ/HIS3 yeast reporter strain. His+/β-gal+ clones were recovered and sequenced. Carboxyl-terminal deletions of the ErbB3 intracellular domain in the same plasmid were cotransformed into yeast with the pGAD plasmid containing no insert or the clone 32 insert (see below), and colonies were scored for β-galactosidase activity.
The ≈650-bp clone 32 from the two-hybrid screen was subcloned into the EcoRI site of pGEX to create a glutathione S-transferase (GST) fusion protein encompassing Nrdp1 residues I135 to I317. Northern blotting was carried out by using clone 32 to probe a filter of adult human tissues (CLONTECH). This clone also was used to screen a λgtll human fetal brain library to obtain ≈3.7-kb and ≈2.2-kb clones, which were partially sequenced. The cDNA sequence and ORF were found to be identical to the unpublished hypothetical protein SBBI03 (GenBank accession no. AF077599) and highly homologous to the mouse FLRF protein (GenBank accession no. AF305730). Four PCR primers were used to generate the full-length protein (M1 to I317), the amino terminal Zn2+-binding domains (M1 to D169), and the clone 32 region, each tagged with the FLAG epitope (DYKDDDDK) at the carboxyl terminus. These were subcloned into the NotI and XbaI sites of mammalian expression plasmid pcDNA3.1 (Invitrogen). A similar PCR approach was used to FLAG tag Nrdp1 at the amino terminus, and this construct was subcloned into baculovirus transfer vector pVL1392 for insect cell expression. cDNAs encoding human ErbB2 and ErbB4 were independently subcloned into expression vector pcDNA3.1, and the vector encoding bovine ErbB3 has been described (10).
A polyclonal rabbit antibody was raised to the bacterially expressed GST fusion protein of clone 32 described above by Research Genetics, Huntsville, AL. Anti-Nrdp1 antibodies were affinity-purified from serum obtained from week 10 bleeds by using the fusion protein immobilized on Affi-Gel 10 beads (Bio-Rad) according to procedures outlined previously (10).
Mammalian Cells, Transfections, and Protein Expression.
COS7 or MCF7 cells from the American Type Culture Collection were maintained in DMEM/10% FCS at 10% CO2. For transfection experiments cells were grown to 50% confluence in 6-well dishes and transfected with 2 μg total DNA and 10 μl Superfect (Qiagen, Chatsworth, CA) according to the protocol outlined by the manufacturer. For stable transfections, cells were split into 100-mm dishes and selected in 0.4 mg/ml G418 (GIBCO). In transient transfection experiments, pcDNA3.1 plasmid was used in mock transfections and cotransfections not requiring a second component. After transfection, cells were allowed to recover for 30 h, and then were treated without or with 2 μM MG132 (Novabiochem) for 18 h. Cells were lysed in an Nonidet P-40-containing lysis buffer (11) or RIPA (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS), or in 1× sample buffer.
For immunoprecipitation experiments, cleared lysates were precipitated with anti-EGF receptor (Ab1), anti-ErbB2 (Ab4), anti-ErbB3 (Ab6), or anti-ErbB4 (Ab1), all from NeoMarkers, Fremont, CA. Blotting antibodies used were: anti-EGF receptor (1005, Santa Cruz Biotechnology), anti-ErbB2 (Ab1, NeoMarkers), anti-ErbB3 (3184; 10), anti-ErbB4 (Ab2, NeoMarkers), and anti-FLAG antibody M2 (Sigma). Chemiluminescent signal from horseradish peroxidase-conjugated secondary antibodies was detected by using an Alpha Innotech (San Leandro, CA) 8000 imaging station with fluorchem software. For Erk stimulation experiments, transfected MCF7 cells were treated without or with various concentrations of NRG1β (12) 15 min before lysis with 1× sample buffer. Lysates were resolved by using 6–10% linear gradient gels and transferred to nitrocellulose. Blots were probed with anti-ErbB3 Ab6, anti-FLAG, anti-hemagglutinin (HA) tag (Roche Molecular Biochemicals), or anti-phospho-Erk1/2 (Cell Signaling Technology, Beverly, MA).
Insect Cell and GST Fusion Protein Pull-Down Experiments.
Baculovirus encoding Nrdp1 tagged at its amino terminus with FLAG epitope was created as described (13). Baculoviruses encoding human EGF receptor, human ErbB2, bovine ErbB3, or human ErbB4 have been described (12). For coimmunoprecipitation experiments Sf9 cells were infected with receptor viruses and coinfected with either wild-type or Nrdp1 baculovirus. Forty eight hours after infection cleared Nonidet P-40 lysates were immunoprecipitated with receptor antibodies and blotted with antibodies directed to receptors or FLAG epitope as described above.
For pull-down experiments receptors were expressed in Sf9 insect cells, cleared lysates were incubated for 1.5 h with 3 μg immobilized GST or GST-32, and beads were washed as described (12). MDA-MB-453 cells were treated for 5 min without or with 10 nM NRG1β, lysed as described (14), and cleared lysates were incubated with GST and GST-32 as above. Precipitated proteins were resolved by 8% SDS/PAGE, transferred to nitrocellulose, and visualized by blotting with antiphosphotyrosine (RC20, Transduction Laboratories, Lexington, KY) or antireceptor antibodies.
