Abstract
Receptors for bone morphogenetic proteins (BMPs), members of the transforming growth factor-β (TGFβ) superfamily, are persistently expressed during cardiac development, yet mice lacking type II or type IA BMP receptors die at gastrulation and cannot be used to assess potential later roles in creation of the heart. Here, we used a Cre/lox system for cardiac myocyte-specific deletion of the type IA BMP receptor, ALK3. ALK3 was specifically required at mid-gestation for normal development of the trabeculae, compact myocardium, interventricular septum, and endocardial cushion. Cardiac muscle lacking ALK3 was specifically deficient in expressing TGFβ2, an established paracrine mediator of cushion morphogenesis. Hence, ALK3 is essential, beyond just the egg cylinder stage, for myocyte-dependent functions and signals in cardiac organogenesis.
Bone morphogenetic proteins (BMPs), named for their first-discovered role in bone differentiation, mediate a diverse spectrum of developmental events, such as dorsal–ventral patterning, mesoderm specification, and the spacing of embryo implantation (1, 2). BMPs are postulated to mediate cardiac development in mammals, by extrapolation from the role of decapentaplegic, a related factor in Drosophila (3), their function in cardiac looping in fish (4) or frogs (5), and, especially, studies of cardiac myogenesis in avians (6, 7). Avian explant studies also implicate BMPs as an essential paracrine signal from myocardium adjacent to the endocardial cushion during the epithelial-mesenchymal transformation and later events that ultimately give rise to the atrio-ventricular (AV) valves (8). This process is known to involve TGFβ2 and TGFβ3 as well (9–12), but the molecular connection between BMPs and TGFβ is uncertain.
Despite inferences from the distribution of BMPs during heart formation (13), and BMP effects on cardiac differentiation in teratocarcinoma cells (14), evidence analogous to that for other species is lacking in mammals themselves. One barrier to using conventional germline deletions to resolve these issues in mice is the greater potential for redundancy among BMP family members than in flies. In the mid-gestation heart, BMP2 and -4 are found, respectively, in myocardial layers of the AV canal and in the AV cushion itself, BMP5, -6, and -7 are initially homogeneous throughout the myocardium, and BMP10, which is heart-specific, is found exclusively in trabeculae (14, 15). This diversity, with overlapping and nonoverlapping programs of expression, suggests the utility of addressing the question via BMP receptors, instead, which are smaller in number.
BMPs bind two serine/threonine kinase receptors, type II (BMPRII) and type I (ALK3/BMPR-IA and ALK6/BMPR-IB), which form a heteromeric signaling complex acting in series, as for other TGFβ family receptors (16). In the presence of ligand, the type II receptor phosphorylates the type I receptors, which activate signaling by intracellular effectors including Smad transcription factors (16). ALK3 is ubiquitous throughout development, whereas ALK6 is absent from the heart at mid-gestation (17). The developing heart also expresses ALK2/ActRIA (5, 18), which can function as a type I BMP receptor with preference for BMP6 and -7 (19). ALK3, ALK2, and BMPR-II are each essential for gastrulation and mesoderm formation (18, 20, 21); mice lacking just BMP4 also fail to progress, typically, beyond the egg cylinder stage (22). In each instance, lethality before mesoderm is formed precludes detecting a role in cardiac development per se. This gap in predicting gene function is highlighted by the existence of human mutations in receptors for BMPs and TGFβ as a cause of clinical cardiovascular disorders including pulmonary hypertension and hereditary hemorrhagic telangiectasia (23, 24).
Even where survival in mice to later stages occurs, extrinsic defects might confound the interpretation of cardiac phenotypes, such as abnormalities of the amnion/chorion, observed with deletion of BMP2 (25). Analogously, deletion of BMP5 plus BMP7 results in death between E9.5 and E10.5, ascribed to cardiac defects (whereas their deletion singly has no apparent effect on the heart), but severe malformations also occurred in neural crest, branchial arches, and, notably, the allantois (26). Hence, the defects or delays in heart morphogenesis might plausibly result, at least in part, from abnormal development external to the heart, including the developing placenta.
Conditional deletion of ALK3, by targeted expression of Cre recombinase (27), provides a means to circumvent early lethality, confounding abnormalities, and, potentially, the greater redundancy among BMP family members than among their receptors. Mechanistically, deleting ALK3—unlike secreted proteins—also would establish unambiguously which cell type in the heart is the direct target for all resulting abnormalities. By conditional deletion of ALK3, we demonstrate an obligatory myocyte-dependent role for ALK3 in AV cushion and septal morphogenesis, functions that cannot be deduced or extrapolated from the conventional null phenotype (which is lethal before cardiac fate determination). Our study provides conclusive evidence that even single mutations in the BMP/ALK3 pathway can disrupt cardiac morphogenesis in mammals, proving that other BMP receptors do not substitute for ALK3 here, and potentially implicates ALK3 signaling in endocardial cushion defects or other congenital heart disorders.
Materials and Methods
Generation of the Conditional ALK3 Allele.
