Abstract
Likelihood estimates of local rates of evolution within proteins reveal that selective constraints on structure and function are quantitatively stable over billions of years of divergence. The stability of constraints produces an intramolecular clock that gives each protein a characteristic pattern of evolutionary rates along its sequence. This pattern allows the identification of constrained regions and, because the rate of evolution is a quantitative measure of the strength of the constraint, of their functional importance. We show that results from such analyses, which require only sequence alignments, are consistent with experimental and mutational data. The methodology has significant predictive power and may be used to guide structure–function studies for any protein represented by a modest number of homologs in sequence databases.
The principle that the rate of molecular evolution is inversely correlated with the strength of selective constraints has long been known (1, 2). The average evolutionary rate of a protein reflects the overall importance of the protein for organismal functions, whereas rate variation within the protein reflects intramolecular differences in structural and functional constraints. Intramolecular rate variation has been the subject of many studies focused on devising more realistic models of sequence evolution that do not assume rate constancy among sites (e.g., refs. 3–6). A more recent application of estimating rate variation within proteins has been the inference of structural and functional constraints (7–9).
To identify evolutionarily constrained regions (ECRs) we devised a general approach to inferring rate variation within proteins. We construct a multiple sequence alignment of orthologs and/or closely related paralogs and build the maximum likelihood tree. Holding the branching structure of the tree fixed, we then calculate the number of substitutions in each window of a fixed width over the entire alignment. The “relative rate” in the window is obtained by dividing the number of substitutions per site in the window by the average of all windows. Plotting the windows' relative rates as a function of their position generates a rate profile (RP), and a heuristic algorithm automatically identifies ECRs and ranks them by their rate of evolution. This approach allows us to infer both the existence of constrained regions in a protein and, because the rate of evolution is a quantitative measure of the strength of the constraint, the relative importance of the identified region.
Our method requires only a multiple sequence alignment and is sufficiently powerful to allow analyses involving a relatively small number of fairly closely related sequences. It enables us (i) to use sequences for which the quality of the alignment over most its length is indisputably robust, and (ii) to use alignments of orthologs and closely related paralogs for which conservation of structural and functional constraints can be reasonably assumed. We show below that the method identifies known domains with a high degree of accuracy, that its rankings are consistent with experimental and mutational data, that it allows the inference of previously unknown constrained domains, and that it can pinpoint the origin of novel functional regions in the evolutionary history of paralogs.
Methods
Overview of Steps of Evolution–Structure–Function (ESF) Analyses.
(i) Choose query protein sequence and identify closely related sequences by similarity search with WUBLASTP (ref. 10; http://blast.wustl.edu). (ii) Build multiple sequence alignment with CLUSTALW (ref. 11; ftp://ftp.ebi.ac.uk/pub/software/unix/clustalw/). (iii) Infer maximum likelihood tree of the sequences with PROTML 2.3 (ref. 12; ftp://ftp.ism.ac.jp/pub/ISMLIB/MOLPHY/). (iv) Estimate local rates of evolution in a sliding window with CODEML (ref. 13; http://abacus.gene.ucl.ac.uk/software/paml.html). (v) Plot rates as a function of position in alignment, detect ECRs, and rank ECRs by slowest evolving window.
Similarity Search and Alignment.
Default parameters are used. Very similar sequences that would only contribute a small number of substitutions are removed to minimize computational time. We attempt to maximize two competing quantities to optimize predictive power: (i) the fraction of the alignment in which positional homology is virtually certain—we discard sequences that disproportionately introduce or extend the areas of uncertain homology around gaps— and (ii) the number of substitutions in the regions where homology is certain—we use proteins for which a reasonably diverse set of homologs (usually orthologs) has been sequenced. A typical alignment contains between 8 and 20 sequences with pairwise differences ranging from 5% to at most 50% and fewer than one-third of the positions residing in areas of dubious homology.
Tree Reconstruction.