Immunofluorescence.
COS cells grown to 50% confluence on cover slips were transfected as described above and fixed in methanol after 48 h. Cover slips were blocked in 1% BSA, 0.2% Nonidet P-40, 5% goat serum, and 0.02% sodium azide, then incubated for 1 h in primary antibody followed by 1 h in secondary antibody. Primary antibodies used were 1/500 anti-FLAG M2, 1/500 anti-ErbB3 Ab6 (NeoMarkers), and 1 μg/ml affinity-purified anti-Nrdp1. The secondary antibodies used were 1/200 FITC-goat anti-mouse IgG and 1/200 Cy3-goat anti-rabbit IgG, both from Jackson Immunologicals, West Grove, PA. Cover slips were mounted onto glass slides by using Fluoromount-G (Southern Biotechnology Associates). Confocal microscopy at ×60 magnification was carried out by using a Bio-Rad MRC 600 confocal microscope with an argon laser. For each transfectant, 24 images were collected corresponding to focal planes encompassing the entire depth of the cell.
Results
Isolation and Characterization of Nrdp1.
The localization of ErbB receptors and the regulation of their levels play critical roles in the fidelity of growth factor signal transduction (6). Proteins involved in these processes are predicted to interact with receptors in an activation-independent manner. Because ErbB3 lacks intrinsic tyrosine kinase and autophosphorylation activities (13), proteins that interact with the intracellular portion of this receptor in a yeast two-hybrid screen bind in a phosphotyrosine- and activation-independent manner. Using the entire intracellular region of ErbB3 as bait, we identified a ≈650-bp cDNA fragment (called clone 32) in a human fetal brain library that encodes an interacting polypeptide. We further identified a 120-aa sequence in the ErbB3 tail region (T1159-E1279) necessary for clone 32 association with bovine ErbB3 (not shown).
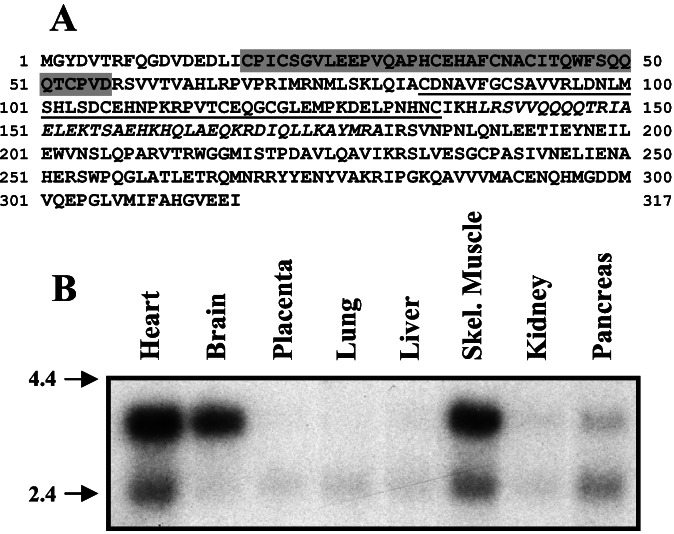
Sequencing of clone 32 revealed that it encodes the carboxyl-terminal 183 aa of human hypothetical protein SBBI03 (GenBank accession no. AF077599) and its mouse ortholog FLRF (GenBank accession no. AF305730), proteins of unknown biochemical or cellular function (15). The complete amino acid sequence of this protein (Fig. 1A) indicates that it is a member of the RBCC (RING, B-box, coiled–coil) subfamily of RING finger-containing proteins. The presence of a putative myristoylation site at its amino terminus implies a membrane function for Nrdp1. On the basis of its homology with RING finger proteins known to be involved in the degradation of cell surface proteins, and on our functional characterization below, we have named the protein Nrdp1 for neuregulin receptor degradation protein-1. Although the message appears to be ubiquitously expressed in adult human tissues, prominent Nrdp1 expression in heart, brain, and skeletal muscle (Fig. 1B) is consistent with the known roles for neuregulin signaling in the development and maintenance of these tissues.
Figure 1.
Molecular characterization of Nrdp1. (A) The amino acid sequence of the full-length RBCC protein Nrdp1 is depicted. The imperfect C3HC4 RING finger domain is shaded, the B-box region is underlined, and the coiled–coil region is italicized. (B) Northern analysis of Nrdp1 expression in adult human tissues.