A neomycin cassette driven by the PGK promoter and flanked by two loxP sites was placed in an EcoRI site in intron 2 of the 129/SvEv ALK3 gene (20) by homologous recombination. The third loxP sequence was placed in one of the two HindIII sites in intron 1, introducing a new NheI site for later diagnosis. The left and right homologous arms were 5.4 and 5.5 kb, respectively. For negative selection, an MC1 herpes simplex virus thymidine kinase cassette was placed at the 5′ end. The vector was linearized for stable transfection by using XhoI and introduced into AB-1 ES cells by electroporation. Correctly targeted clones were injected into blastocysts for chimera production. Heterozygous mice were mated with CMV-Cre transgenic mice (28) for site-specific recombination between the loxP motifs. Progeny that executed recombination between the second and third loxP motifs, resulting in an expressed but loxP-tagged allele, were identified by Southern blot (ALK3F/+). The mating strategy is detailed in Fig. 1.
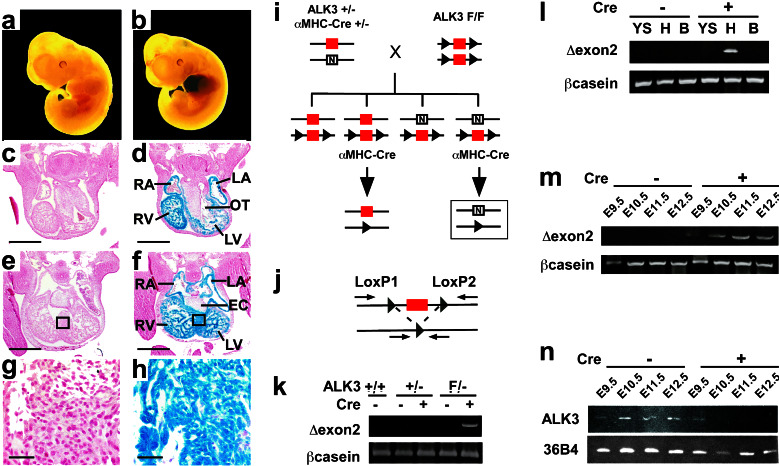
Figure 1.
Cardiac-restricted recombination of the floxed ALK3 allele. (a–h) The prevalence and specificity of early recombination using αMHC-Cre was tested at E11.5 in ROSA26 Cre-reporter embryos, by whole-mount β-galactosidase staining (a and b) and by coronal sections at the level of the outflow tract (c and d) and endocardial cushion (e and f). The box in e and f denotes the region of interventricular septum shown at higher magnification in g and h. (a, c, e, and g) ROSAF/−; (b, d, f, and h) ROSAF/−;αMHC-Cre+/−. Scale bar, 500 μm (c–f) or 30 μm (g and h); RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; OT, outflow tract; EC, endocardial cushion. (i–n) Cardiac-specific disruption of ALK3. (i) Homozygous floxed ALK3 mice (F/F) were bred with αMHC-Cre mice that are hemizygous for ALK3 (+/−), to produce a cardiac-specific deletion of exon 2 (boxed). red, wild-type exon 2; arrowhead, LoxP site; N, null allele (PGK-neo cassette). (j) Deletion of the 2.2-kb DNA fragment containing exon 2 is detected by PCR, using primers LoxP1 and LoxP2, which amplify a 170-bp product. (k–m) PCR analysis of ALK3 recombination (Δexon2) and a control gene (βcasein) at E11.5. (k) Recombination in myocardium was contingent on co-inheritance of αMHC-Cre and the floxed ALK3 allele. (l) Recombination was detected in heart (H) of ALK3F/−;αMHC-Cre+/− embryos, but not the yolk sac (YS) or body (B). (m) Recombination increased progressively in myocardium of ALK3F/−;αMHC-Cre+/− embryos from E9.5-E11.5. (n) RT-PCR analysis of ALK3 and a control gene (36B4) expression in myocardium of ALK3F/− embryos, in the presence (+) or absence (−) of αMHC-Cre. Note the absence of ALK3 elicited by Cre from E10.5 and onward.
PCR and Reverse Transcription (RT)-PCR.
To genotype progeny by PCR, DNA was isolated from yolk sacs of embryos or tail biopsies of weaned mice. The specificity and timing of DNA recombination were tested using primers 5′ and 3′ to the first and second loxP motifs. RNA was prepared from five embryonic hearts by using TRIzol (GIBCO). Expression of MEF2A, MEF2C, ANF, αMHC, and βMHC was measured by real-time quantitative RT-PCR (29), and corrected for 36B4, encoding a constitutive ribosomal phosphoprotein. For other genes studied, amplification was performed under conditions of linearity. Primer sequences are available on request.
Histology.
Embryos were fixed in 10% neutral formalin and embedded in paraffin. Serial sections (6 μm) were stained with hematoxylin and eosin for morphology studies. Apoptosis was monitored using a terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay and fluorescein-conjugated sheep polyclonal antibody against digoxigenin (Intergen, Purchase, NY). Sections were counterstained for sarcomeric MHC by using MF20 (Hybridoma Bank, University of Iowa, Iowa City, IA), Texas Red-conjugated goat IgG against mouse IgG (Molecular Probes), and Hoechst 33258. To detect Cre-dependent β-galactosidase expression, embryos were harvested at E11.5, processed for whole-mount staining, photographed, and sectioned (30).
In Situ Hybridization.
