Abstract
The most impressive property of outer hair cells (OHCs) is their ability to change their length at high acoustic frequencies, thus providing the exquisite sensitivity and frequency-resolving capacity of the mammalian hearing organ. Prestin, a protein related to a sulfate/anion transport protein, recently has been identified and proposed as the OHC motor molecule. Homology searches of 1.5 kb of genomic DNA 5′ of the coding region of the prestin gene allowed the identification of a thyroid hormone (TH) response element (TRE) in the first intron upstream of the prestin ATG codon. PrestinTRE bound TH receptors as a monomer or presumptive heterodimer and mediated a triiodothyronine-dependent transactivation of a heterologous promotor in response to triiodothyronine receptors α and β. Retinoid X receptor-α had an additive effect. Expression of prestin mRNA and prestin protein was reduced strongly in the absence of TH. Although prestin protein typically was redistributed to the lateral membrane before the onset of hearing, an immature pattern of prestin protein distribution across the entire OHC membrane was noted in hypothyroid rats. The data suggest TH as a first transcriptional regulator of the motor protein prestin and as a direct or indirect modulator of subcellular prestin distribution.
Keywords: transcriptional regulation‖outer hair cell‖postnatal development‖ electromotility‖thyroid hormone receptor
Outer hair cells (OHCs) exhibit a unique motile phenotype that operates at acoustic frequencies (1). This electromotility is supposed to be based on voltage-dependent conformational changes of motor proteins that are densely packed in the lateral plasma membrane (2, 3). By using a cDNA library subtraction procedure, prestin was identified as a putative motor protein (4). In rats and gerbils, structural and functional maturation of the organ of Corti (5, 6) and appearance of OHC electromotility (7) occurred within the first 2 weeks after birth parallel to the onset of prestin expression (3, 8). Studying the transcriptional regulation of this gene as well as its up- and downstream cascades may reveal new insights into the causes of congenital or acquired hearing impairments and deafness. Analysis of the 5′-flanking region of the rat prestin gene as well as the analysis of prestin mRNA and protein expression in the absence and presence of thyroid hormone (TH) performed in this study identified TH as a first critical determinant for the regulation of prestin.
Materials and Methods
Animals and Drug Administration.
Wistar rats were used in this study. The antithyroid drug (1-methyl-2-mercaptoimidazole, 0.02%), described to safely suppress plasma TH levels in both animals and humans (9, 10), was administered in the drinking water of dams before estrus day 17 after conception. This administration ensures that TH levels were suppressed to <0.5 μg/dl l-thyroxine (T4) and <15 ng/dl triiodothyronine (T3) from the onset of fetal thyroid gland function onward (11, 12), which in rats takes place on estrus day 17 or 18. The treatment was continued after birth until the animal was killed. The control of hypothyroidism was measured after T4 or T3 determination in postnatal pups as described (11). Hypothyroidism was counteracted after s.c. injection of the animals with 0.3 μg/g body weight T4 (Sigma) at indicated ages. Animal experiments followed the approved institutional protocols at the University of Tübingen.
Tissue Preparation.
Cochleae of postnatal untreated (control) and methylmercaptoimidazole-treated (hypothyroid) animals were isolated and dissected as described (12). Cochleae were fixed, decalcified, and cryosectioned as described (11). For Northern blot, tissues were frozen immediately in liquid nitrogen and stored at −70°C before use.
Cloning and Sequencing of a Rat Prestin Homologue and the 5′-Flanking Region.
mRNA from rat cochleae of different ages was isolated by using the Dynabeads mRNA Direct kit (Dynal, Great Neck, NY) and reverse-transcribed with SuperScript II reverse transcriptase (GIBCO). By using degenerated oligonucleotide primers deduced from the original Meriones prestin sequence (GenBank accession no. AF230376) and a human baculovirus clone (GenBank accession no. RG107G13), a 282-bp fragment was amplified with rat cochlea cDNA as template. Another 723-bp clone was derived from the rat prestin cDNA sequence (GenBank accession no. AJ303372). We used prestin gene-specific PCR primers (PreGSP1, 5′-GAGTCTGTGACTTTGTCCTTGACTTGCAGC-3′, and PreGSP2, 5′-CCTGGAGGACCGGATGACTGAAGATAGG-3′) deduced from the 282-bp sequence together with adapter primers (GenomeWalker Kit, CLONTECH) to clone a stretch of the 5′-flanking region of the prestin gene as recommended by the manufacturers. 5′-Rapid amplification of cDNA ends PCR (P12) was performed by using the GeneRacer kit (Invitrogen) and prestin gene-specific primers under conditions recommended by the manufacturers.
Riboprobe Synthesis and in Situ Hybridization.
Prestin cDNA cloned into pCRII TOPO vector (Invitrogen) was used for in vitro transcription. Complementary strands for sense and antisense were transcribed from either SP6 or T7 promoter sites in the presence of digoxigenin-labeling mix (Roche Diagnostics, Mannheim, Germany). In situ hybridization was performed as described (11–13).
Preparation of Prestin Antiserum.
To produce antibodies against prestin, two rabbits were immunized (Charles River Breeding Laboratories) with synthetic peptides corresponding to the C-terminal domain (amino acids 725–745, TVLPPQEDMEPNATPTTPEA, GenBank accession no. AJ303372) of rat prestin. The prestin antiserum was validated by the successful detection of the recombinant protein expressed in Chinese hamster ovary cells (data not shown). The antiserum labeled exclusively OHCs in the rodent organ of Corti and did not crossreact with brain tissue. No immunofluorescence labeling of the OHCs was seen when the primary antibody was omitted (data not shown).
Immunohistochemical Staining for Fluorescence and Laser Scanning Confocal Microscopy.
For immunohistochemistry, rat cochlear sections from control or hypothyroid rats were stained as described (11). Laser scanning confocal microscopy was performed as described (13). Prestin antibody was diluted 1:5,000 and synaptophysin antibody (The Binding Site, Birmingham, U.K., PH510) 1:500. Primary antibodies were visualized with either Cy3-conjugated goat anti-rabbit Ig (Jackson IRL) or Alexa Green-conjugated goat anti-mouse antibodies (Molecular Probes).