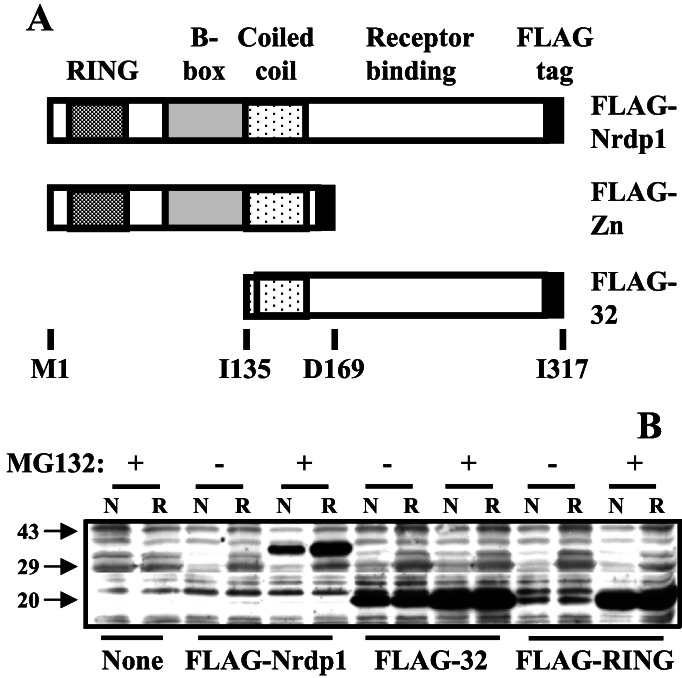
To begin to characterize the function of the Nrdp1 protein we expressed epitope (FLAG)-tagged forms of the full-length protein and its two halves FLAG-RING and FLAG-32 (see Fig. 2A) in a variety of mammalian cell lines. The FLAG tag was placed at the carboxyl terminus of each construct to avoid disrupting myristoylation of the amino terminus. No colonies were obtained after G418 selection when cells were stably transfected with either FLAG-Nrdp1 or FLAG-32. Therefore, we used transient transfection approaches for protein expression to examine the properties of the Nrdp1 protein. No deleterious effects were observed in any transiently transfected cells after 48 h of expression.
Figure 2.
Biochemical properties of Nrdp1 and its domains. (A) Illustration of FLAG-tagged constructs used in these studies. (B) Stability and detergent extractability of Nrdp1. MCF7 cells were transfected with the FLAG-tagged constructs illustrated in A, and cells were treated without or with MG132 for 18 h as indicated. Cells were lysed in RIPA (R, 0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) or an Nonidet P-40-containing buffer (N). Proteins in lysates were immunoblotted with anti-FLAG epitope.
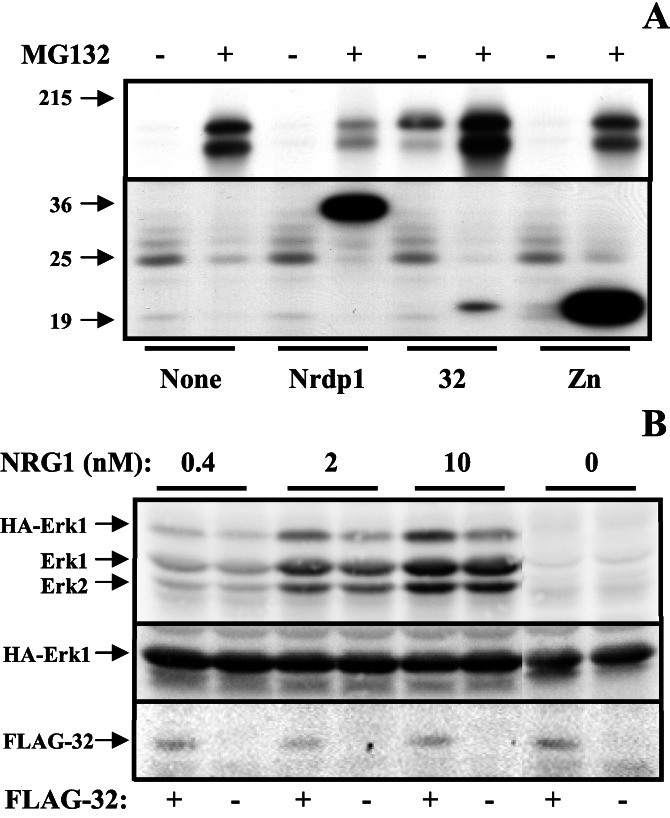
In the experiment shown in Fig. 2B we examined the expression and solubility of FLAG-Nrdp1 forms in transiently transfected MCF7 human mammary tumor cells. We observed that detection of FLAG-Nrdp1 by immunoblotting markedly depended on the presence of the proteasome inhibitor MG132. Other proteasome inhibitors, but not lysosome or calpain inhibitors, also stabilized FLAG-Nrdp1 (unpublished observations). The N-terminal region (called FLAG-Zn), including the RING finger, B-box, and coiled–coil domains, was also significantly stabilized by MG132. In MCF7 cells, the carboxyl-terminal portion (FLAG-32) was constitutively expressed and only modestly stabilized by the drug. Similar results were obtained when numerous other mammalian cell lines were transfected with Nrdp1 constructs (unpublished observations). These observations indicate that Nrdp1 is an intrinsically unstable protein in mammalian cells and suggest that its instability is mediated by the amino terminal region of the protein. The full-length protein, but neither of the two halves expressed at similar levels, was more easily extracted from cells under partially denaturing conditions than in an Nonidet P-40 lysis buffer, suggesting that some of the intact Nrdp1 protein may be associated with insoluble cellular components.