Embryos were fixed in 4% paraformaldehyde and embedded in paraffin. Serial sections (6 μm) were incubated with 35S-labeled riboprobes, processed for emulsion autoradiography, counterstained with Hoechst, and visualized by dark-field and epifluorescence microscopy (31). Probes were kindly provided by M. C. Jones, Institute of Cancer Research, London (BMP2); B. Hogan, Vanderbilt University Medical Center, Nashville, TN (BMP4); V. Rosen, Genetics Institute, Cambridge, MA (BMP10); R. J. Schwartz, Baylor College of Medicine, Houston (Nkx2.5); M. A. van Rooijen, Hubrecht Laboratory NIOB, Utrecht, The Netherlands (Smad6); and H. L. Moses, Vanderbilt–Ingram Cancer Center, Nashville, TN (TGFβ3).
Statistics.
TUNEL assays were compared by analysis of variance and the unpaired two-tailed t test, using a significance level of P < 0.05. RT-PCR results were compared by analysis of variance and Scheffé's test, using a significance level of P < 0.05.
Results
Conditional, Cardiac-Specific Deletion of ALK3.
We previously reported transgenic mice expressing Cre under the control of the cardiac αMHC promoter, which elicit efficient DNA recombination in myocardium (30). To establish the extent of recombination during cardiac organogenesis, we bred the αMHC-Cre mice to a Cre-dependent lacZ knock-in at the ubiquitously transcribed locus (32), and harvested embryos at E11.5. In agreement with the transient expression of endogenous αMHC throughout myocardium at that stage (33), whole-mount X-Gal staining revealed localized induction of lacZ in the heart, contingent on the presence of Cre (Fig. 1 a and b). Recombination in atrial and ventricular myocytes was virtually homogeneous, in coronal sections at the level of the outflow tract (Fig. 1 c and d) and AV cushion (Fig. 1 e–h). No promiscuous recombination was seen elsewhere or in nonmyocyte components of the heart, and there was no effect on cardiac development from the Cre transgene alone.
To produce the conditional mutation of ALK3, exon 2 (encoding 35% of the extracellular ligand-binding domain) was flanked by loxP sites, using homologous recombination. Deletion of exon 2 was sufficient to inactivate ALK3 function completely (Y.M. and R.R.B., unpublished results). Homozygous floxed ALK3 mice (F/F) were bred to αMHC-Cre mice that were hemizygous for ALK3 (+/−) (Fig. 1i). DNA recombination was detected as a 170-bp PCR product that results from deletion of the 2.2-kb fragment containing exon 2 (Fig. 1j). Recombination occurred in the heart of E11.5 embryos that were ALK3F/−;αMHC-Cre+/−, contingent on co-inheritance of Cre and the floxed ALK3 allele (Fig. 1k). Recombination was restricted to the heart, and was not detected in the yolk sac or body (Fig. 1l). Recombination of the floxed ALK3 allele was faintly detectable at E9.5 and maximal by E11.5 (Fig. 1m). RT-PCR confirmed the Cre-dependent loss of ALK3 expression in myocardium from E10.5 onwards (Fig. 1n). ALK6, the type IB BMP receptor, is not expressed in the heart at this stage (17).
ALK3 Is Required for Cardiac Septation and Normal Cushion Morphogenesis.
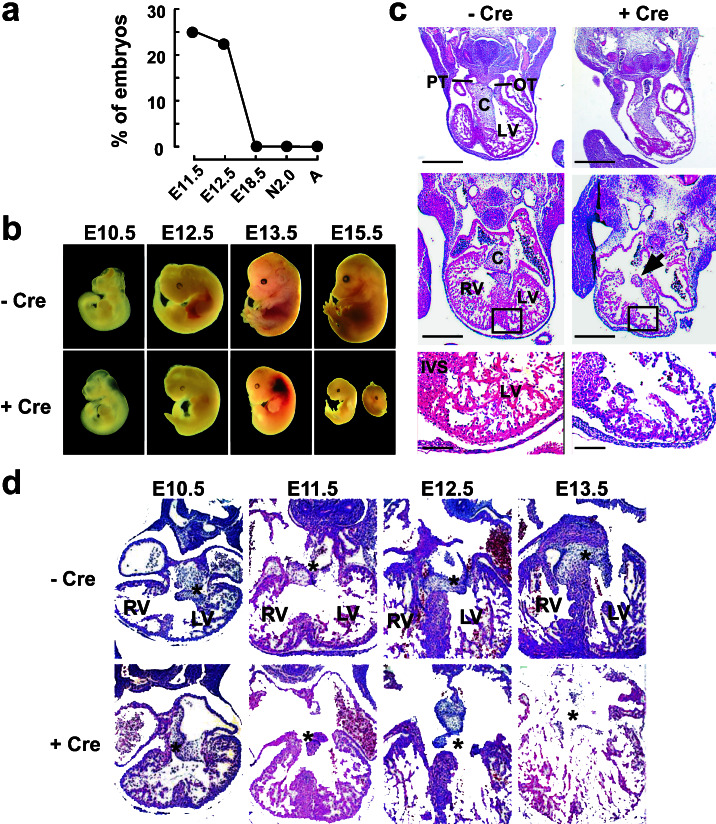
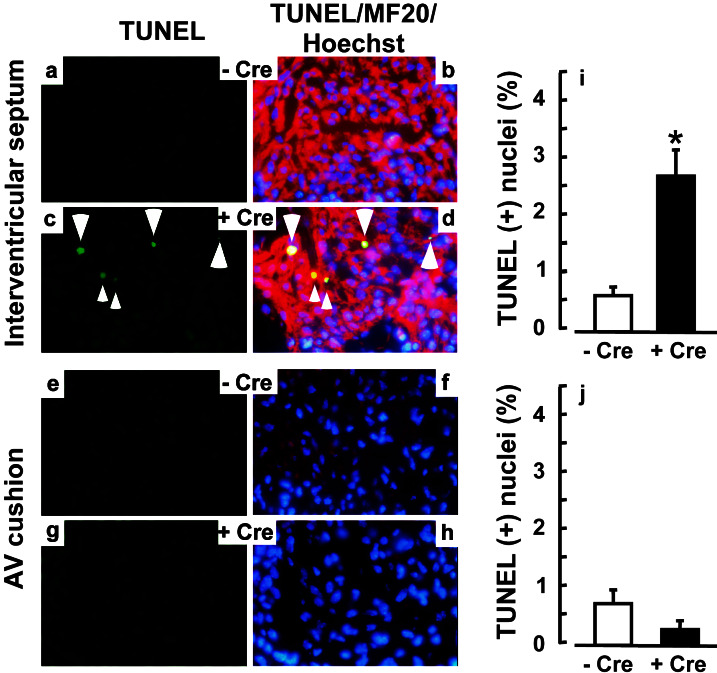