Electrophoretic Mobility-Shift Assay (EMSAs).
Protein–DNA complexes were formed in 10–15-μl reaction mixtures containing either Roche or Promega binding buffer. Reaction mixtures contained ≈10 μg of HeLa cell nuclear extract protein (Promega) or 1.5 μg of recombinant protein cTRα (Santa Cruz Biotechnology), depending on the assay. Nuclear extracts from cochleae were prepared as described (12, 14). Radiolabeled components were detected by autoradiography.
Oligonucleotides and Labeling.
In EMSAs, the double-stranded synthetic oligonucleotides used were: TH response element (TRE) (TREPrest), 5′-ACGTGAGGTCAGTTTAAGGACAATCTTA-3′; TREmutA, 5′-ACGTGAGAACAGTTTAAGGACAATCTTA-3′; TREmutB, 5′-ACGTGAGGTCAGTTCGCAGACAATCTTA-3′; TREmutC, 5′-ACGTGGTTCGCAGACAATCTTA-3′; TREmutD, 5′-ACGTGGTTTAAGGACAATCTTA-3′; TREmutE, 5′-ACGTGAGGTCAGTTATCTTA-3′; T3 receptor (TR) (DR-4), 5′-AGCTTCAGGTCACAGGAGGTCAGAGAG-3′ (Santa Cruz Biotechnology); transcription factor IID, 5′-GCAGAGCATATAAGGTGAGGTAGGA-3′ (Promega); and Oct-2A, 5′-GTACGGAGTATCCAGCTCCGTAGCATGCAAATCCTGG-3′ (Roche Diagnostics). Oligonucleotides (3.85 pmol) were labeled with [γ-32P]ATP [3,000 Ci/mmol (1 Ci = 37 GBq); NEN Life Science Products] by phosphorylation with T4 polynucleotide kinase (Promega).
DNA Construction.
The TREPrest oligonucleotide was cloned into the multiple cloning site of the pGL3-promoter vector (Promega) after the addition of a KpnI site (CCATGG) at the 5′ end and an XhoI site (GAGCTC) at the 3′ end of TREPrest (TREPrest-pGL3-promoter). The cDNA sequences of human retinoid X receptor (RXR)-α (GenBank accession no. XM011778), rat TRα (GenBank accession no. X12744), and rat TRβ (GenBank accession no. J03819) were subcloned into the multiple cloning site of the pTargeT mammalian expression vector (Promega). The sequence and correct orientation of each insert was confirmed by sequence analysis.
Cell Culture and Transfections.
HEK293 cells were grown in DMEM (GIBCO) supplemented with 10% FCS. Cells were plated at a density of 106 cells per 6-cm dish on the day before transfection. Transfection was performed by using the PolyFect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Each 6-cm dish received 500 ng of the reporter construct (TREPrest-pGL3-promoter), 500 ng of the internal control plasmid RSV-LacZ, and 800 ng of the appropriate expression vector. Twelve hours after DNA addition the transfection was stopped, and the cells were incubated for another 48 h in fresh medium containing serum depleted of T3 and retinoic acid (15) before harvesting. T3 (Sigma) or all-trans-retinoic acid (Sigma) were added to a final concentration of 200 nM and 1.5 μM, respectively, as indicated. Cell lysates were prepared by using the luciferase assay system (Promega). The protein concentration of each lysate was measured by using Bradford reagent (Sigma), and the transfection efficiency was determined by using the β-galactosidase enzyme assay system (Promega). Receptor expression was verified on Western blot (12) by using primary antibodies to TRα1, TRβ1, and RXRα (Santa Cruz Biotechnology SC-772, SC-738, and SC-774, respectively). Luciferase activity was measured by using an Orion microplate luminometer (Berthold Detection Systems, Pforzheim, Germany) and corrected for transfection efficiency and protein concentration with their respective values.
Northern Blot Analysis.
Northern blot analysis was performed as described (12, 16). The effect of TH on mRNA levels was evaluated and semiquantified by using mRNAs isolated from a similar number of cochleae as described (16).
Results
Prestin Localization.
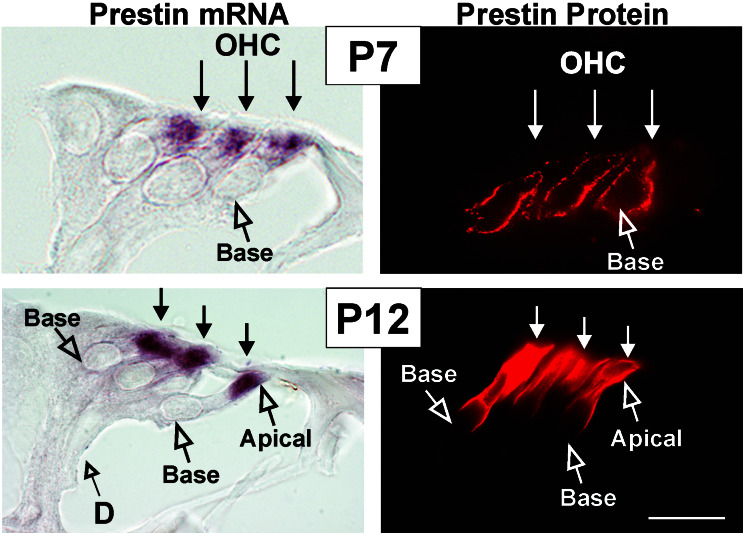
We used the 723-bp cDNA clone to synthesize digoxigenin-labeled riboprobes and localized prestin mRNA and protein in cross sections of P7 (before the onset of hearing in rats) and P12 (onset of hearing in rats; Fig. 1). From P2 onward, prestin mRNA was restricted to the apical cell pole of the OHC, shown for the midbasal turn at P7 and P12 in Fig. 1. No mRNA signals were detected with the corresponding sense riboprobes (data not shown). In agreement with previously published data (8), antiprestin antibodies labeled the lateral plasma membrane of the OHCs. During the first days of expression, prestin protein was distributed across the entire OHC membrane (shown for P7, Fig. 1). Between P7 and P12, prestin protein became redistributed to the lateral region of the OHC membrane (shown for P12, Fig. 1). The prestin protein-free basal region of the OHC membrane declined from the basal to the apical cochlear turn (data not shown).