Specificity of Nrdp1 Interaction with ErbB Receptors.
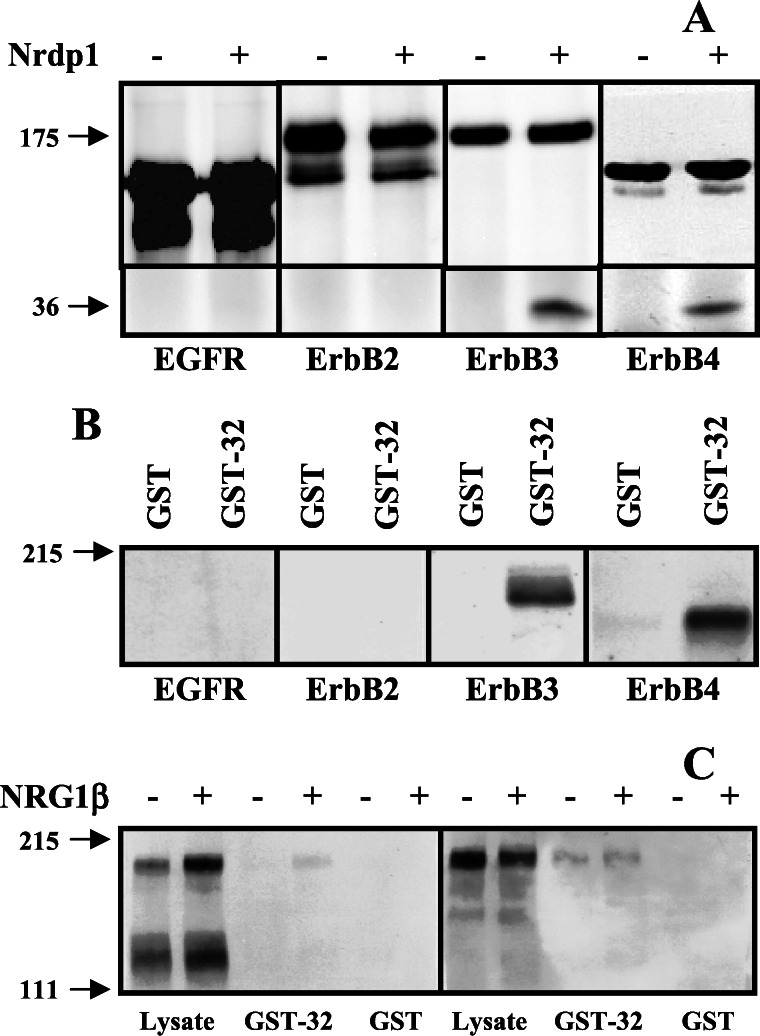
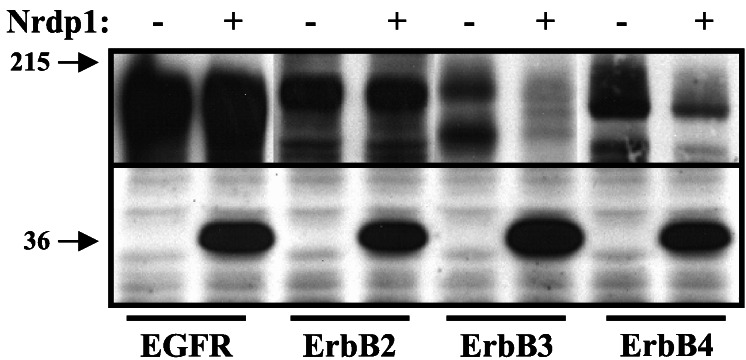
To determine whether Nrdp1 interacts specifically with ErbB3 or might also interact with other members of the ErbB family, we took advantage of the stability of the Nrdp1 protein in Sf9 insect cells. We have observed that in contrast with numerous mammalian cell lines, Nrdp1 protein is stably expressed (unpublished observations) and does not suppress ErbB3 levels in these cells (see below). In the experiment shown in Fig. 3A, we independently expressed each of the known mammalian ErbB receptors without or with FLAG-Nrdp1 in Sf9 cells, immunoprecipitated receptors, and blotted precipitates with antibodies to receptors and anti-FLAG epitope. We observed that Nrdp1 could be coimmunoprecipitated with the neuregulin receptors ErbB3 and ErbB4, but could not be coprecipitated with either ErbB2 or EGF receptor.
Figure 3.
Association of Nrdp1 with neuregulin receptors. (A) FLAG-Nrdp1 selectively interacts with ErbB3 and ErbB4 in insect cells. Sf9 insect cells were infected with baculovirus encoding each of the ErbB receptors or coinfected with receptor and FLAG-Nrdp1 viruses, as indicated. Cleared lysates were immunoprecipitated with antireceptor antibodies and blotted with antibodies to receptors (Upper) or FLAG-epitope (Lower). (B) Association of GST-32 with neuregulin receptors. Cleared lysates from Sf9 cells infected with the indicated receptor viruses were incubated with immobilized GST or GST-32 and bound proteins were detected by immunoblotting with antireceptor antibodies. (C) Nrdp1 binds ErbB3 in an activation-independent manner. MDA-MB-453 cells were treated without or with NRG1β, and lysates were incubated with immobilized GST or GST-32. Bound proteins were immunoblotted with antiphosphotyrosine (Left) or anti-ErbB3 (Right).