No newborn or weaned pups were found with the ALK3F/−;αMHC-Cre+/− genotype, of 15 litters examined (Fig. 2a). All three other potential genotypes were present in a Mendelian ratio (not shown). Myocyte-restricted deletion of ALK3 (ALK3F/−;αMHC-Cre+/−) was invariably lethal before E18.5, with marked resorption at E15.5 and growth retardation or internal hemorrhages at E12.5–13.5 (Fig. 2b). To seek potentially direct effects of the null mutation, and not secondary or tertiary consequences, we analyzed E11.5 embryos: at that stage DNA recombination is maximal, yet no early lethality or change in gross morphology is seen. In control embryos (ALK3F/−), endocardial cushions have developed in the outflow tract and AV canal, and the ventricles show a typical pattern of trabeculation (Fig. 2c Left; see also Fig. 1). Cardiac-specific deletion of ALK3 (ALK3F/−;αMHC-Cre+/−) invariably produced heart defects, involving the interventricular septum, trabeculae, and AV cushion (Fig. 2c Right). No defects were seen in the outflow tract. All mutant embryos showed the same phenotype, with some variation in severity. We tested whether this ensemble of defects might result from enhanced apoptosis in ventricular myocytes, AV cushion, or both. At E11.5 a 5-fold increase was seen in TUNEL-positive myocytes in the interventricular septum of ALK3F/−;αMHC-Cre+/− mice (Fig. 3 a–d and i), but no change in the AV cushion (Fig. 3 e–h and j). Thus, an ALK3-mediated pathway is required for cardiac myocyte survival here, consistent with the antiapoptotic function of BMPs in some settings (26).
Figure 2.
ALK3 is essential for cardiac septation and AV cushion morphogenesis. (a) A myocyte-dependent function of ALK3 is required for survival beyond mid-gestation. Prevalence of the ALK3F/−;αMHC-Cre+/− genotype in litters from the breeding strategy in Fig. 1i, at E10.5-E18.5, 2 days after birth (N2.0) or after weaning (A). (b) Morphology of ALK3F/− embryos during development, in the presence (Lower) or absence (Upper) of αMHC-Cre. (c) Hematoxylin- and eosin-stained coronal sections through the heart of E11.5 ALK3F/− embryos, in the presence (Right) or absence (Left) of αMHC-Cre. Top, at the level of the outflow tract; Middle, at the level of the AV cushion; Bottom, higher magnification of trabeculae in the left ventricle. Note the defects in the endocardial cushion (arrow), interventricular septum, and trabeculae of ALK3 mutant embryos. PT, pulmonary trunk; OT, outflow tract; C, endocardial cushion; LV, left ventricle; RV, right ventricle; IVS, interventricular septum. [Scale bar, 500 μm (Top and Middle) or 50 μm (Bottom).] (d) Time course of AV cushion development in ALK3F/− embryos in the absence or presence of αMHC-Cre. Asterisks denote progressive fusion of the two AV cushions if ALK3 is intact, and the defect in fusion if deleted in cardiac myocytes.
Figure 3.
ALK3 is essential for cardiac myocyte survival. (a–h) Apoptosis was quantified by TUNEL analysis at E11.5 in the interventricular septum (a–d) and AV cushion (e–h). Green, TUNEL reaction; red, MF20 antibody to sarcomeric MHC (Texas red); blue, Hoechst dye 33258. (i and j) Mean ± SE for the interventricular septum (i) and endocardial cushion (j). − Cre, control embryos (ALK3F/−); + Cre, mutant embryos (ALK3F/−;αMHC-Cre+/−); *, P < 0.05.
ALK3 Is Required for Cardiac Expression of TGFβ2.
