Abstract
The development of Drosophila requires the function of the CREB-binding protein, dCBP. In flies, dCBP serves as a coactivator for the transcription factors Cubitus interruptus, Dorsal, and Mad, and as a cosuppressor of Drosophila T cell factor. Current models propose that CBP, through its intrinsic and associated histone acetyltransferase activities, affects transient chromatin changes that allow the preinitiation complex to access the promoter. In this report, we provide evidence that dCBP may regulate the formation of chromatin states through interactions with the modulo (mod) gene product, a protein that is thought to be involved in chromatin packaging. We demonstrate that dCBP and Modulo bind in vitro and in vivo, that mutations in mod enhance the embryonic phenotype of a dCBP mutation, and that dCBP mutations enhance the melanotic tumor phenotype characteristic of mod homozygous mutants. These results imply that, in addition to its histone acetyltransferase activity, dCBP may affect higher-order chromatin structure.
The CREB-binding protein (CBP) is one of a growing family of homologous proteins that function as signal-responsive transcriptional coactivators by serving as a physical link between signal-activated transcription factors and the basal transcriptional machinery.
Since the initial cloning of CBP and its homologue p300, much work has been dedicated to defining the role of CBP/p300 in integrating the transcriptional events that are stimulated by diverse signaling pathways. CBP/p300 interacts with and stimulates the activity of a variety of signal-responsive transcription factors, including nuclear hormone receptors, CREB, MyoD, STAT2, and p53 (1). In Drosophila, dCBP can stimulate the activity of the signal-responsive transcriptional activators Mad and Dorsal (2, 3), and it is required for the activity of the hedgehog transcriptional activator Cubitus interruptus (4). The coactivator function of CBP/p300 can be modulated by interactions with different classes of proteins, such as kinases (p90rsk) and viral proteins (E1A, Tax, large T antigen), and histone acetyltransferases (HATs), such as P/CAF and SRC-1 (1). CBP/p300 has been shown to possess an intrinsic HAT activity (5) that is required for transactivation in cell culture (6). Thus, the CBP coactivator function may involve the remodeling of the promoter chromatin structure as well as the bridging of transcription factors to the basal transcriptional complex.
We have used yeast two-hybrid screens to identify the proteins that interact with different domains of Drosophila dCBP, and thus to define the signaling cascades and transcriptional processes that require dCBP function. One of the screens identified Modulo, a protein originally described as a modifier of position effect variegation (PEV), as an interacting partner of dCBP. PEV is a model system in Drosophila for the study of functional differences between heterochromatin and euchromatin (7–9). Modulo appears to be a highly pleiotropic protein. Mutations in modulo (mod) are dominant suppressors of PEV (10), which suggests that mod is either directly or indirectly involved in the packaging of DNA into heterochromatin. In support of the role of mod in chromatin packaging, Mod colocalizes with condensed chromatin and heterochromatin; immunohistochemical analysis of polytene chromosomes shows that Mod binds to regions of centric heterochromatin as well as to specific bands in the euchromatic chromosomal arms (11). Further support for Mod function in euchromatin is the observation that Polycomb group complexes contain Mod (12). Mod also colocalizes with the nucleolus and ribonucleoprotein complexes, suggesting that mod is a regulator of rRNA synthesis and/or function (11, 13). The analysis of Mod in both the chromatin and nucleolus fractions of cell extracts suggests that the phosphorylation of Mod mediates the transition from chromatin binding to nucleolus association (13).
In this report, we show that Modulo and dCBP can interact in vitro and in vivo, and can colocalize to embryonic cell nuclei and specific bands in polytene chromosomes. However, they do not appear to colocalize to the nucleolus. We also demonstrate that mod mutations can enhance the segmentation defects detected in dCBP mutant embryos, and that mutations in dCBP can enhance the melanotic tumor phenotype seen in the third-instar larvae of mod mutants. These results demonstrate that dCBP is involved with a different class of chromatin-binding protein than previously described and suggest that, in addition to modulating the chromatin surrounding promoter sequences, dCBP may function in complexes that remodel chromatin into heritably stable, higher-order states.
Materials and Methods
Plasmid Constructions.