To confirm the specificity of the interaction we used a GST fusion of clone 32 in receptor pull-down experiments. A GST fusion of clone 32 was used instead of full-length Nrdp1 because we observed that GST-Nrdp1 is insoluble in bacteria. In the experiment illustrated in Fig. 3B, we expressed each of the ErbB family members in Sf9 cells (12), incubated cleared lysates with immobilized GST or GST-32, and analyzed associated receptors by blotting precipitates with receptor-specific antibodies. We again observed that clone 32 is capable of physically interacting with ErbB3 and ErbB4, but not EGF receptor or ErbB2. Taken together these results indicate that Nrdp1 interacts specifically with the neuregulin-binding ErbB3 and ErbB4 receptors relative to the other family members and suggest that the GST-32 construct accurately recapitulates Nrdp1 binding to receptors.
To assess whether the interaction of Nrdp1 with receptors depends on receptor activation state, we examined the association of ErbB3 with GST-32 before and after NRG1β stimulation of MDA-MB-453 human breast cancer cells. Stimulation of these cells with NRG1β results in the rapid tyrosine phosphorylation of ErbB2 and ErbB3 as revealed by blotting with antiphosphotyrosine antibodies (11), and a tyrosine-phosphorylated band may be pulled down from lysates of NRG1β-treated MDA-MB-453 cells with GST-32 (Fig. 3C). Reprobing with anti-ErbB3 revealed that similar amounts of this receptor associated with GST-32 whether or not cells were stimulated with NRG1β. These results are consistent with the original yeast two-hybrid screen where the clone 32-encoded polypeptide recognized ErbB3 in its inactive state. Similar results were obtained with ErbB4 expressed in Sf9 cells, where binding of GST-32 to this receptor was independent of NRG1β stimulation (not shown). These results indicate that Nrdp1 is capable of interacting with neuregulin receptors independent of receptor activation state, suggesting that it could play a role in constitutive receptor activities such as routing or localization. Attempts to coimmunoprecipitate Nrdp1 and neuregulin receptors from coexpressing mammalian cells were unsuccessful because Nrdp1 expression very potently stimulates the loss of neuregulin receptors from cells (see below).
Colocalization of ErbB3 and Nrdp1.
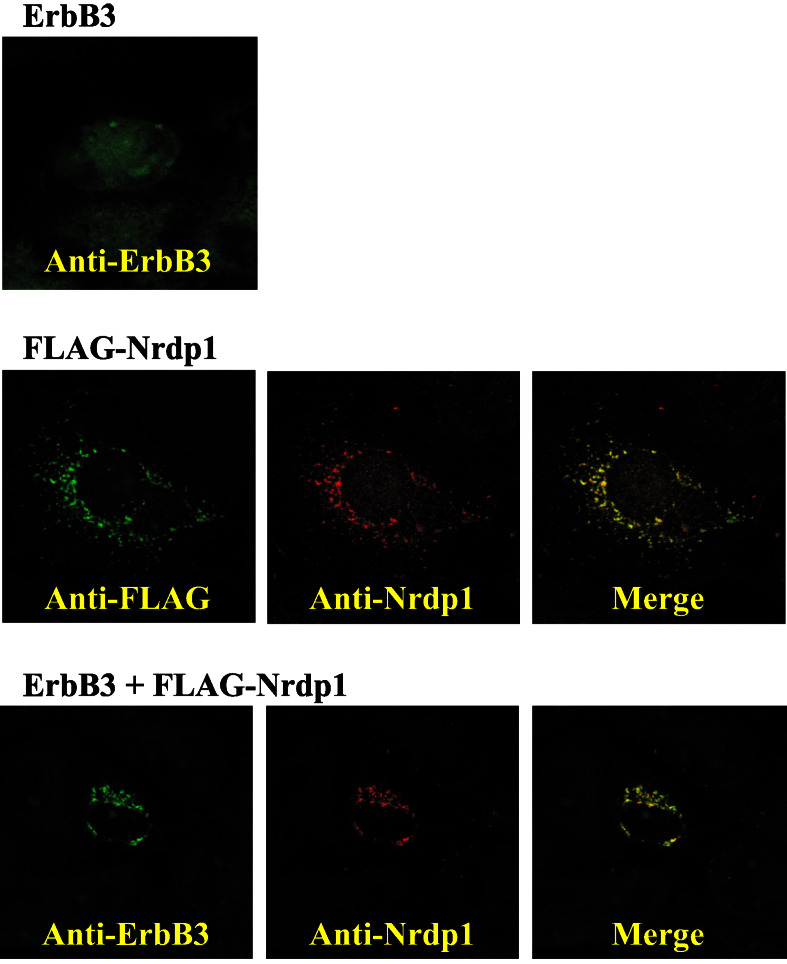
Despite their inability to be coimmunoprecipitated, confocal immunofluorescence microscopy revealed that ErbB3 and Nrdp1 are very extensively colocalized in transfected COS7 cells and that Nrdp1 changes the cellular location of ErbB3. In the experiments depicted in Fig. 4, COS7 cells were transfected with ErbB3 alone, FLAG-Nrdp1 alone, or the two proteins together, and then were treated with 2 μM MG132 to stabilize protein expression to facilitate visualization. The localization of ErbB3 and Nrdp1 was then examined by using indirect immunofluorescence microscopy with anti-ErbB3, anti-Nrdp1, and anti-FLAG antibodies.