Defects in organization of the AV cushion, where αMHC-Cre was not expressed, indicate that ALK3-dependent crosstalk from cardiac myocytes to the cushion is essential for normal AV valve formation in mammals, as is known to be true for avians (9–12). To establish more precisely the earliest step(s) in cushion morphogenesis affected by disruption of ALK3 in the myocytes, cushion development was examined at E10.5–13.5 (Fig. 2d). In normal mice, the two AV cushions formed by epithelial-to-mesenchymal transformation (E10.5) fuse at E11.5–12.5 to form the AV valves (E13.5). Despite conditional disruption of ALK3, the AV cushions appeared normal at E10.5 in all mutant embryos, suggesting that the initial epithelial-to-mesenchymal transformation occurs normally. By E11.5–12.5, however, the two cushions were reduced in size in more than 60% of embryos, often misaligned (not seen in the same short-axis plane of section), and unfused even when in contiguity. Fusion of the cushions and leaflet formation were deficient even at E13.5, when myocardial necrosis precludes further analyses. Thus, continued expression of ALK3 in cardiac myocytes is indispensable for subsequent development of the cushion and, hence, the AV valves.
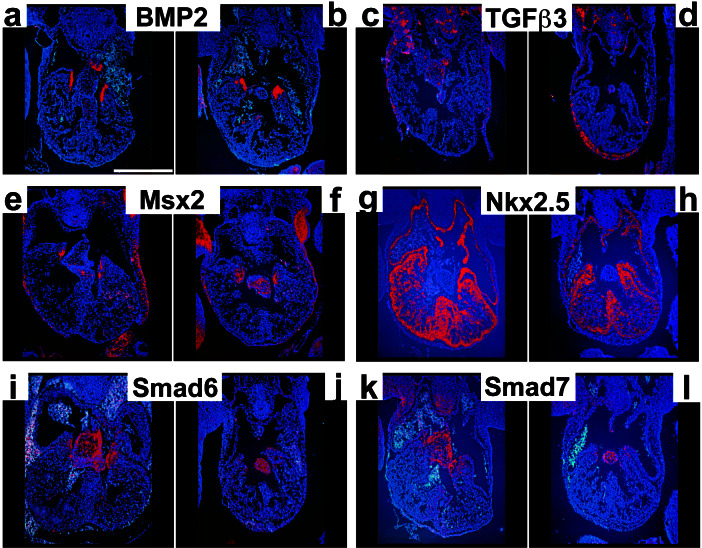
Because TGFβ2 and -β3 are both essential paracrine factors for cushion development in avians (9–11), we tested whether ALK3 was required for their expression in mice. By quantitative RT-PCR (Fig. 4a), TGFβ2 expression was normal at E9.5, when disruption of ALK3 was incipient, but decreased 75% at E10.5, thus preceding the first detected changes in cushion morphogenesis. Myocyte-restricted deletion of ALK3 decreased expression of TGFβ2 specifically in myocardium adjacent to the AV canal, whereas sites outside the heart were unaffected (Fig. 4 b–e). As measured by quantitative RT-PCR, or in situ hybridization, genes for other TGFβ/BMP signaling proteins that may participate in heart valve formation were unchanged: TGFβ3, BMP2, BMP4, BMPR-II, the Hox gene Msx2, and the inhibitory Smads, Smad6 and -7 (Figs. 4 d and e and 5, and unpublished results). Time-points later than E11.5 were not analyzed; older mutant embryos were typically friable.
Figure 4.
ALK3 regulates TGFβ2, ANF, and BMP10 in myocardium. (a and f) Quantitative RT-PCR analysis (mean ± SE) for TGFβ2 (a) and ANF (f). Expression was unaffected until E10.5. White, control (ALK3F/−); black, mutant (ALK3F/−;αMHC-Cre+/−); *, P < 0.05. (b–e and g–j) In situ hybridization for TGFβ2 (b–e), ANF (g and h), and BMP10 (i and j). Hearts were sectioned at E11.5, at the level of the AV cushion. b, d, g, and i, Control (ALK3F/−); c, e, h, and j, mutant (ALK3F/−;αMHC-Cre+/−). (d and e) Higher magnification of AV canal corresponding to b and c, respectively. Expression patterns with the hemizygous floxed allele did not differ from those reported for these genes in a wild-type background. TGFβ2 expression was defective in the ventricle surrounding the AV canal (arrows) but normal in noncardiac tissue (arrowheads). Asterisks denote the adjacent AV cushions. ANF and BMP10 were unchanged in the ventricle (arrowheads), but expressed precociously in the atria (arrow). [Scale bar, 500 μm (b, c, and g–j) or 50 μm (d and e).]
Figure 5.
Cardiac gene expression after conditional, myocyte-restricted deletion of ALK3. (a–l) In situ hybridization analysis of E11.5 hearts sectioned at the level of the AV cushion. a, c, e, g, i, and k, Control embryos (ALK3F/−); b, d, f, h, j, and l, mutant embryos (ALK3F/−;αMHC-Cre+/−). a and b, BMP2; c and d, TGFβ3; e and f, Msx2; g and h, Nkx2.5; i and j, Smad6; k and l, Smad7. (Scale bar, 500 μm.)