A BamHI–EcoRI and a BamHI–NotI restriction fragment of pBSK-dCBP (14) were inserted into the bait vector pBTM116 (16) to give pLexA-dCBP-2278–2678 and pLexA-dCBP-2278–3191, respectively. Other dCBP fragments were inserted by PCR cloning into the bait vector to give pLexA-dCBP-2278-2476, pLexA-dCBP-2471-2677, and pLexA-dCBP-2679-3191. pACT-Mod-219-544 was the clone obtained from the yeast two-hybrid screen. Other Mod fragments were inserted by PCR cloning into the prey vector pACT to give pACT-Mod-219-493, pACT-Mod-305-493 and pACT-Mod-305-544. The BamHI–EcoRI fragment of dCBP was inserted into the pGEX-KG vector (Amersham Pharmacia) to generate pGST-dCBP-2278-2678. Modulo fragments were inserted into a pCITE vector (Novagen) for in vitro translation.
Yeast Two-Hybrid Assays.
The yeast strain L40 (15), which includes the reporter genes HIS3 and lacZ, was used. All transformations were performed by using a variation of the lithium acetate method (16). For the screening, L40 yeast expressing LexA-dCBP-2278-2678 were transformed with a 0- to 6-h Drosophila embryo pACT cDNA library (provided by Greg Golling, State University of New York, Stony Brook), and transformants were plated on selective minimal medium supplemented with 30 mM 3-aminotriazole. From 2.0 × 106 estimated transformants, approximately 100 were histidine and β-galactosidase (β-gal) positive. pACT plasmids from these colonies were extracted, transformed into Escherichia coli M1066, and sequenced. β-gal activity was determined by using a β-gal filter assay (17).
In Vitro Binding Assays.
Glutathione S-transferase (GST) fusion proteins were produced in E. coli BL21 and purified by affinity chromatography on glutathione-agarose beads according to the Pharmacia protocol. In vitro translated [35S]methionine-labeled proteins were incubated with immobilized GST fusion proteins for 2 h at room temperature in Harlow buffer (50 mM Hepes, pH 7.5/100 mM NaCl/0.2 mM EDTA/0.01 mM NaF/1 mM DTT/0.5% Nonidet P-40) with 5 mg/ml BSA. After four washes in Harlow buffer (containing 200 mM NaCl), bound proteins were eluted by boiling in SDS-loading buffer, resolved by SDS/PAGE, and visualized by using a phosphorimager (Molecular Dynamics).
Coimmunoprecipitation Assay.
Kc cells were lysed in low-stringency (LS) buffer (PBS/0.1% Nonidet P-40/1 mM DTT) containing protease inhibitors [10 μg/ml leupeptin/10 μg/ml pepstatin A/10 μg/ml antipain/10 μg/ml E64/1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride] and cell extracts were prepared as described (18). After preclearing for 30 min with 50 μl of protein A-Sepharose (Pharmacia), supernatants were incubated at 4°C for 90 min with 10 μl of preimmune or anti-dCBP (rat antiserum directed against the CBD domain of dCBP), followed by incubation at 4°C for 30 min with protein A-Sepharose beads. Beads were washed four times in LS buffer, and immunoprecipitated proteins were eluted by boiling in SDS-loading buffer, resolved by SDS/PAGE, and examined by Western blotting with the Mod LA9 antibody (19) at 20 μg/ml.
Drosophila Strains.
The mod allele L8 is considered to be a null allele of mod (10), and was kindly provided by Jacques Pradel (Centre National de la Recherche Scientifique, Marseilles). The y1 w1 nej3 recombinant chromosomes were generated by standard genetic techniques. Generation of the nej1 and nej3 alleles has been described (14); and are deletions of the dCBP gene. The nej1 deletion includes the second exon of dCBP, whereas the nej3 deletion is somewhat larger and includes part of the third exon. By antibody staining, both the nej1 and nej3 alleles are molecular nulls. The balancer chromosomes FM7c, TM2 and TM6B, Tb that are used in these studies can be found in Lindsley and Zimm (20). The TM3, ftz-lacZ was generated by S.M.S. The FM7c, P{w + mC = GAL4-Kr.C}DC1, P{w+mC = UAS-GFP.S65T}DC5, sn+ (FM7c-GFP in the text) and TM3, P{w + mC = GAL4–Kr.C}DC2, P{w+mC = UAS-GFP.S65T}DC10, Sb1 (TM3–GFP in the text; GFP, green fluorescent protein) chromosomes were obtained from the Bloomington Stock Center. The expression of GFP from these chromosomes is not maternal. The wild-type strain used in these studies is Canton-S. The progeny of crosses were reared at 25°C on standard cornmeal yeast-extract source media.