Figure 1.
Localization of prestin mRNA and protein in cross sections of rat cochleae at P7 and P12 using in situ hybridization and immunohistochemistry. Note the restricted localization of mRNA to the apical part of OHC, whereas prestin protein becomes redistributed from the entire OHC membrane (P7) to the lateral plasma membrane (P12). D, Deiters cell. (Bar, 20 μm.)
Identification of TRE.
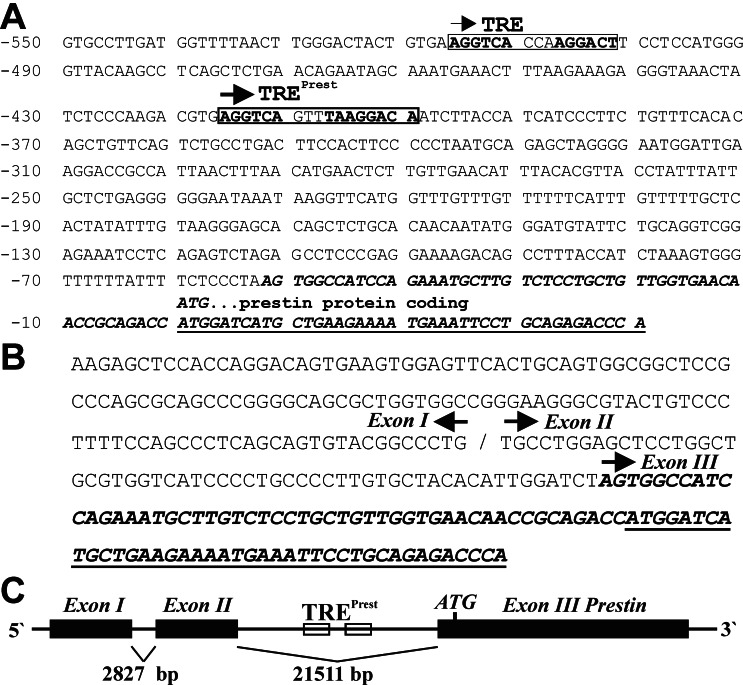
Rat genomic DNA (1.5 kb) upstream of the putative translation initiation codon (ATG) of the prestin gene was cloned and sequenced by using linker PCR. By using MATINSPECTOR software (version 2.2, http://transfac.gbf.de/TRANSFAC), we identified a putative response element (TREPrest) for nuclear receptors starting at position −416 (AGGTCAGTTTAAGGACA) relative to the ATG codon (Fig. 2A), which has a human prestin gene homologue in approximately the same position. Another presumptive TRE was noted at position −516. We focused on the TREPrest, which contains two hexamers that are almost identical to the consensus sequence AGGTCA (Fig. 2A). The hexamers are organized in a direct repeat configuration (6DR), separated by a 5-bp spacer (6DR5; Fig. 2A). According to the 3-4-5 rule, this configuration is expected to be a retinoid acid response element recognized by RXR and retinoic acid receptor (17). Taking two nucleotides at the 5′-flanking region of the downstream hexamer into consideration, this configuration can be interpreted also as an octamer (TA AGGACA) that is separated by three nucleotides from an upstream hexamer (AGGTCA) (6/8DR3). Such motifs have been described as optimal binding sites for TRs (TRE) with no spacing limitations as implied by the 3-4-5 rule (18). To determine the relative position of TREPrest we identified the transcriptional start site of the prestin gene performing a 5′ rapid amplification of cDNA ends PCR with mRNA isolated from P12 rat cochlea. We did not observe any similarity between the 5′ end of the obtained cDNA and the genomic rat DNA upstream of the ATG codon over a stretch of 188 bp (shown in Fig. 2B) of the cDNA. Similarity searches performed with the ENSEMBL human genome server detected two similar fragments on chromosome 7 that matched the missing 188 bp. This sequence covers a presumptive first and second exon, which are separated from each other by 2,827 bp and from a third exon by 21,511 bp. Considering this human prestin sequence in light of the rat sequence information, data are consistent with the presence of two untranslated exons (exons I and II) upstream of the exon containing the ATG codon (exon III). Thus, rat TREPrest probably is located 52 bp upstream of the boundary between the second intron and third exon (Fig. 2C) and therefore constitutes an element downstream of the transcriptional start site.
Figure 2.
Position of TREPrest within the gene. (A) Nucleotide sequence of the rat prestin gene containing the first coding exon (in italics). The protein-coding region is underlined. The putative translation initiation codon (ATG) is referred to as +1. A TRE at position −416, referred to as TREPrest, is boxed. Another putative TRE (6DR3) is located at position −516. (B) 5′ rapid amplification of cDNA ends PCR generated from P12 cochlear mRNA detected a 188-bp sequence (indicated as putative exons I and II), which does not match the exon containing the protein-coding region (exon III). (C) Based on homologous human genomic sequence, a model was designed proposing the position of TREPrest within the exon-intron map of the prestin gene. Note its position downstream of exons I and II and upstream of exon III.