Figure 4.
Colocalization of Nrdp1 and ErbB3. COS7 cells were transfected with ErbB3 alone (Top), FLAG-Nrdp1 alone (Middle), or both (Bottom). Cells were stained with the indicated antibodies, and ErbB3 and Nrdp1 localizations were visualized by confocal microscopy. The focal plane in Top corresponds to the top of the cell, while the focal planes in Middle and Bottom correspond to the middle of the cell.
Fig. 4 Top shows a focal plane corresponding to the surface of cells expressing ErbB3 alone. ErbB3 in these cells exhibited a diffuse, predominantly cell surface distribution, and no significant immunoreactivity was observed in focal planes corresponding to internal structures (not shown). In Fig. 4 Middle, cells transfected with FLAG-Nrdp1 alone were costained with mouse anti-FLAG (green) and rabbit anti-Nrdp1 (red). Focal planes corresponding to the cell interior revealed a largely punctate perinuclear distribution of FLAG-Nrdp1 when visualized with either antibody. Relatively little Nrdp1 was found at the cell surface in these cells (not shown). An untagged version of Nrdp1 exhibited an identical localization pattern in transfected COS7 cells (not shown). These observations suggest that Nrdp1 accumulates in intracellular organelles in MG132-treated cells, and the merged images indicate that the anti-Nrdp1 antibody accurately reflects the distribution of the expressed protein. Fig. 4 Bottom shows the distribution of ErbB3 (green) and FLAG-Nrdp1 (red) in cotransfected cells. Cotransfectants exhibited the same perinuclear staining of both proteins as observed with FLAG-Nrdp1 alone. Identical results were obtained when the experiment was performed in the absence of MG132, and experiments using transfected ErbB2 revealed that Nrdp1 expression had no effect on its localization (not shown). These observations indicate that Nrdp1 expression specifically induces a redistribution of ErbB3 from the cell surface to intracellular Nrdp1-containing compartments.
Nrdp1 Suppresses Cellular Neuregulin Receptor Levels.
In the immunofluorescence experiments we noted that the number of ErbB3-expressing cells was reduced by 75–90% when ErbB3 was coexpressed with FLAG-Nrdp1 relative to when expressed on its own (not shown). Moreover, it has been demonstrated that RING finger domain-containing proteins serve as E3 ubiquitin ligases that mediate the conjugation of ubiquitin to specific cellular targets (16–20). Two RING finger proteins, c-cbl and Siah, are known to influence cellular levels of plasma membrane proteins by marking them for degradation through ubiquitination. Hence, one possible functional consequence of the interaction between Nrdp1 and neuregulin-binding ErbB receptors could be the suppression of receptor levels.
To determine whether Nrdp1 might influence cellular neuregulin receptor levels, we used the COS7 cell system to transiently coexpress FLAG-Nrdp1 and ErbB receptors. COS7 cells express abundant EGF receptor, but little or no ErbB2, ErbB3, or ErbB4. In the experiment depicted in Fig. 5, we transiently transfected COS7 cells with each of these receptors or cotransfected cells with each of the receptors together with FLAG-Nrdp1. The experiment was carried out in the presence of 2 μM MG132, which stabilizes receptor expression in these cells (see below). Lysates of each transfection were blotted with antireceptor and anti-FLAG antibodies. We observed that FLAG-Nrdp1 expression had no discernable impact on the steady-state levels of endogenous EGF receptor, as expected, or on transfected ErbB2, but markedly reduced levels of transfected ErbB3 and ErbB4. This result was not caused by an effect of Nrdp1 on receptor transcription because ErbB2 was expressed by using the same expression plasmid as ErbB4. Moreover, Nrdp1 had no effect on transfected EGF receptor in MCF7 cells (not shown), a cell line that contains no endogenous EGF receptor. These results are consistent with the specific association of Nrdp1 and clone 32 with ErbB3 and ErbB4 and suggest that the functional outcome of Nrdp1/receptor interaction could be to suppress levels of neuregulin-binding receptors in cells.
Figure 5.
Suppression of steady-state neuregulin receptor levels by Nrdp1. COS7 cells were transfected with cDNAs encoding FLAG-Nrdp1, ErbB2, ErbB3, or ErbB4, as indicated. Lysates from MG132-treated cells were blotted with antibodies to endogenous EGF receptor or expressed ErbB receptors (Upper) and with anti-FLAG epitope (Lower). Ponceau S staining of filters revealed similar levels of total lysate proteins in each lane.
Potentiation of Neuregulin Signaling by a Putative Dominant-Negative Nrdp1.