Cuticle Preparation of Embryos.
Eggs were collected after a 24-h period and aged for an additional 48 h at 25°C. The cuticles of the unhatched embryos were prepared as described (21), visualized by dark-field optics, and photographed with Tech Pan 50 film.
Confocal Analysis of Embryos and Polytene Chromosomes.
The immunohistochemistry of whole-mount embryos was carried out as described (22). Polytene chromosomes were fixed and squashed for immunohistochemistry as described (23). Modulo was detected by the mAb LA9 at 80 μg/ml, dCBP by a chicken antiserum raised against the CBD domain of dCBP (24) at a dilution of 1:800, and β-gal by affinity-purified rabbit antibodies (ICN/Cappel) at a dilution of 1:200. Fluorescein anti-mouse (Vector), Texas Red anti-rabbit (Vector), and rhodamine anti-chicken (Jackson ImmunoResearch) secondary antibodies were used at a dilution of 1/200e. Embryos were examined with a confocal laser scan microscope (Bio-Rad 1024 ES/Nikon Eclipse TE300) and the polytene chromosomes were visualized with a Leica DMR8 microscope equipped with a Hamamatsu charge-coupled device camera C5985.
Phenotypic and Statistical Analysis of the Melanotic Tumor Phenotype of Third-Instar Larvae.
Eggs were laid for 24 h and incubated in fly bottles for 7 days at 25°C. During this time, larvae of both populations entered the third-instar stage and started to develop melanotic tumors. At this critical stage, we selected and analyzed the wandering larvae that were nontubby females. Three independent larval populations for the cross were assayed. For statistical analysis, numbers from these experiments were pooled. Because our data were ordinal, we used the nonparametric test of Kolmogorov–Smirnov (25). We calculated the χ2 value for a degree of freedom (df) of 2, from the D values, which represent the maximum differences for pairs of groups. A D value of 0.457 was obtained for this experiment. Larvae were heat-fixed at 55°C for 30 min in PBS/50% glycerol, mounted in the same buffer, and photographed with Ecktachrome 64 film using indirect light.
Results and Discussion
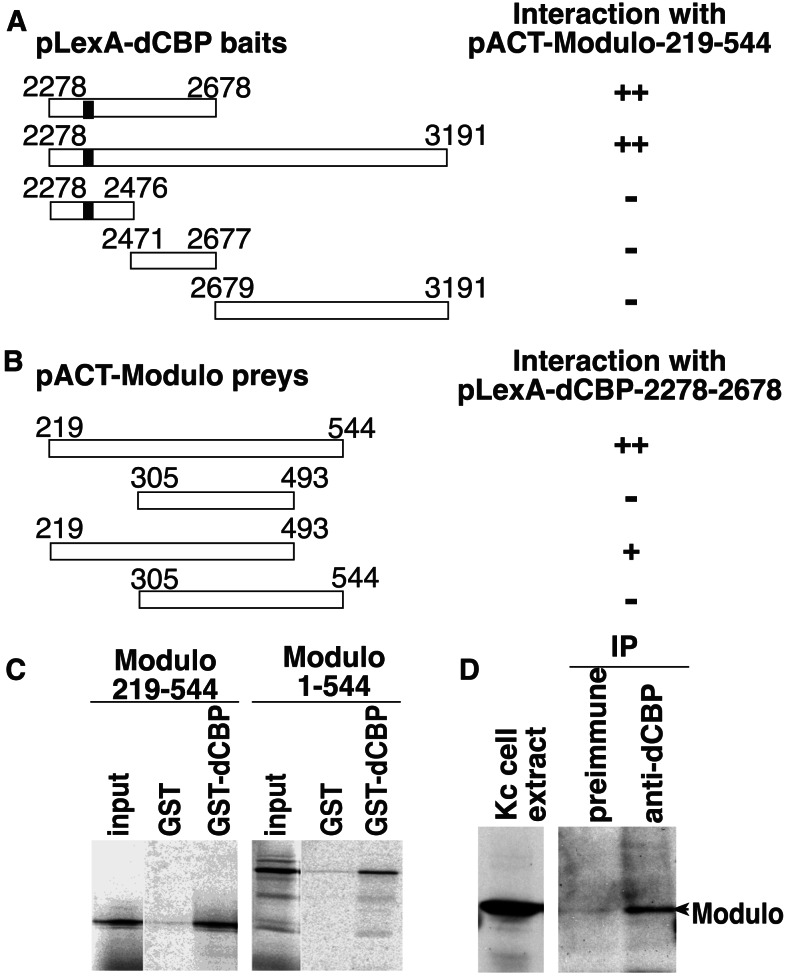
We performed a yeast two-hybrid screen of a Drosophila cDNA library to assess binding of the well-conserved dCBP C-terminal C/H3 domain to nuclear factors. Three positive clones encoded Modulo, a DNA-binding protein that appears to be involved in the compaction of chromatin in Drosophila (10, 11, 13). Both the C/H3 domain and the N-terminal region of the dCBP activation domain are necessary for Modulo binding (Fig. 1A). We also found that amino acids 219–493 of Modulo are sufficient to bind dCBP (Fig. 1B). Binding between dCBP and Modulo was confirmed in vitro by using a bacterially