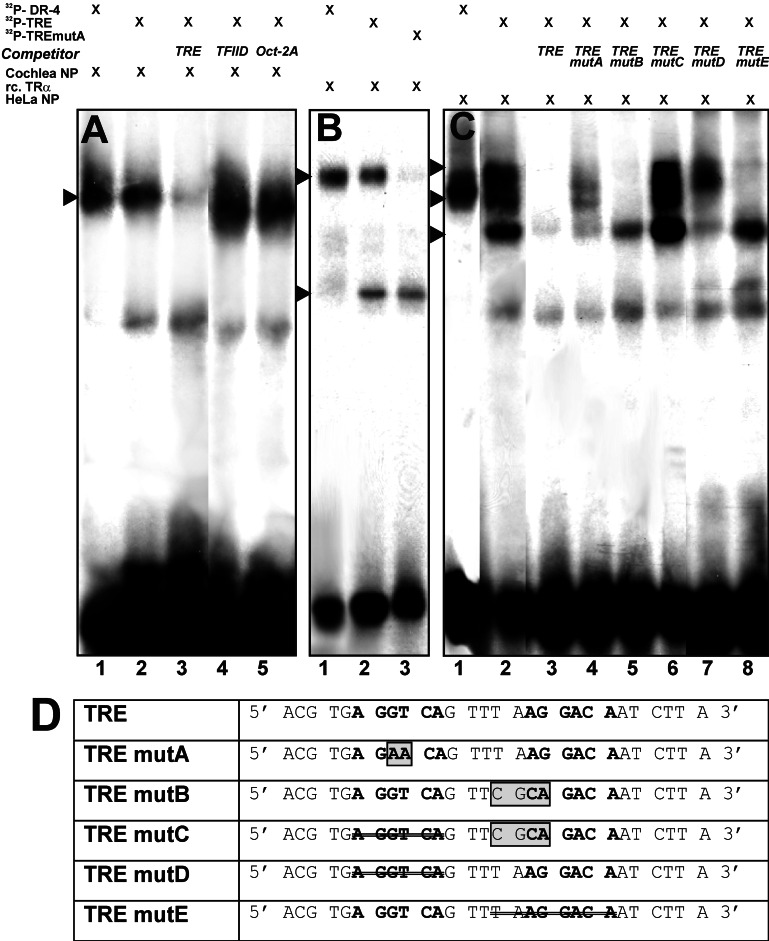
We performed EMSA to study the protein binding properties of TREPrest. In cochlear extracts polypeptides readily shifted 32P-labeled TREPrest oligomers (Fig. 3A, lane 2) to a position similar to that of a 32P-labeled control oligomer (DR-4; Fig. 3A, lane 1), which typically binds TR (17). To test further whether this interaction could be attributed to TR we performed EMSA with a commercially available recombinant TRα (Fig. 3B). [32P]TREPrest (Fig. 3B, lane 2) and control oligomer ([32P]DR-4, Fig. 3B, lane 1) both were shifted by TRα to a higher homodimer band, and in addition [32P]TREPrest was shifted by TRα to a lower presumptive monomer band. To further specify TREPrest we used a HeLa cell extract known to contain TR as well as putative interaction partners, e.g., RXR and retinoic acid receptor. The HeLa cell extract shifted the [32P]TREPrest to three different positions (Fig. 3C, lane 2, three arrowheads), which were interpreted as monomer-, homodimer-, and heterodimer-receptor interactions (19). The monomer binding position equaled that shifted by recombinant TRα, as verified by competition EMSA using mutated versions of TREPrest (data not shown). The shift of [32P]TREPrest with cochlear extract (Fig. 3A, lane 3), HeLa cell extract (Fig. 3C, lane 3), and TRα (data not shown) no longer was observed when an unlabeled double-stranded oligomer containing TREPrest was added as a specific competitor. Other classic regulator binding motifs specific for transcription factor IID or Oct-2A did not compete with the shift of [32P]TREPrest, shown for the cochlear extract (Fig 3A, lanes 4 and 5). Unlabeled double-stranded oligonucleotides containing various mutations within TREPrest were used as competitors for authentic [32P]TREPrest shifted by the HeLa cell extract (Fig. 3C, lanes 4–8). The sequences of the mutated oligomers, named TREmutA–E, are shown in Fig. 3D. Nucleotide exchanges or deletions introduced in the 5′ half-site (hexamer) of TREPrest either significantly (Fig. 3C, lane 4, mutA) or completely (Fig. 3B, lane 7, mutD) reduced the capacity to act as competitor for binding presumptive homodimer–heterodimer complexes but left the capacity for binding monomers nearly intact. No putative dimeric complex was observed with TRα when mutations were introduced in the 5′ half-site of [32P]TREPrest; however, the monomer interaction was not affected (Fig. 3B, lane 3, mutA). In contrast, mutations (Fig. 3C, lane 5, mutB) or deletions (Fig. 3C, lane 8, mutE) in the 3′ octamer of TREPrest did not alter its properties in the competition assays with the homodimer–heterodimer complex but suppressed its ability to act as an efficient competitor for binding TR monomers. When the complete 5′ hexamer of TREPrest was deleted in addition to the exchange of four (Fig. 3C, lane 6, mutC) or two (data not shown) upstream nucleotides within the 3′ octamer, TREPrest sequences totally lost the capacity to compete with monomer and dimer complex binding of [32P]TREPrest. To summarize, the binding characteristics support the notion of a preferential role of the 3′ octamer for monomer binding and necessity of the 5′ hexamer for homodimer–heterodimer binding.
Figure 3.
DNA binding of TRs to TREPrest analyzed by EMSA. (A) Cell extract from the cochlea at P8 shifts 32P-labeled TREPrest oligomers (lane 2) to a similar position to that of control [32P]DR-4 (lane 1). The interaction is reduced in the presence of an excess of unlabeled TREPrest competitor oligomers (lane 3) but not in the presence of unlabeled oligomers specific for transcription factor IID or Oct-2A (lanes 4 and 5, respectively ). (B) Recombinant (rc.) TRα shifts [32P]DR-4, [32P]TREPrest, and [32P]TREmutA to only one or both positions consistent with monomer and homodimer binding, respectively. (C) HeLa cell extract shifts [32P]TREPrest to positions that resembled monomer, homodimer, and heterodimer receptor interaction (lane 2, three arrowheads); the presumptive heterodimeric interaction is similar to that observed with [32P]DR-4 (lane 1). An excess of unlabeled TREPrest reduces the bands in lane 3. Modifications introduced in the first or second half-site of TREPrest influence either monomer or homodimer and heterodimer binding in competition assays (see Results). A schematic representation of the modified oligomers used as unlabeled competitors is given in D.