We use the likelihood implementation in PROTML with the JTT stochastic model (14). Alignments with fewer than 12 sequences allow an exhaustive search of all trees; for those with more, a heuristic search is used. Positions of uncertain homology are removed before analysis, in accordance with standard practice in phylogenetic reconstruction.
Estimating Local Relative Rates.
For every window of nineteen amino acids in the alignment, we calculate the number of substitutions per site by likelihood, using the JTT model and the branching pattern inferred from the step above. The relative rate in the window is obtained by dividing the number of substitutions per site in the window by the average of all windows. The window width was chosen to maximize the signal-to-noise ratio for the detection of constraints on regions, not individual amino acids, and to reduce the statistical uncertainty associated with estimates of the number of substitutions in slowly evolving regions. Window widths could be narrowed for other applications. Gaps in the alignment are almost always part of regions of uncertain positional homology, and are therefore filled with alanines. This results in high relative rates whose values can be regarded as arbitrary.
Detection of ECRs.
Relative rates for all windows are smoothed using a ten-position-wide moving window arithmetic average and then plotted as a function of alignment position in a two-dimensional array. Scanning the array from the bottom (minimum) to top (maximum) yields a ranked list of local minima. Each minimum defines an ECR whose maximum extent is bounded by the first positions to the left and to the right where the first derivative is zero—i.e., by the peaks neighboring the trough.
Difference in Rate Plots of Independent Datasets.
As a measure of difference between rate plots X and Y, we define the distance ρ as ρ(X,Y) = (1/n) Σ | log(Xi/Yi) |, where Xi and Yi are the rates calculated in the window centered at position i for plots X and Y, respectively, and i ranges from positions 1 to n of the plots. ρ satisfies the formal criteria for a distance metric. We use Xi/Yi as the relative rate of Xi with respect to Yi because Xi/Yi is the factor by which the rate calculated at position i in plot X differs from that calculated at position i in plot Y. The value | log(Xi/Yi) | is a symmetric measure of the size of this difference, and ρ(X,Y) computes the average of these differences across the whole of the plots.
Note that for c > 1, | log(cx/x) | = log c. It follows that ρ(cX,X) = log c is the calculated distance between plot X and the rate plot obtained from scaling X by a factor of c. We invert this relationship to relate calculated distances to a scaling factor, c, with c = eρ(X,Y). In the case of hemoglobin, we calculated the distance between plots of the α and β chains as ρ(α,β) = 0.12. An equivalent distance is ρ(cα,α), where scaling factor c = e0.12 = 1.13. In other words, the RP of alpha hemoglobin exhibits a difference to that of beta hemoglobin as if each relative rate differed by 13%.
Results
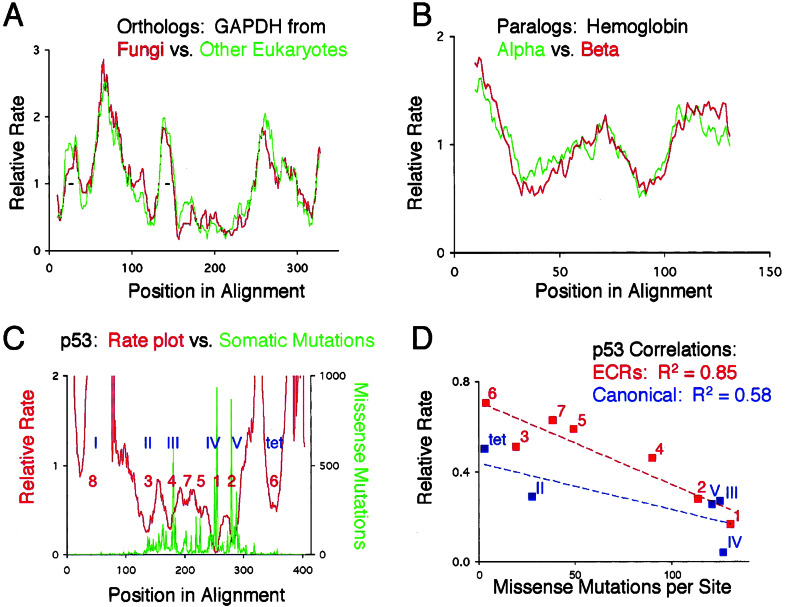
A critical test of the methodology we are proposing is whether local rates of evolution are quantitatively stable, and whether likelihood inference of relative rates is reproducible. If so, independent data sets of the same protein should give similar RPs regardless of taxonomic sampling. Because we plot the relative rate of evolution, RPs derived from different paralogs that evolve at different overall rates or from orthologs representing diverse taxonomic sampling should be directly comparable. We chose two test cases, eukaryotic glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and vertebrate hemoglobins. For GAPDH, one alignment contained sixteen sequences from fungi, and the other had sixteen non-fungal sequences mostly from plants and animals. Because inference is done on unrooted trees, the evolutionary history of the two datasets is independent. The sequences span a divergence of 1,718 point substitutions accumulated over a total of several billion years, with the fungal dataset having a slightly larger number of substitutions than the other eukaryotes. We find that the RPs track each other closely (Fig. 1A).