expressed dCBP fusion protein (GST-dCBP-2278-2678) that contains the C/H3 domain and the first 200 aa of the glutamine-rich activation domain. An in vitro translated Modulo polypeptide (amino acids 219–544) corresponding to the fragment isolated with the two-hybrid screen, as well as the full-length Modulo, binds strongly to GST-dCBP-2278-2678 (Fig. 1C). However, no binding of Modulo is observed to GST fusion proteins containing the dCBP CREB-binding domain or the bromo-zinc finger domain (data not shown). We used whole-cell extracts from Drosophila Kc cells, and showed that an antibody directed against dCBP specifically immunoprecipitates Modulo, indicating that both proteins are part of the same complex in vivo (Fig. 1D).
Figure 1.
dCBP interacts with Modulo. Two-hybrid interactions between different pLexA-dCBP baits and the pACT-Modulo clone isolated by the two-hybrid screen (A) or between different pACT-Modulo preys and the pLexA-dCBP-2278–2678 bait (B). The dCBP baits and the Modulo preys used are represented, the numbers show the amino acid positions, and the black box in dCBP represents the C/H3 domain. The results of the two-hybrid interactions obtained by using a β-gal filter assay are represented on the right. ++, Strong interaction; +, weak interaction; −, no interaction. (C) In vitro binding between dCBP and Modulo. Equimolar amounts of immobilized GST and GST-dCBP-2278-2678 were incubated with in vitro translated 35S-labeled Modulo proteins. Some (10%) of the total reaction was loaded in the input lanes, and all samples were run on the same gel. (D) Coimmunoprecipitation of the dCBP-Modulo complex from Kc cell extracts by using a dCBP antisera. Some (2%) of the unprecipitated extract was loaded in the input lane and run on the same gel as the immunoprecipitated complexes.
We then investigated the functional significance of this interaction in the fly. We determined the phenotype of embryos hemizygous for the dCBP allele nej1 (14) and either homozygous or heterozygous for the mod allele L8 (10). These phenotypes were then compared with those of nej1/+; L8 and nej1/+; L8/+ control embryos. We analyzed the cuticles of 300–400 unhatched embryos per genotype. We found that most of the nej1 hemizygotes develop to the end of embryogenesis and are phenotypically wild type. Approximately 13% of the embryos have head defects and 4.5% have fusions of the segmental denticle belts, primarily between the denticle belts of the fourth and the fifth abdominal segments (Fig. 2a). We also found that most, over 90%, of the L8 homozygous embryos survive to hatching. Approximately 27% of the nonhatching embryos show head defects, tail defects, or both as described (26). The denticle belts