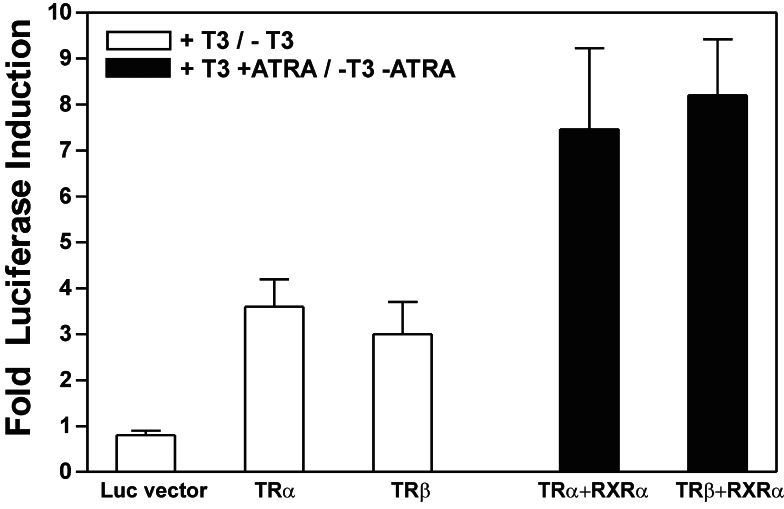
TREPrest Shows T3-Dependent Transactivation of a Reporter Gene in Transient Transfections.
A series of transfections was performed in HEK293 cells to examine the T3 responsiveness of a TREPrest luciferase reporter plasmid carrying the simian virus 40 promoter, which by itself showed no T3 responsiveness (Fig. 4, Luc vector). The average amount of total protein and the expression of a cotransfected RSV-LacZ control plasmid was unaffected by the depletion of T3 and all-trans-retinoic acid. Luciferase expression was induced by transfection of TRα and TRβ. In addition, we used RXRα as one of the most important heterodimer partners of TR (20) to exemplarily investigate the role of heterodimers for transactivation. The expression of TRα, TRβ, or RXRα after transfection of HEK293 cells was verified by Western blotting (data not shown). Fig. 4 illustrates an observed 3.6-fold TRα-T3-dependent and 3-fold TRβ-T3-dependent increase in reporter gene activation. RXRα and all-trans-retinoic acid enhanced the T3-induced transactivation to 7.5-fold for TRα and to 8.2-fold for TRβ. Cotransfection of TRα or TRβ with RXRα thus had an additive effect on T3-dependent reporter gene activation.
Figure 4.
TREPrest confers T3 responsiveness to a heterologous promoter. TREPrest was inserted upstream of the simian virus 40 promoter of a luciferase reporter plasmid, which in itself was not T3-responsive. HEK293 cells were cotransfected with this reporter plasmid together with TRα, TRβ, or RXRα expression vectors. Luciferase activity was determined after 48 h of incubation in the presence or absence of T3 and T3 + all-trans-retinoic acid (ATRA), respectively. Luciferase values were corrected for transfection efficiency and variations in total protein concentration. The data are the mean ± SD of luciferase activity obtained from at least three independent transfections, each measured in triplicate.
Effect of TH on Prestin Expression and Prestin Distribution.
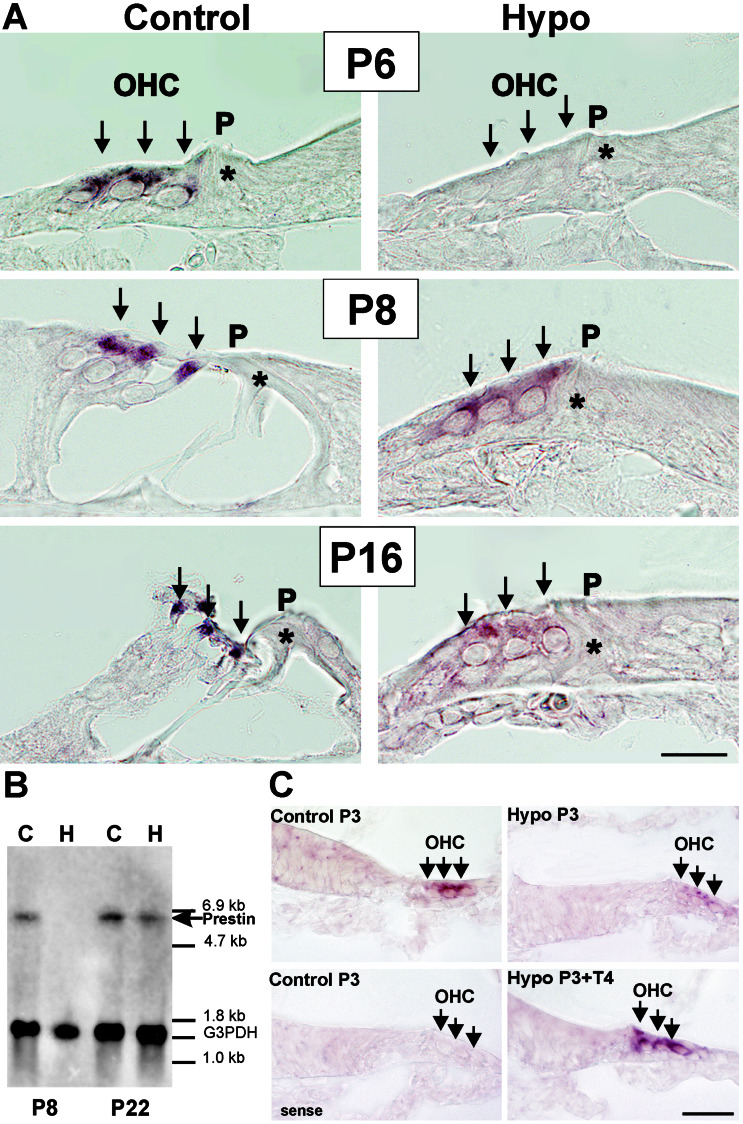
The presence of an active TRE within the prestin gene would suggest an influence of TH on gene expression. When we analyzed prestin mRNA in the presence (control) and absence (hypo) of TH by using in situ hybridization (Fig. 5A), a severe reduction of mRNA was noted in the absence of TH at P6, P8, and P16 (Fig. 5A, Hypo) in comparison to controls. By using the Northern blot technique, we furthermore noted a significant reduction of the 6.4-kb prestin mRNA in the absence of TH at P8 (Fig. 5B, P8), whereas toward the third/fourth postnatal week (Fig. 5B, P22) mRNA gradually approached control levels. The blots were probed with glyceraldehyde-3-phosphate dehydrogenase to ensure the loading of similar amounts of mRNA per lane. When hypothyroid rats were injected with T4 on P3 and killed 12 h later, the suppression of prestin mRNA, typically noted in the absence of TH (Fig. 5C, Hypo P3) in comparison to the control (Fig. 5C, Control P3), was overcome (Fig. 5C, Hypo P3+T4). No hybridization signals were detected when corresponding sense probes were used (Fig. 5C, sense).