Figure 1.
(A and B) Overlays of RPs from homologous proteins. x axis, position in sequence alignment; y axis, relative rate calculated from the ratio of substitutions within each window, divided by the total number of substitutions in the alignment. Rates were first calculated and normalized independently, and plots were then overlaid in register with the sequence alignment. Small insertions in single sequences were removed before analysis. (C) Overlay of the rate plot of p53 (eleven sequences) and the frequency of missense mutations isolated from somatic tumors. Blue Roman numerals denote the canonical domains as described in the p53 literature (17); tet, tetramerization domain. Red numbers are the inferred ECRs, ranked according to the rate of the slowest evolving window. (D) Correlation of the average rate of evolution with the density of point mutations in the canonical domains (blue) or ECRs (red).
RPs from closely related paralogs are also quite similar, as evidenced by the analyses of alpha and beta hemoglobin (Fig. 1B). These RPs were generated from the same 14 organisms, which allowed us to calculate the difference in the average rate between the two globins without having to make assumptions about divergence times. Alpha evolves 1.2 times faster than beta, and yet the RPs superimpose.
To estimate how different superimposed RPs are, we devised a distance measure (see Methods). The mean distances in relative rates between the RPs of GAPDH and hemoglobin are 0.22 and 0.13, respectively. These are small values considering that the rates within an RP can differ by as much as two orders of magnitude.
Having shown that constraints are reproducibly detected we turned to four tests designed to elucidate the biological significance of RPs and ECRs. We first asked whether there is a correlation between a protein's RP and the degree of functional impairment by point mutations. We used the frequency distribution of 11,360 missense mutations in p53 isolated from somatic tumors (15) as an indirect measure of functional impairment of a protein. (The tumors in which these mutations were identified represent one phenotypic effect of somatic mutations in p53, whereas the rate of evolution is a function of the severity of any phenotypic effect of germline mutations. We would have preferred to use a database of germline mutations whose phenotypic effect had been measured but we are not aware that such a database currently exists.) In agreement with a previous study of p53 that correlated a different measure of evolutionary rates with mutational density (16), Fig. 1C shows qualitatively that missense mutations are more likely to promote the development of tumors when they occur in slowly evolving regions. To quantify the inverse correlation between density of point mutations and rate of evolution, we compared the number of missense mutations in the ECRs and in the domains as described in the literature (17), with their rates of evolution. We detect eight ECRs in p53, six that mostly overlap with the domains and two that reside between canonical domains III and IV. The average relative rate of evolution and number of mutations per site are more strongly correlated for the ECRs than for the canonical domains (Fig. 1D). This result validates the ECR as a meaningful concept in studying protein function.