Conversely, by quantitative RT-PCR, expression of ANF in the mutant embryos increased 3-fold at E10.5 (Fig. 4f). By in situ hybridization, baseline ANF expression was greatest in the ventricular trabeculae at this stage, whereas the increase localized to the atria (Fig. 4 g and h). We therefore tested a second trabeculae-restricted gene, BMP10 (13). In mutant embryos, trabecular expression of BMP10 was preserved, but with precocious expression in the atria, where it would normally be found only at later time points (ref. 13; Fig. 4 i and j). Hence, ALK3 inhibits or delays the appearance of ANF and BMP-10 in the atria: these findings argue against a generalized maturation arrest as the basis for cardiac phenotypes resulting from the absence of ALK3.
The cardiogenic transcription factors Nkx2.5, eHAND, Tbx2, and Pitx2 each may require endogenous BMPs for their induction in avian embryos (6, 7, 31, 34). However, expression of these four transcription factors was not affected by heart-specific deletion of ALK3 (Fig. 5 g and h and unpublished results). Hence, either the initial expression of ALK3 suffices, or an alternative signal. However, necrosis and friability would interfere with assessing ALK3-dependent genes even 48 h after heart-specific disruption of the gene. Small (10–30%) yet consistent defects occurred in expression of MEF2A, MEF2C, MLC2v, and βMHC, and other markers of cardiac differentiation—i.e., αMHC, dHAND, GATA4, and FOG2—were unchanged (not shown).
Discussion
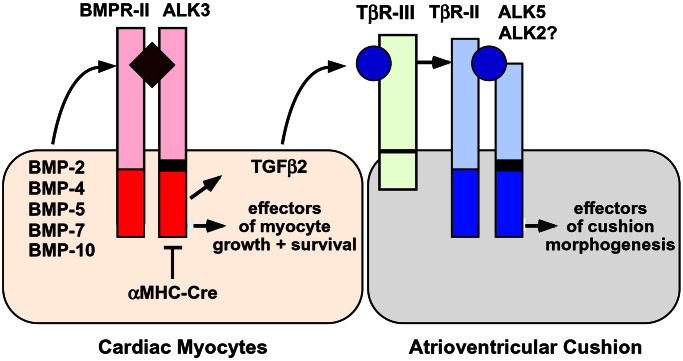
The mechanistic focus of this study was to determine—by conditional, lineage-restricted deletion of ALK3—if an essential role is served by the continued expression of this BMP receptor in cardiac myocytes subsequent to onset of the cardiac “fate.” Disruption of ALK3 in differentiated myocytes unmasked an essential requirement for ALK3 in heart morphogenesis, beyond myocyte determination (6, 7, 14, 35), involving the ventricular septum, trabeculae, compact myocardium, and endocardial cushion. Second, although BMP signaling via ALK3 may be required to maintain (as well as induce) Nkx2.5 in Xenopus (35), ALK3 appears dispensable for its expression in mice, and for other transcription factors tested, once the cardiac fate is established. Third, given the prevalence of AV cushion defects and proven cross-talk in avians between myocardium and the developing cushion, we tested whether disruption of ALK3 in myocytes caused defective expression of established paracrine signals for cushion morphogenesis. ALK3 was essential for cardiac myocyte expression of TGFβ2, suggesting a myocyte-dependent pathway in forming the AV cushion and the resulting cardiac valves (Fig. 6).
Figure 6.
ALK3 is essential for cardiac myocyte expression of TGFβ2 and AV cushion morphogenesis. Genetic interactions between BMPs and TGFβ signaling are shown schematically. In mid-gestation, cardiac myocytes express BMP2 and BMP4 adjacent to the AV cushion, BMP5 and BMP7 more homogeneously, and BMP10 in trabeculae (in addition to BMP production by other sources). Deletion of ALK3 in cardiac muscle cells, using αMHC-Cre, results in defective AV cushion morphogenesis, associated with the lack of TGFβ2 produced by cardiac myocytes, a proven paracrine signal for this process in avians (10). The type III TGFβ receptor (TβR-III) is needed for TGFβ2 binding to TβR-II (39) and, in avian explants, AV cushion morphogenesis (11). In avians, ALK2 also mediates cushion formation, not the type I TGFβ receptor ALK5 (36), but given early lethality for both null alleles, it is unknown whether this is true in mice (18, 40). Myocyte-restricted deletion of ALK3 disinhibits the expression of ANF and BMP10 in the atria, but causes defects in the ventricular septum, trabeculae, and compact myocardium.
Germline mutations of ALK3 and BMPR-II lead to death before cushion formation or even cardiac specification. For this reason, the conventional null alleles cannot be used readily to test whether, where, and how BMP signals function during cardiac morphogenesis. By contrast, the conditional loss of ALK3 in mid-gestation myocardium quickly leads to multiple cardiac defects, which coincide with the distribution of BMPs in the heart: BMP2 and BMP4 adjacent to the AV cushion, BMP5 and BMP7 more homogenously, and BMP10 in the trabeculae. Deletion of BMP5 and -7 in tandem results in cardiac defects that involve the AV cushion, septum, free wall, and trabeculae—resembling heart-specific deletion of ALK3—but with extensive abnormalities of the allantois, neural crest, and vasculature that make a role for BMPs within the heart less straightforward to assert (26). Myocyte-restricted deletion of ALK3 enabled us, with one mutation, to overcome the confounding problems of redundancy among multiple BMP ligands, systemic and local confounding effects, and the ambiguous identity of the key target cell.