of L8 homozygous embryos are virtually wild type (Fig. 2b), and only 3% have segmental fusions. To analyze nej1 hemizygous embryos in an L8 background, nej1/FM7-GFP; L8/TM3-GFP female flies were crossed to FM7-GFP; L8/TM3-GFP males and cuticles were prepared from the unhatched, fluorescent, and nonfluorescent embryos. The nonfluorescent embryos are nej1; L8 male embryos and the fluorescent embryos are nej1; L8/TM3-GFP or nej1; TM3-GFP male embryos. Fifteen percent (73 of 487) of the nej1; L8 embryos have fusions of the segmental denticle belts. These fusions occur between the second and the third, the third and the fourth, or the fourth and the fifth abdominal segments (Fig. 2 c and d). In contrast, only 3% (13 of 467) of the nej1; L8/TM3-GFP embryos have denticle belt fusions. Thus, nej and mod mutations appear to have a synergistic effect on denticle belt fusions (Fig. 2 c and d). We also observed that 24.2% (118 of 487) of the nej1; L8 embryos have head defects, whereas 12% (56 of 467) of the nej1; L8/TM3-GFP embryos have head defects. The percentage of head defects observed in the nej1; L8 double mutants might be additive if the two genes affected head development through two independent pathways. The percentage of head defects observed in nej1; L8 double mutants is neither enhanced nor suppressed, and is the same as that seen in the L8 homozygotes, suggesting that mod might be epistatic to dCBP in head development. Similar results were obtained with the nej3 allele (data not shown). To control for the possibility that the L8 chromosome carried modifiers of dCBP function, we repeated our experiment by using the L8 parental A4-4 chromosome (27). We crossed nej1/+; A4-4/+ females to A4-4/A4-4 males. Only 5.5% of the unhatched embryos have denticle belt fusions, and 10% of the unhatched embryos have head defects, indicating that the effect seen in the nej1; L8 double mutant is specific to the mod mutation. Taken together, these results indicate that a synergistic interaction between dCBP and Mod is required during embryogenesis for proper segmentation. Furthermore, because the nej1; L8 double mutants have an enhanced phenotype, rather than a nej1 or L8 phenotype, we suggest that Modulo and dCBP may be members of a larger regulatory complex that is sensitive to changes in their relative dosages.
Figure 2.
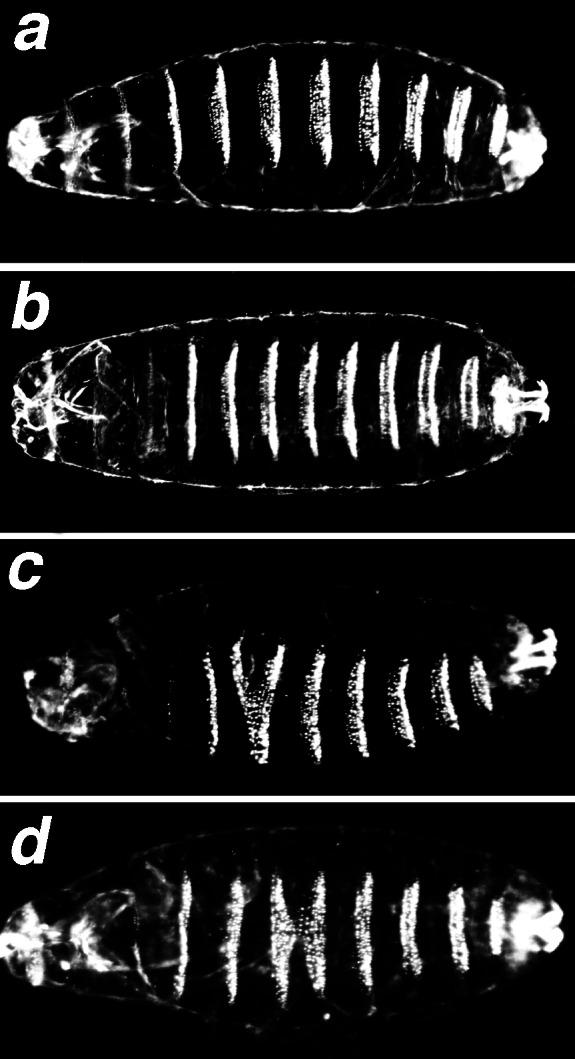
Denticle belt fusion phenotype of nej1; L8 double mutant embryos. Cuticle preparation of late-stage embryos photographed with dark-field optics; anterior is to the left. (a) nej1/Y embryo. (b) L8 embryo. (c and d) nej1/Y; L8 double mutant embryos showing head defects and fusions of segment denticle belts: fusion A2–A3 (c), fusion A3–A4 (d).