Figure 5.
Expression of prestin mRNA in rat cochleae using in situ hybridization (A and C) and Northern blot (B). (A) At P6, P8, and P16, in hypothyroid animals (Hypo) prestin mRNA is reduced severely in comparison to controls (Control). (B) At P8, the 6.4-kb prestin mRNA band is reduced significantly in the absence of TH, whereas mRNA approaches nearly control levels toward P22. Blots were probed with glyceraldehyde-3-phosphate dehydrogenase to ensure similar loading of mRNA per lane. (C) Expression of prestin mRNA in rat cochleae at P3 in the presence (Control P3) and absence (Hypo P3) of TH. Twelve hours after injecting hypothyroid rats with T4, prestin mRNA is up-regulated (Hypo P3+T4). No mRNA was detected when correspondent sense probes were used (Control P3, sense). P, Pillar cell. (Bar, 20 μm.)
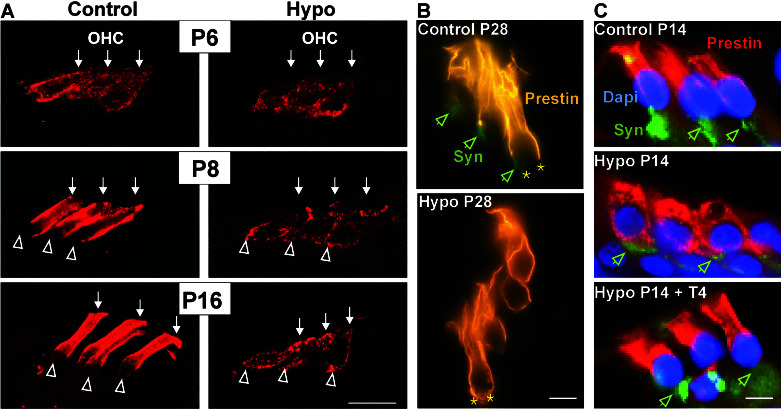
Significant reduction in prestin protein concentration also was observed when cochleae of same-aged animals were analyzed by using laser confocal microscopy (Fig. 6A). It was striking to note that from ≈P8 onward, prestin protein was concentrated in the lateral region of the membrane in the more basal cochlear turns. No labeling of the supranuclear region was observed in the OHC base at higher ages (Fig. 6A, Control P8, P16, open arrows), whereas the protein persisted in an immature distribution along the entire membrane in hypothyroid animals (Fig. 6A, Hypo P8, P16, open arrows). When prestin protein distribution was analyzed immunohistochemically in the cochleae of hypothyroid animals at P28 using double labeling of antiprestin and antisynaptophysin, an immature prestin protein distribution across the entire OHC membrane still was noted in the absence of TH (Fig. 6B, Hypo P28). This distribution occurs in conjunction with a decline of synaptophysin-positive nerve endings, a typical feature of OHC maturing under hypothyroid conditions (21). The aberrant distribution of prestin protein in the absence of TH was analyzed in animals at P14 after parallel staining of prestin (red), cell nuclei with 4′,6-diamidino-2-phenylindole (blue), and synaptophysin (green; Fig. 6C). A complete recovery of the normal prestin distribution was observed after injection of T4 every second day from P4 onward until P14 (Fig. 6C, Hypo P14+T4).
Figure 6.
Prestin protein expression and distribution in rat cochleae using laser confocal microscopy (A) and immunohistochemistry (B and C). (A) In hypothyroid animals (Hypo) a significant reduction of prestin protein is observed. Note the aberrant prestin distribution in hypothyroid P8 and P16 sections (open arrows). (Bar, 20 μm.) (B) Cochlea sections of control and hypothyroid animals at P28 were costained for prestin and synaptophysin (Syn). Note the immature prestin distribution across the entire OHC membrane including the synaptic pole of the cell (Hypo P28, asterisks). The synaptophysin expression in hypothyroid animals is reduced in comparison to controls (open arrows). (Bar, 10 μm.) (C) Prestin protein distribution in control and hypothyroid animals is compared with hypothyroid animals treated with T4 after parallel staining of prestin (red), 4′,6-diamidino-2-phenylindole (DAPI)-stained cell nuclei (blue), and synaptophysin (green) at P14. (Bar, 10 μm.)
Discussion
The data in the present study suggest TH as a first transcriptional regulator of the prestin gene and point at an as yet undiscovered developmental redistribution of prestin toward the lateral membrane under the control of TH. A TRE was detected in the rat prestin gene upstream of the translation initiation codon (ATG). Rapid amplification of cDNA ends PCR with rat mRNA and similarity searches within the human prestin gene suggested the presence of two untranslated exons upstream of the exon containing the ATG codon. A similar prestin gene configuration was suggested recently for mice.§ Thus, the rat TREPrest is expected to be localized in the second intron, in which an additional putative TRE was localized further upstream. Because multiple TREs are known to act in concert (22), we cannot exclude that others than the described TRE at position −416 are functional in vivo. Interestingly, additional putative TREs are located also within the first 600 bp of the homologous intron of the human prestin gene. Although most of the reported TREs exert a positive or negative effect at various positions upstream of the transcriptional start site (23–26), the number of TREs found in introns currently is increasing. Examples have been described for RGH (27), NCAM (28), Pep2 (29), NRGN (30), and CPT-1 (31).