These functions of ALK3 in cardiac morphogenesis are essential and not supplanted by other type I receptors. By contrast, cardiac-lethal effects of deleting BMP7 plus BMP6 preferentially involve the outflow tract (15), a region unaffected by the loss of ALK3. Although ALK6 is not expressed in the mid-gestation heart and thus is extraneous here, ALK2/ActRIA is expressed in the mouse heart at this stage and binds a subset of BMP family members (18). Notably, for Xenopus and avians, ALK2 has reported functions during cardiac morphogenesis, in laterality and AV cushion transformation, respectively (5, 36). Mice lacking ALK2 die in early gastrulation, before even the earliest steps in cardiac specification, and a requirement for ALK2 in extraembryonic tissues complicates even the use of chimeric mice to address its possible function in the mammalian heart (18). Notwithstanding possible redundancy between ALK3 and ALK2 in other circumstances, ALK2 does not overcome the cardiac-lethal phenotype unmasked by conditional deletion of ALK3, as with the essential role for each in gastrulation. Whether this, in turn, reflects distinctions in ligand preference or downstream effectors is presently conjectural.
Dependence of the AV cushion on ALK3—despite the absence of Cre from this site—indicates conclusively that crosstalk from myocytes to the cushion, in some form, is obligatory for AV valve formation in mice. Down-regulation of TGFβ2 by the disruption of ALK3 in cardiac muscle cells suggests that a paracrine circuit, as in avian explants (9–12), might also apply to mammalian cushion morphogenesis (Fig. 6). Although TGFβ2 is one of many potential downstream candidate genes that might contribute to developmental defects in ALK3-deficient hearts, it is noteworthy that mice lacking TGFβ2 die soon after birth with cardiac defects including malformations of the ventricular septum and AV valves (37).
In humans, congenital heart defects occur with a prevalence of at least 1% in newborns, and are even more common in death before term (38). Most frequent are defects in septation and the cardiac valves, and few single-gene etiologies are known. The invariable defects in myocardium and AV cushion resulting from conditional deletion of ALK3 provide strong support for its assessment as a candidate gene in human congenital heart disease.
Acknowledgments
We thank P. Soriano for ROSA26 Cre-reporter mice; D. Anderson for CMV-Cre mice; P. Davies, G. Shipley, and G. Eichele for access to instrumentation; B. Hogan, M. Jones, H. L. Moses, V. Rosen, R. J. Schwartz, and M. A. van Rooijen for reagents; and J. Pike and A. Francis for technical help. This work was supported by the National Institutes of Health (M.D.S., R.R.B., and V.G.), the American Heart Association (V.G.), the March of Dimes (V.G.), the Fund for Scientific Research–Flanders (A.Z., D.H.), a collaboration agreement between the Flanders Interuniversity Institute for Biotechnology and Innogenetics (D.H.), the University of Leuven (D.H.), the RGK Foundation (M.D.S.), and the M. D. Anderson Foundation Professorship (M.D.S.).
Abbreviations
- AV
atrio-ventricular
- BMP
bone morphogenetic proteins
- MHC
myosin heavy chain
- TGFβ
transforming growth factor-β
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- RT
reverse transcription
Note Added in Proof.
Construction of the floxed ALK3 mice and their use to show the requirement for ALK3 in limb development have been described since the submission of our report (41, 42).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ray R P, Wharton K A. Cell. 2001;104:801–804. doi: 10.1016/s0092-8674(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 2.Paria B C, Ma W, Tan J, Raja S, Das S K, Dey S K, Hogan B L. Proc Natl Acad Sci USA. 2001;98:1047–1052. doi: 10.1073/pnas.98.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frasch M. Nature (London) 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen J N, van Eeden F J, Warren K S, Chin A, Nusslein-Volhard C, Haffter P, Fishman M C. Development (Cambridge, UK) 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- 5.Ramsdell A F, Yost H J. Development (Cambridge, UK) 1999;126:5195–5205. doi: 10.1242/dev.126.23.5195. [DOI] [PubMed] [Google Scholar]
- 6.Schultheiss T M, Burch J B E, Lassar A B. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 7.Schlange T, Andree B, Arnold H H, Brand T. Mech Dev. 2000;91:259–270. doi: 10.1016/s0925-4773(99)00311-1. [DOI] [PubMed] [Google Scholar]
- 8.Yamagishi T, Nakajima Y, Miyazono K, Nakamura H. J Cell Physiol. 1999;180:35–45. doi: 10.1002/(SICI)1097-4652(199907)180:1<35::AID-JCP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 9.Potts J D, Dagle J M, Walder J A, Weeks D L, Runyan R B. Proc Natl Acad Sci USA. 1991;88:1516–1520. doi: 10.1073/pnas.88.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer A S, Ayerinskas I I, Vincent E B, McKinney L A, Weeks D L, Runyan R B. Dev Biol. 1999;208:530–545. doi: 10.1006/dbio.1999.9211. [DOI] [PubMed] [Google Scholar]