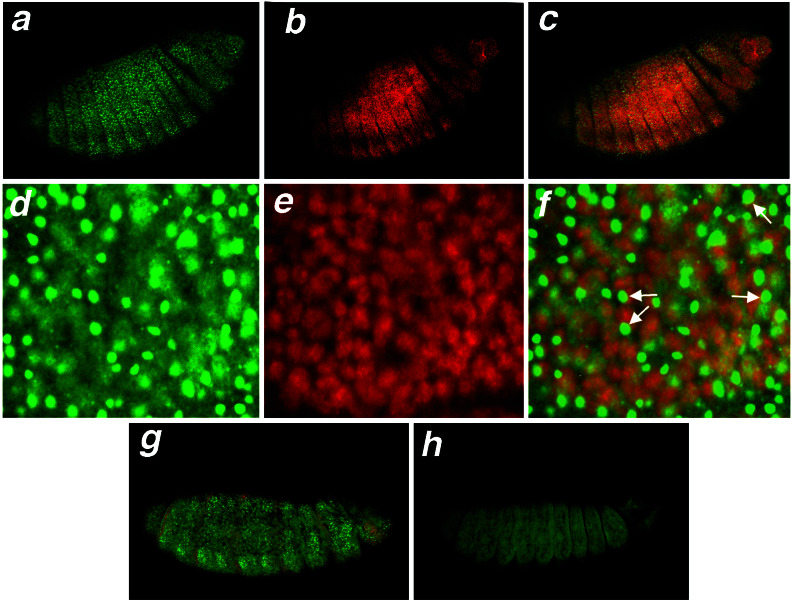
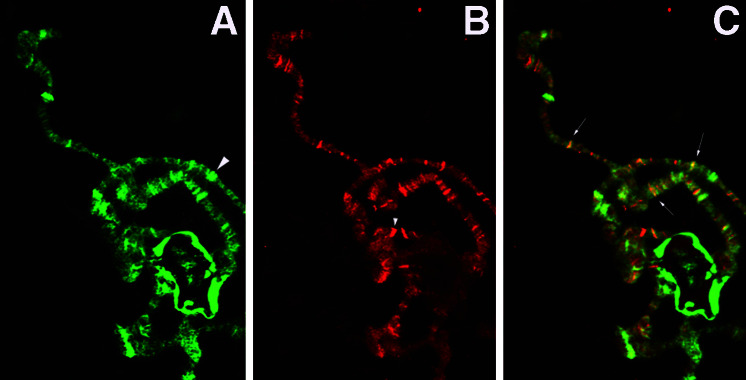
Because we obtained segmental fusion phenotypes in the nej1; L8 double mutants, we determined whether dCBP and Modulo were co-expressed during embryonic development. We used confocal microscopy to show that both proteins are ubiquitously expressed during embryogenesis (Fig. 3 a, b, and c, and data not shown). Furthermore, Modulo is not detected in the epidermis of the L8 homozygous mutants (Fig. 3 g and h). Modulo and dCBP are expressed in the nuclei of all segments; however, the subnuclear colocalization is difficult to determine because dCBP is distributed ubiquitously within the nucleus. dCBP is not localized to the nucleolus, where Mod is predominantly located (Fig. 3 d–f). Although a band of colocalization can sometimes be seen at the periphery of the nucleolus, it is not possible to assess whether this is significant. To determine whether dCBP and Mod colocalized to specific loci in chromatin, we performed an immunohistochemical analysis of polytene chromosomes with antibodies directed against dCBP and Mod (Fig. 4). As shown in Fig. 4, there are some loci that bind only Mod, others that bind only dCBP and a large subset of bands that bind both dCBP and Mod. Both dCBP and Mod bind to heterochromatin however the Mod signal is considerably more intense (data not shown). The fact that dCBP and Mod coimmunoprecipitate from cell extracts and colocalize on polytene chromosomes reinforces the idea of a functional interaction between dCBP and Modulo during embryonic development.
Figure 3.
Laser confocal microscopy of Modulo and dCBP in the epidermis of stage 14 embryos. (a–f) Colocalization in wild-type embryos. Mod immunostaining with FITC (a and d), dCBP immunostaining with rhodamine (b and e), merged image from a and b (c), and merged image from d and e (f). Arrows in f indicate potential colocalization of Mod and dCBP at the periphery of the nucleolus. A 20× objective was used for a–c and a 60× objective with lasersharp to zoom 3× for d–f. (g and h) In green fluorescence, Mod immunostaining of L8 heterozygous embryos (g) or L8 homozygous embryos (h). The third chromosome balancer TM3, Sb Ser carrying a ftz-lacZ gene was used to distinguish the two populations of embryos, and the red stripes represent the β-gal staining of the balanced heterozygous animals (g). Anterior is to the right.