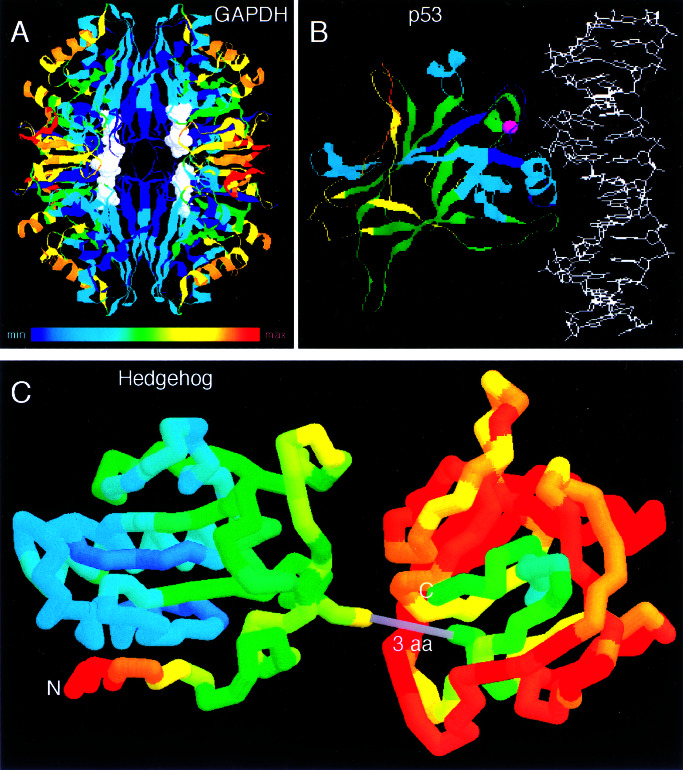
Second, we asked whether the RPs are in general agreement with accepted principles of protein folding and function. In GAPDH (Fig. 2A), buried amino acids that are responsible for folding and stabilization of the tetrameric interface, NAD+ cofactor binding, and enzyme catalysis evolve more slowly (blue) than the solvent exposed residues (red). This pattern is in striking contrast to proteins that bind other proteins or DNA, and whose functionally most important residues reside on the surface. In p53, the most highly constrained region is a surface patch that forms the DNA binding interface (Fig. 2B). In Hedgehog (Fig. 2C), the C-terminal domain, which is not directly involved in signaling (20, 21), evolves 4-fold faster than the N-terminal domain, the bioactive part of the protein (22). Within the C-terminal domain, the core exhibits the slowest rates of evolution. Within the N-terminal domain, the rate of evolution is much lower in the part that is oriented away from the C terminus. This constraint is likely due to the presence of surface residues that bind the receptor, Patched.
Figure 2.
Visualization of relative rates in structures using a color spectrum; blue represents the slowest rates, orange and red the highest. This figure was prepared using RASMAC Version 2.6 (ref. 18; http://www.umass.edu/microbio/rasmol/) and modified Protein Data Bank (PDB) files (ref. 19; ht) in which the temperature field was substituted with the relative rate scaled linearly to span the range of available colors. (A) GAPDH tetramer (PDB ID code 1GD1). (B) The p35 core bound to DNA (PDB ID code 1TUP). (C) Virtual fusion of the N terminus of mouse Sonic Hedgehog (PDB ID code 1VHH) with the C terminus of Drosophila Hedgehog (PDB ID code 1AT0). Only three amino acids (3 aa) in the alignment separate the structures of the two domains.