The TREPrest detected in the prestin gene contains two half-sites that have been reported as having been recognized by TR and related receptors of the steroid hormone receptor family (20, 32). These half-sites are known to bind TR as monomeric or homodimeric and heterodimeric complexes with other proteins such as RXR and retinoic acid receptor (20). Although supershift studies will be required to finally prove the identity of homodimers and heterodimers binding to the distinct TREPrest in the present study, the characterization of TREPrest using EMSA competition assays of wild-type and mutated sequence motifs suggested the high priority of the 3′ octamer sequence for TR when binding as a monomer. Comparable monomer binding properties have been reported recently (22, 33). Furthermore, the integrity of the 5′ hexamer motif of TREPrest was shown to be a prerequisite for presumptive homodimer or heterodimer receptor binding.
In transactivation studies, TREPrest showed T3 responsiveness with either TRα or TRβ, supporting the notion of a potential monomeric transactivation by TRα and TRβ. Gene activation through an octamer element has been described to be initiated by either monomeric TR independently of RXR (22) or heterodimeric TR complexes with human RXRα (34). In this context, the observation that cotransfection of TR and RXR increased reporter gene activity may point to a transactivation mechanism analogous to that described in ref. 34. Both myelin basic protein (MBP) gene expression (16) and prestin gene expression (present study) are up-regulated during postnatal periods, in parallel to the rise of TH plasma levels during a peak of TRβ expression. TRα, known to be present before and during postnatal periods in rodents (12, 35, 36), may occupy TREPrest binding sites in OHC before prestin expression, similar to what has been suggested for the MBP gene (34). The increase of T3 during postnatal periods in parallel to a subsequent up-regulation of TRβ expression (12, 36) then might cause a dissociation of TRα and a replacement by TRβ because of its higher concentration. According to MBP gene regulation, a strong TRα monomer binding was correlated inversely with the transactivation by TRβ/RXR heterodimers, the latter inducing a higher transactivation potential in comparison to TRα receptors alone (34). Considering the similarities between MBP and prestin, we may suggest a T3-dependent transactivation of the prestin gene caused by TRβ with either RXR or an as yet uncharacterized heterodimer partner. This suggestion also would be in line with the observed fact that nuclear pellet extracts of the postnatal cochlea shifted TREPres to a presumptive heterodimer position. Considering that TRβ mutant mice are deaf despite normal cochlear morphology (36), it may be challenging to analyze whether in addition to obvious changes in K+ channel properties (37) prestin expression may be altered in TRβ or TRβ/TRα double mutant mice.
Although examinations in the present study clearly show that the levels of prestin mRNA and protein were reduced severely in the absence of TH during the first postnatal week, prestin gene expression eventually achieved the levels in controls at the end of the fourth postnatal week as do other TH-regulated genes (16, 25). This phenomenon might be attributed either to transcription factors (25) or local compensation of T3 levels by type 2 deiodinase (38). The absence of TH for more than 3 weeks, however, still was associated with an immature prestin protein distribution across the entire plasma membrane. It remains to be clarified whether prestin redistribution to the lateral region of the membrane is guided primarily by scaffolding of prestin protein itself or secondarily by scaffolding of other membrane proteins. Such membrane proteins may be ion channels, e.g., KCNQ4 potassium channel proteins, which gradually occupy the basal part of OHCs (39). Thus, a striking question in a hypothyroid in vivo model may be to analyze the role of compactness of prestin protein in OHC for motile function.
Acknowledgments
We thank M. Meyer-Ficca, R. Meyer, and A. Rodriguez-Peña for helpful discussions and assistance during the reporter gene assays. The RSV- LacZ control plasmid and an RXR-encoding plasmid were gifts from M. Meyer-Ficca. We also thank R. J. Koenig and M. A. Lazar for the gift of TRα- and TRβ-encoding plasmids. This work was supported by Federal Ministry of Education and Research Grant 01KS9602, a grant from the Interdisciplinary Center of Clinical Research Tübingen, and Deutsche Forschungsgemeinschaft Sonderforschungsbereich Grant 430/Kni-B3.
Abbreviations
- OHC
outer hair cell
- TH
thyroid hormone
- T4
l-thyroxine
- T3
triiodothyronine
- P
postnatal day
- EMSA
electrophoretic mobility-shift assay
- TR
T3 receptor
- TRE
TH response element
- RXR
retinoid X receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ428404).
Long, K. Zheng, J., Dallos, P. & Madison, L. D. (2001) J. Assoc. Res. Otolaryngol. 158, 561 (abstr.).