Figure 4.
Microscopy of wild-type polytene chromosomes. (A) Portion of chromosome arm showing localization of Mod. (B) Chromosome arm in A showing localization of dCBP. (C) Merged image of A and B. Large arrowhead indicates locus that binds Mod and not dCBP. Small arrowhead indicates band that binds dCBP and not Mod. Small arrows indicated loci that colocalize both dCBP and Mod, and are shown in yellow. Images were taken with a 40× objective.
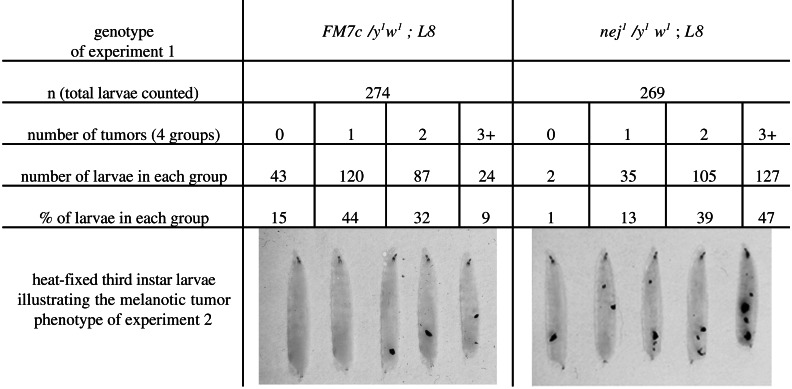
As third-instar larvae, L8 homozygous mutants develop melanotic tumors (10), which form as a result of an immune response to the presence of abnormal target tissues (28, 29). The melanotic tumor phenotype of L8 is characterized by incomplete penetrance and variable expressivity of the tumor phenotype (ref. 10 and our observations). The nej1 and nej3 heterozygous larvae never develop melanotic tumors. We determined whether the dosage of dCBP could affect the melanotic tumor phenotype of mod-deficient animals. In this experiment, y1w1; L8/TM6B,Tb males were crossed to females from a nej1/FM7c; L8/TM6B,Tb stock. This cross allowed us to compare nej1/y1w1; L8/L8 (black mouth hooks) to FM7c/y1w1; L8/L8 (yellow mouth hooks) third-instar females. For each population, a large number of third-instar larvae were analyzed and divided, by tumor number, into four different groups (Fig. 5). The nej1/y1w1; L8/L8 population has a greater number of tumors than the FM7c/y1w1; L8/L8 population. The statistical nonparametric test of Kolmogorov and Smirnov gave a χ2 = 113.34, P < 0.001 and shows that the L8/L8 population containing the nej allele are significantly different form the L8/L8 population. We observed the same results with the nej3 allele (data not shown). This result suggests that when associated with the chromatin-binding factor Modulo, dCBP can affect the immune response needed to suppress the formation of melanotic tumors in fly larvae.
Figure 5.
Phenotypic and statistical analysis of y1w1/FM7c; L8 and nej1/y1w1; L8 third-instar larvae.
In this report, we demonstrate a genetic interaction between dCBP and Modulo. While the genetic interactions suggest that Modulo and dCBP act in the same or parallel developmental pathways, the biochemical and colocalization data provide evidence supporting the idea that the two proteins interact directly. Interestingly, like mutations in mod (10), mutations in dCBP act as dominant suppressors of PEV (B. Newman and S.M.S., unpublished data). Thus, although the molecular mechanism of the interaction between dCBP and Modulo is not known, it probably involves the direct or indirect modification of chromatin structure.
Previous studies have shown that CBP interacts with components of the preinitiation complex and histone acetyltransferases. Our results indicate that CBP interacts with an additional class of chromatin-associated proteins as well and suggests that CBP effects on chromatin may not be limited to the level of the nucleosome and may also involve the modulation of higher-order chromatin structure.