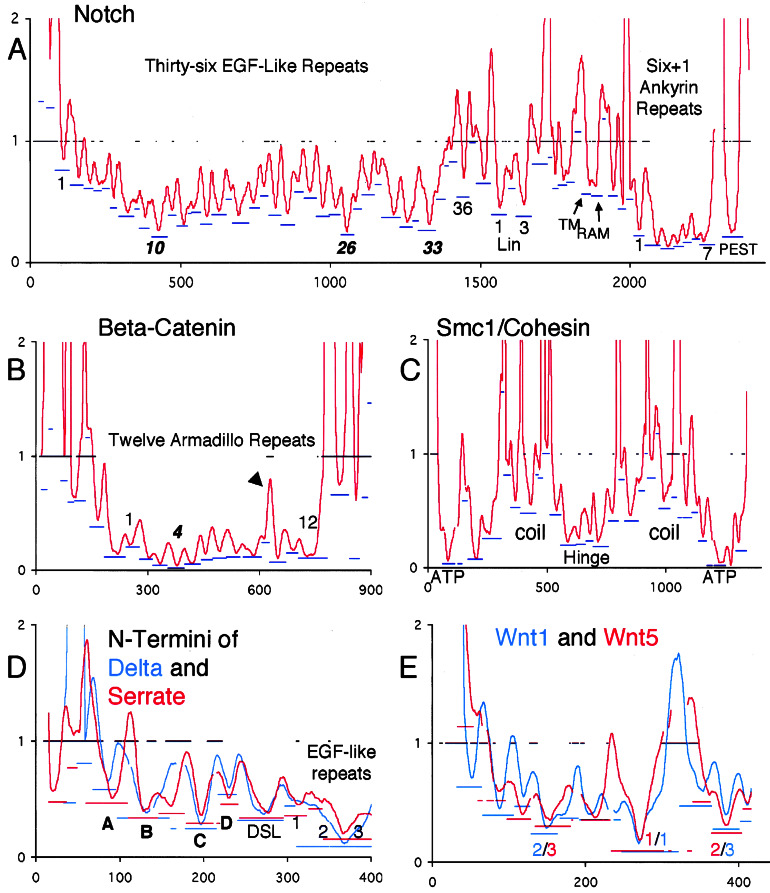
In a third test, we asked whether there is a consistent correspondence of the location of ECRs and that of known structural domains. In Notch (Fig. 3A), for example, 34 of 36 EGF-like repeats and all Lin/Notch and Ankyrin repeats are accurately inferred in the sense that each contains one local minimum bordered by local maxima. Similarly, in β-catenin (Fig. 3B), all twelve armadillo repeats are detected, with repeat number 10, which contains a quickly evolving insertion, having two minima.
Figure 3.
RPs for five case studies. x axis, position in sequence alignment; y axis, smoothed relative rates for emphasis on detection of ECRs. Bars at y = 1 indicate regions of uncertain alignment. Blue and/or red bars underneath ECRs indicate the rate of the most slowly evolving window in the ECR (position on y axis) and the extent of the inferred ECR (extent along x axis). ECRs whose troughs are entirely contained with a region of uncertain alignment should be disregarded, but note that the resolution of the graphic is limited. (A) Notch; ECRs at the start and end of the repeat regions are labeled with the number of the repeat to which they correspond. Most-slowly evolving repeats are in italics. The gap just N-terminal to the PEST ECR corresponds to 180 highly divergent positions. (B) β-catenin/armadillo; arrowhead points to quickly evolving insertion in repeat 10. (C) SMC1/cohesin with the five known regions labeled. Note that each region contains several ECRs. (D) Overlay of the N-terminal RPs of Delta and Serrate prepared as described in Fig. 1. Gaps in the plots are due to alignment of the RPs to each other. Novel predicted domains are labeled in bold. (E) Overlay of Wnt1/wg and Wnt5a/b plots. The ranks of the three most slowly evolving ECRs are in the color corresponding to the paralog to which they belong. The sequences from human Wnt1 that correspond to the most slowly evolving windows in ECRs 1, 2, and 3 are, respectively, CKCHGMSGSCTVRTCWMRL, VNRGCRETAFIFAITSAGV, and CNSSSPALDGCELLCCGRG.
Having established that ECRs consistently correspond to structural domains in modular proteins, we tested whether constraints imposed by function could be detected by ranking the ECRs according to their rate. In Notch, the most slowly evolving domains (Fig. 3A) are the Ankyrin repeats (transcriptional coactivation), followed by the PEST sequence (degradation), and then EGF repeat number 10 (binding the ligands, Delta, and Serrate). Other ECRs that correspond to regions of known function include the Lin repeats and the RAM region (23–25). In β-catenin, the armadillo repeat region, which extensively interacts with TCF and E-cadherin (26, 27), evolves most slowly. The strongest constraint resides in repeat four, in which Lys-312 has been shown to be essential for E-cadherin binding (26, 27). In Mybs, the most slowly evolving ECRs are the Myb repeats (data not shown).