References
- 1.Dallos P, Evans B N. Science. 1995;267:2006–2009. doi: 10.1126/science.7701325. [DOI] [PubMed] [Google Scholar]
- 2.Kalinec F, Holley M C, Iwasa K H, Lim D J, Kachar B. Proc Natl Acad Sci USA. 1992;89:8671–8675. doi: 10.1073/pnas.89.18.8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver D, Fakler B. J Physiol. 1999;519:791–800. doi: 10.1111/j.1469-7793.1999.0791n.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng J, Shen W, He D Z, Long K B, Madison L D, Dallos P. Nature (London) 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 5.Pujol R, Carlier E, Lenoir M. Hear Res. 1980;2:423–430. doi: 10.1016/0378-5955(80)90078-7. [DOI] [PubMed] [Google Scholar]
- 6.Rubel E W, Lippe W R, Ryals B M. Ann Otol Rhinol Laryngol. 1984;93:609–615. doi: 10.1177/000348948409300614. [DOI] [PubMed] [Google Scholar]
- 7.He D Z, Evans B N, Dallos P. Hear Res. 1994;78:77–90. doi: 10.1016/0378-5955(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 8.Belyantseva I A, Adler H J, Curi R, Frolenkov G I, Kachar B. J Neurosci. 2000;20:RC116. doi: 10.1523/JNEUROSCI.20-24-j0002.2000. :1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becks G P, Burrow G N. Med Clin N Am. 1991;75:121–150. doi: 10.1016/s0025-7125(16)30475-8. [DOI] [PubMed] [Google Scholar]
- 10.Lind P. Acta Med Austriaca. 1997;24:157–158. [PubMed] [Google Scholar]
- 11.Knipper M, Zinn C, Maier H, Praetorius M, Rohbock K, Köpschall I, Zimmermann U. J Neurophysiol. 2000;83:3101–3112. doi: 10.1152/jn.2000.83.5.3101. [DOI] [PubMed] [Google Scholar]
- 12.Knipper M, Gestwa L, Ten Cate W J, Lautermann J, Brugger H, Maier H, Zimmermann U, Rohbock K, Köpschall I, Wiechers B, Zenner H P. J Neurobiol. 1999;38:338–356. [PubMed] [Google Scholar]
- 13.Wiechers B, Gestwa G, Mack A, Carroll P, Zenner H P, Knipper M. J Neurosci. 1999;19:3033–3042. doi: 10.1523/JNEUROSCI.19-08-03033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz H L, Strait K A, Ling N C, Oppenheimer J H. J Biol Chem. 1992;267:11794–11799. [PubMed] [Google Scholar]
- 15.Pombo P M, Barettino D, Espliguero G, Metsis M, Iglesias T, Rodriguez-Peña A. J Biol Chem. 2000;275:37510–37517. doi: 10.1074/jbc.M006440200. [DOI] [PubMed] [Google Scholar]
- 16.Knipper M, Bandtlow C, Gestwa L, Köpschall I, Rohbock K, Wiechers B, Zenner H P, Zimmermann U. Development (Cambridge, UK) 1998;125:3709–3718. doi: 10.1242/dev.125.18.3709. [DOI] [PubMed] [Google Scholar]
- 17.Umesono K, Murakami K K, Thompson C C, Evans R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz R W, Koenig R J. J Biol Chem. 1993;268:19392–19397. [PubMed] [Google Scholar]
- 19.Demczuk S, Harbers M, Vennström B. Proc Natl Acad Sci USA. 1993;90:2574–2578. doi: 10.1073/pnas.90.7.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Lazar M A. Annu Rev Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 21.Uziel A, Pujol R, Legrand C, Legrand J. Brain Dev. 1983;283:295–301. doi: 10.1016/0165-3806(83)90186-4. [DOI] [PubMed] [Google Scholar]
- 22.Katz R W, Subauste J S, Koenig R J. J Biol Chem. 1995;270:5238–5242. doi: 10.1074/jbc.270.10.5238. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y T, Nadal-Ginard B. Mol Cell Biol. 1989;1:1839–1849. doi: 10.1128/mcb.9.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madison L D, Ahlquist J A, Rogers S D, Jameson J L. Mol Cell Endocrinol. 1993;94:129–136. doi: 10.1016/0303-7207(93)90060-w. [DOI] [PubMed] [Google Scholar]
- 25.Pombo P M, Barettino D, Ibarrola N, Vega S, Rodriguez-Peña A. Brain Res Mol Brain Res. 1999;64:92–100. doi: 10.1016/s0169-328x(98)00311-8. [DOI] [PubMed] [Google Scholar]
- 26.Lin K H, Shieh H Y, Hsu H C. Endocrinology. 2000;141:2540–2547. doi: 10.1210/endo.141.7.7570. [DOI] [PubMed] [Google Scholar]
- 27.Sap J, de Magistris L, Stunnenberg H, Vennström B. EMBO J. 1990;9:887–896. doi: 10.1002/j.1460-2075.1990.tb08186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iglesias T, Caubin J, Stunnenberg H G, Zaballos A, Bernal J, Munoz A. EMBO J. 1996;15:4307–4316. [PMC free article] [PubMed] [Google Scholar]
- 29.Hagen S G, Larson R J, Strait K A, Oppenheimer J H. J Mol Neurosci. 1996;7:245–255. doi: 10.1007/BF02737062. [DOI] [PubMed] [Google Scholar]
- 30.de Arrieta C M, Morte B, Coloma A, Bernal J. Endocrinology. 1999;140:335–343. doi: 10.1210/endo.140.1.6461. [DOI] [PubMed] [Google Scholar]
- 31.Jansen M S, Cook G A, Song S, Park E A. J Biol Chem. 2000;275:34989–34997. doi: 10.1074/jbc.M001752200. [DOI] [PubMed] [Google Scholar]
- 32.Desvergne B, Wahli W. Endocr Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 33.Katz R W, Koenig R J. J Biol Chem. 1994;269:18915–18920. [PubMed] [Google Scholar]
- 34.Jeannin E, Robyr D, Desvergne B. J Biol Chem. 1998;273:24239–24248. doi: 10.1074/jbc.273.37.24239. [DOI] [PubMed] [Google Scholar]
- 35.Bradley D J, Towle H C, Young W S. Proc Natl Acad Sci USA. 1994;91:430–443. doi: 10.1073/pnas.91.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forrest D, Erway L C, Ng L, Altschuler R, Curran L. Nat Genet. 1996;13:354–357. doi: 10.1038/ng0796-354. [DOI] [PubMed] [Google Scholar]
- 37.Rüsch A, Erway L C, Oliver D, Vennström B, Forrest D. Proc Natl Acad Sci USA. 1998;95:15758–15762. doi: 10.1073/pnas.95.26.15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campos-Barros A, Amma L L, Faris J S, Shailam R, Kelley M W, Forrest D. Proc Natl Acad Sci USA. 2000;97:1287–1292. doi: 10.1073/pnas.97.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kharkovets T, Hardelin J P, Safiedienne S, Schweizer M, Amraoul A E, Petit C, Jentsch T J. Proc Natl Acad Sci USA. 2000;97:4333–4338. doi: 10.1073/pnas.97.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]