- 11.Brown C B, Boyer A S, Runyan R B, Barnett J V. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 12.Mjaatvedt C, Yamamura H, Wessels A, Ramsdell A, Turner D, Markwald R R. In: Heart Development. Harvey R P, Rosenthal N, editors. San Diego: Academic; 1999. pp. 159–177. [Google Scholar]
- 13.Neuhaus H, Rosen V, Thies R S. Mech Dev. 1999;80:181–184. doi: 10.1016/s0925-4773(98)00221-4. [DOI] [PubMed] [Google Scholar]
- 14.Monzen K, Shiojima I, Hiroi Y, Kudoh S, Oka T, Takimoto E, Hayashi D, Hosoda T, Habara-Ohkubo A, Nakaoka T, et al. Mol Cell Biol. 1999;19:7096–7105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim R Y, Robertson E J, Solloway M J. Dev Biol. 2001;235:449–466. doi: 10.1006/dbio.2001.0284. [DOI] [PubMed] [Google Scholar]
- 16.Massague J, Blain S W, Lo R S. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 17.Dewulf N, Verschueren K, Lonnoy O, Moren A, Grimsby S, Vandespiegle K, Miyazono K, Huylebroeck D, ten Dijke P. Endocrinology. 1995;136:2652–2663. doi: 10.1210/endo.136.6.7750489. [DOI] [PubMed] [Google Scholar]
- 18.Gu Z Y, Reynolds E M, Song J H, Lei H, Feijen A, Yu L A, He W W, MacLaughlin D T, van den Eijnden-van Raaij J, Donahoe P K, Li E. Development (Cambridge, UK) 1999;126:2551–2561. doi: 10.1242/dev.126.11.2551. [DOI] [PubMed] [Google Scholar]
- 19.Aoki H, Fujii M, Imamura T, Yagi K, Takehara K, Kato M, Miyazono K. J Cell Sci. 2001;114:1483–1489. doi: 10.1242/jcs.114.8.1483. [DOI] [PubMed] [Google Scholar]
- 20.Mishina Y, Suzuki A, Ueno N, Behringer R R. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 21.Beppu H, Kawabata M, Hamamoto T, Chytil A, Minowa O, Noda T, Miyazono K. Dev Biol. 2000;221:249–258. doi: 10.1006/dbio.2000.9670. [DOI] [PubMed] [Google Scholar]
- 22.Winnier G, Blessing M, Labosky P A, Hogan B L M. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 23.Scott J. Nat Genet. 2000;26:3–4. doi: 10.1038/79148. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D W, Berg J N, Baldwin M A, Gallione C J, Marondel I, Yoon S J, Stenzel T T, Speer M, Pericak-Vance M A, Diamond A, et al. Nat Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H B, Bradley A. Development (Cambridge, UK) 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- 26.Solloway M J, Robertson E J. Development (Cambridge, UK) 1999;126:1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 27.Rossant J, McMahon A. Genes Dev. 1999;13:142–145. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- 28.Arango N A, Lovell-Badge R, Behringer R R. Cell. 1999;99:409–419. doi: 10.1016/s0092-8674(00)81527-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhang D, Gaussin V, Taffet G E, Belaguli N S, Yamada M, Schwartz R J, Michael L H, Overbeek P A, Schneider M D. Nat Med. 2000;6:556–563. doi: 10.1038/75037. [DOI] [PubMed] [Google Scholar]
- 30.Agah R, Frenkel P A, French B A, Michael L H, Overbeek P A, Schneider M D. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada M, Revelli J P, Eichele G, Barron M, Schwartz R J. Dev Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- 32.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 33.Zammit P S, Kelly R G, Franco D, Brown N, Moorman A F, Buckingham M E. Dev Dyn. 2000;217:75–85. doi: 10.1002/(SICI)1097-0177(200001)217:1<75::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 34.Zhu L, Marvin M J, Gardiner A, Lassar A B, Mercola M, Stern C D, Levin M. Curr Biol. 1999;9:931–938. doi: 10.1016/s0960-9822(99)80419-9. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Katsev S, Cai C, Evans S. Dev Biol. 2000;224:226–237. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- 36.Lai Y T, Beason K B, Brames G P, Desgrosellier J S, Cleggett M C, Shaw M V, Brown C B, Barnett J V. Dev Biol. 2000;222:1–11. doi: 10.1006/dbio.2000.9698. [DOI] [PubMed] [Google Scholar]
- 37.Sanford L P, Ormsby I, GittenbergerdeGroot A C, Sariola H, Friedman R, Boivin G P, Cardell E L, Doetschman T. Development (Cambridge, UK) 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman J I. Pediatr Cardiol. 1995;16:155–165. doi: 10.1007/BF00794186. [DOI] [PubMed] [Google Scholar]
- 39.Blobe G C, Schiemann W P, Pepin M C, Beauchemin M, Moustakas A, Lodish H F, O'Connor-McCourt M D. J Biol Chem. 2001;276:24627–24637. doi: 10.1074/jbc.M100188200. [DOI] [PubMed] [Google Scholar]
- 40.Larsson J, Goumans M J, Sjostrand L J, van Rooijen M A, Ward D, Leveen P, Xu X, ten Dijke P, Mummery C L, Karlsson S. EMBO J. 2001;20:1663–1673. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An K, Mishina Y, Hanks M C, Behringer R R, Crenshaw E B., III Development (Cambridge, UK) 2001;128:4449–4461. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- 42.Mishina, Y., Hanks, M. C., Miura, S., Tallquist, M. D. & Behringer, R. R. (2002) Genesis32, in press. [DOI] [PubMed]