We observed that the presence of very modest levels (2 μM) of MG132 dramatically stabilized ErbB3 in transiently transfected COS7 cells (Fig. 6A), pointing to the existence of a proteasome-dependent process that regulates receptor steady-state levels. The MG132-mediated stabilization of ErbB3 could be overcome by the expression of FLAG-Nrdp1, suggesting that this protein can complement the existing system for ErbB3 removal. Identical results were obtained by using transfected MCF7 human mammary tumor cells and 293 human embryonic kidney cells (unpublished observations). Interestingly, FLAG-32 stabilized the presence of ErbB3 both in the absence and presence of MG132, whereas FLAG-Zn had no effect on receptor steady-state levels. These observations indicate that the polypeptide encoded by clone 32 interferes with the mechanism of ErbB3 removal, perhaps by acting as a dominant-negative inhibitor of the process.
Figure 6.
Potentiation of ErbB3-mediated NRG1 signaling by dominant-negative Nrdp1. (A) Effect of Nrdp1 domains on ErbB3 expression. COS7 cells were transfected with the indicated FLAG-tagged constructs and treated without and with MG132. Lysates were blotted with antibodies to ErbB3 (Upper) and FLAG epitope (Lower). (B) MCF7 cells were transfected with HA-Erk1, and the indicated samples were cotransfected with FLAG-32. Cells were then treated with increasing levels of NRG1β. Lysates were blotted with antiphospho-Erk (Top), anti-HA epitope (Middle), or anti-FLAG epitope (Lower).
The stabilizing effect of clone 32 on ErbB3 levels permitted us to examine the effect of Nrdp1 on neuregulin signaling in a transient transfection assay in MCF7 cells. These cells express modest levels of ErbB2 and ErbB3 and respond in an all-or-nothing manner to treatment with NRG1β. In the experiment depicted in Fig. 6B, cells were transfected with epitope (HA)-tagged Erk1, a downstream effector of neuregulin stimulation in receptor-expressing cells, and cotransfected with either plasmid alone or FLAG-32. Cells were then treated with increasing amounts of NRG1β, and lysates were blotted with antiphosphoErk1/2 to assess the impact of clone 32 expression on Erk activity. Fig. 6B Top shows that NRG1β potently stimulated the phosphorylation of both endogenous Erk1/2 and HA-tagged Erk1. The presence of 32 only very modestly enhanced the response of endogenous Erk1/2 to growth factor, but significantly enhanced (2- to 2.5-fold after quantification) the response of HA-Erk1 to NRG1β. Hence, as expected because of the low transfection efficiency (10–20%) transiently transfected 32 had a marginal effect on the total cellular Erk1/2 population and preferentially acted on the transfected population. Blotting with anti-HA antibodies showed similar levels of the transfected protein in lysates, and blotting with anti-FLAG showed FLAG-32 expression only in transfected cells. These results indicate that the polypeptide encoded by clone 32 potentiates NRG1β signaling through receptors.
Discussion
The maintenance of a narrow range of cell surface growth factor receptor tyrosine kinases at specific sites of signal reception ensures sufficient signaling for a proper cellular response, while preventing oversignaling that could lead to disease (6). In this study we identify and characterize a protein that may be involved in this process. We observed that Nrdp1 expression elicits the redistribution of neuregulin receptors from the cell surface to punctate perinuclear intracellular compartments and induces a loss of receptor expression in cells. These processes are independent of growth factor binding and receptor activation. Moreover, a putative dominant-negative form enhances receptor expression in cells and potentiates NRG1-stimulated Erk1 activity, strongly suggesting that Nrdp1 functions to regulate steady-state cell surface receptor levels and neuregulin signaling efficiency.
Nrdp1 is a member of a subfamily of RING finger domain-containing proteins called the tripartite motif family (21) or RBCC for RING, B-box, coiled–coil. This subfamily consists of more than 24 proteins thought to be involved in a variety of developmental and cellular processes, and mutation or rearrangement of some RBCC genes is associated with human disease (22). RING fingers are zinc-binding domains thought to mediate a variety of protein–protein interactions and are found in a subclass of E3 ubiquitin ligases (20). B-boxes, also called TRAF-type zinc fingers, are zinc-binding domains of unknown function. Coiled–coil regions also mediate protein–protein interactions and often mediate the homotypic dimerization of a protein. Taken together with its structure, our results are most consistent with a model where Nrdp1 mediates the ubiquitination and trafficking of neuregulin receptors to intracellular degradative compartments.
It is tempting to draw parallels between the activity of Nrdp1 toward ErbB3 and ErbB4 and the activity of c-cbl toward EGF receptor. Upon ligand stimulation, EGF receptor is internalized through clathrin-coated pits and is delivered to intracellular degradative compartments. It has been proposed that EGF stimulation of the EGF receptor results in the autophosphorylation of a specific tyrosine residue, which mediates the recruitment of c-cbl to the receptor through its tyrosine kinase binding domain. c-cbl then mediates the ubiquitination of the receptor, which in turn routes the receptor to lysosomes for degradation (19). In contrast, Nrdp1 appears to act on receptors independent of their activation state. In this regard, it is interesting that the neuregulin target receptors ErbB2, ErbB3, and ErbB4 exhibit impaired ligand-induced internalization, down-regulation, and degradation (8, 9). In the absence of down-regulation, mechanisms that regulate receptor levels could play a major role in defining receptor signaling activity.