Acknowledgments
We thank J. Pradel and L. Perrin for the L8-mod mutant, mod cDNA, and Mod LA9 monoclonal antibody; G. Golling for the yeast two-hybrid cDNA library; A. Snyder (Molecular Microbiology and Immunology Department, Oregon Health Sciences University, and the Oregon Hearing Research Center) for confocal analysis; and J. Broadbent for helping us with the statistical analysis. This work was partly supported by grants from the Association pour la Recherche contre le Cancer and the National Institutes of Health.
Abbreviations
- CBP
CREB-binding protein
- dCBP
Drosophila CBP
- β-gal
β-galactosidase
- GST
glutathione S-transferase
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Shikama N, Lyon J, Thangue N B L. Trends Cell Biol. 1997;7:230–236. [Google Scholar]
- 2.Akimaru H, Hou D-X, Ishii S. Nat Genet. 1998;17:211–214. doi: 10.1038/ng1097-211. [DOI] [PubMed] [Google Scholar]
- 3.Waltzer L, Bienz M. EMBO J. 1999;18:1630–1641. doi: 10.1093/emboj/18.6.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Goodman R H, Smolik S M. Mol Cell Biol. 2000;20:1616–1625. doi: 10.1128/mcb.20.5.1616-1625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogryzko V, Schiltz R, Russanova V, Howard B, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henikoff S. BioEssays. 1996;18:401–409. doi: 10.1002/bies.950180510. [DOI] [PubMed] [Google Scholar]
- 8.Wakimoto B T. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 9.Zhimulev I F. Adv Genet. 1998;37:422–434. doi: 10.1016/s0065-2660(08)60341-7. [DOI] [PubMed] [Google Scholar]
- 10.Garzino V, Pereira A, Laurenti P, Graba Y, Lewis R W, Parco Y L, Pradel J. EMBO J. 1992;11:4471–4479. doi: 10.1002/j.1460-2075.1992.tb05548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrin L, Demakova O, Fanti L, Kallenbach S, Saingery S, Mal'ceva N I, Pimpinelli S, Zhimulev I, Pradel J. J Cell Sci. 1998;111:2753–2761. doi: 10.1242/jcs.111.18.2753. [DOI] [PubMed] [Google Scholar]
- 12.Saurin A J, Shao Z, Erdjument-Bromage H, Tempst P, Kingston R E. Nature (London) 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- 13.Perrin L, Romby P, Laurenti P, Bérenger H, Kallenbach S, Bourbon H-M, Pradel J. J Biol Chem. 1999;274:6315–6323. doi: 10.1074/jbc.274.10.6315. [DOI] [PubMed] [Google Scholar]
- 14.Akimaru H, Chen Y, Dai P, Hou D-X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Nature (London) 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 15.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 16.Gietz D, Jean A S, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bantignies F, Rousset R, Desbois C, Jalinot P. Mol Cell Biol. 1996;16:2174–2182. doi: 10.1128/mcb.16.5.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 19.Krejci E, Garzino V, Mary C, Bennani N, Pradel J. Nucleic Acids Res. 1989;17:8101–8115. doi: 10.1093/nar/17.20.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 21.Lewis E B. Nature (London) 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 22.Rose R E, Gallaher N M, Andrew D J, Goodman R H, Smolik S M. Genetics. 1997;146:595–606. doi: 10.1093/genetics/146.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zink B, Paro R. Nature (London) 1989;337:468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]
- 24.Bantignies F, Goodman R H, Smolik S M. Mol Cell Biol. 2000;20:9317–9330. doi: 10.1128/mcb.20.24.9317-9330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel S. Nonparametric Statistics for the Behavioral Sciences. New York: McGraw-Hill; 1956. [Google Scholar]
- 26.Graba Y, Laurenti P, Perrin L, Aragnol D, Pradel J. Developmental Biology. 1994;166:704–715. doi: 10.1006/dbio.1994.1349. [DOI] [PubMed] [Google Scholar]
- 27.Hazelrigg T, Lewis R, Rubin G M. Cell. 1984;36:469–481. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- 28.Riski T M, Riski R M. In: Hemocytic and Humoral Immunity in Arthropods. Gupta A P, editor. New York: Wiley; 1986. pp. 157–190. [Google Scholar]
- 29.Watson K L, Johnson T K, Denell R E. Dev Genet. 1991;12:173–187. doi: 10.1002/dvg.1020120302. [DOI] [PubMed] [Google Scholar]