Our observations more closely parallel the impact of the RING finger protein Siah on cell surface levels of the membrane protein DCC (deleted in colorectal cancer), a transmembrane receptor for the axon guidance and neuronal migration factor netrin-1. Siah is localized to cytoplasmic particles and mediates the ubiquitination and degradation of DCC in a ligand-independent manner (23). Similar to Nrdp1, Siah is also intrinsically unstable and a putative dominant-negative form stabilizes DCC (24). Hence, a family of functionally related proteins could mediate the cell surface expression of a variety of proteins.
Future studies should be aimed at more precisely defining the mechanism of receptor degradation, including the roles of receptor and Nrdp1 ubiquitination. It will also be of interest to determine whether Nrdp1 might influence the levels of other proteins, particularly in cardiac, neural, and skeletal muscle tissues. Finally, tissue-specific knockout studies should uncover the physiological role of Nrdp1 in regulating the development of neuregulin-dependent tissues.
Acknowledgments
We gratefully acknowledge Frank Ventimiglia for invaluable assistance in confocal microscopy. This work was supported by National Institutes of Health Grant CA71702 (to K.L.C.). C.S. was supported in part by a fellowship from the Massachusetts Department of Public Health Breast Cancer Research Program and a grant from the California Breast Cancer Research Program.
Abbreviations
- Nrdp1
neuregulin receptor degradation protein-1
- EGF
epidermal growth factor
- NRG1
neuregulin-1
- HA
hemagglutinin
- GST
glutathione S-transferase
References
- 1.Burden S, Yarden Y. Neuron. 1997;18:847–855. doi: 10.1016/s0896-6273(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 2.Carraway K L, III, Cantley L C. Cell. 1994;78:5–8. doi: 10.1016/0092-8674(94)90564-9. [DOI] [PubMed] [Google Scholar]
- 3.Alroy I, Yarden Y. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 4.Riese D J, II, Stern D F. BioEssays. 1998;20:41–48. doi: 10.1002/(SICI)1521-1878(199801)20:1<41::AID-BIES7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.Olayioye M A, Neve R M, Lane H A, Hynes N E. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carraway K L, III, Sweeney C. Curr Opin Cell Biol. 2001;13:125–130. doi: 10.1016/s0955-0674(00)00188-5. [DOI] [PubMed] [Google Scholar]
- 7.Simske J S, Kaech S M, Harp S A, Kim S K. Cell. 1996;85:195–204. doi: 10.1016/s0092-8674(00)81096-x. [DOI] [PubMed] [Google Scholar]
- 8.Baulida J, Kraus M H, Alimandi M, Di Fiore P P, Carpenter G. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 9.Baulida J, Carpenter G. Exp Cell Res. 1997;232:167–172. doi: 10.1006/excr.1997.3515. [DOI] [PubMed] [Google Scholar]
- 10.Carraway K L, III, Soltoff S P, Diamonti A J, Cantley L C. J Biol Chem. 1995;270:7111–7116. doi: 10.1074/jbc.270.13.7111. [DOI] [PubMed] [Google Scholar]
- 11.Crovello C S, Lai C, Cantley L C, Carraway K L., III J Biol Chem. 1998;273:26954–26961. doi: 10.1074/jbc.273.41.26954. [DOI] [PubMed] [Google Scholar]
- 12.Carraway K L, III, Weber J L, Unger M J, Ledesma J, Yu N, Gassmann M, Lai C. Nature (London) 1997;387:512–516. doi: 10.1038/387512a0. [DOI] [PubMed] [Google Scholar]
- 13.Guy P M, Platko J V, Cantley L C, Cerione R A, Carraway K L., III Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweeney C, Lai C, Riese D J, 2nd, Diamonti A J, Cantley L C, Carraway K L., III J Biol Chem. 2000;275:19803–19807. doi: 10.1074/jbc.C901015199. [DOI] [PubMed] [Google Scholar]
- 15.Abdullah J M, Li X, Nachtman R G, Jurecic R. Blood Cells Mol Dis. 2001;27:320–333. doi: 10.1006/bcmd.2001.0390. [DOI] [PubMed] [Google Scholar]
- 16.Joazeiro C A, Weissman A M. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 17.Jackson P K, Eldridge A G, Freed E, Furstenthal L, Hsu J Y, Kaiser B K, Reimann J D. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 18.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 19.Levkowitz G, Waterman H, Ettenberg S A, Katz M, Tsygankov A Y, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 20.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borden K L. Biochem Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 22.Torok M, Etkin L D. Differentiation. 2001;67:63–71. doi: 10.1046/j.1432-0436.2001.067003063.x. [DOI] [PubMed] [Google Scholar]
- 23.Hu G, Zhang S, Vidal M, Baer J L, Xu T, Fearon E R. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu G, Fearon E R. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]