Functional domains need not belong to known structural modules to be detected. In the Mybs, the next-ranking ECRs are the acidic domain in A- and C-myb (transactivation), and the N-terminal part of the negative regulatory domain. In p53, the two arginines that make DNA contact are in the middle of the two most slowly evolving ECRs (Fig. 1C). In Smc1/cohesin (Fig. 3C), the most slowly evolving ECRs comprise the N- and C-terminal ATPase domains. The hinge, which lacks discernable sequence motifs but is known to be important for function, has the next-highly ranking ECRs. Finally, the coiled coils, in which only a small subset of residues are important for binding, comprise the most quickly evolving ECRs. The striking symmetry of the cohesin RP matches the model that has been proposed from functional studies (28), but the multitude of detected ECRs also suggests that each of the five known domains is comprised of several smaller regions of functional importance.
The results from our tests support two major conclusions. First, divergent evolution of homologs that are alignable over most of their length follows stable patterns that manifest themselves in quantitatively similar relative rates of evolution even when the overall rate of evolution varies between the homologs. Second, estimating the local relative rates of evolution by likelihood analyses, and ranking inferred ECRs by their rate, is predictive of functional importance. We now turn to analyses that show the full range of hypotheses that can be generated.
Of all EGF repeats in Notch, number 26 is the second-most slowly evolving (Fig. 3A). It contains a conserved fucosylation consensus sequence (29) that may be the site of modification by Fringe, the glycosyltransferase that modulates Notch's sensitivity to its ligands. Consistent with this prediction is the fact that the abruptex mutations in Notch, which genetically interact with alleles of Fringe, cluster between repeats 24 and 29 (30).
In the N termini of Delta and Serrate, four ECRs that do not correspond to previously identified domains stand out as evolving sufficiently slowly to have important functional roles (Fig. 3D). Two of them, B and C, evolve just as slowly as the DSL domain, which is the defining domain of this gene family. In addition, EGF repeats 2 and 3 of both Delta and Serrate evolve about twice as slowly as the DSL domain, with the greatest constraint neatly centered on their border. This constraint is unlikely to be due to structural requirements because the relative rate of evolution in most other EGF repeats of Notch, Delta, and Serrate is much higher, with the minima much less skewed toward one end of the domain. We predict that this region and probably the novel domains as well are important for the main function of these ligands, their interaction with the Notch receptor.
As an example for predicting the functional importance of ECRs in nonmodular proteins we analyzed the Wnts, which are signaling molecules that bind to and activate the Frizzled receptors (31). It is currently unknown which regions in the Wnts are responsible for receptor binding but, given the results from our tests above, the most slowly evolving ECRs are good candidates for bearing this important function. As a test of the consistency of our predictions, we independently analyzed two ancient paralogous Wnts that had been shown to evolve at 2-fold different average rates (32), Wnt1/wg and Wnt5a/b. Overlay of the RPs and ranking of their ECRs shows that the most slowly evolving ECRs of both paralogs are close in location and have similar relative rates (Fig. 3E). We predict that these regions contain residues that make the most important contacts with their cognate Frizzled receptors.
The results from the comparisons involving ancient paralogs such as Delta/Serrate and Wnt1/Wnt5 show that the location of ECRs and their relative evolutionary rates can be well conserved. Conversely, different patterns in related proteins provide the opportunity to define the evolutionary origin of a new constraint. We chose the Myb gene family as a test case because despite fairly high levels of sequence similarity, a functional difference has been hypothesized between homologs: A- and C-myb can act as transcriptional activators, whereas B-myb and the single invertebrate myb probably do not (33).
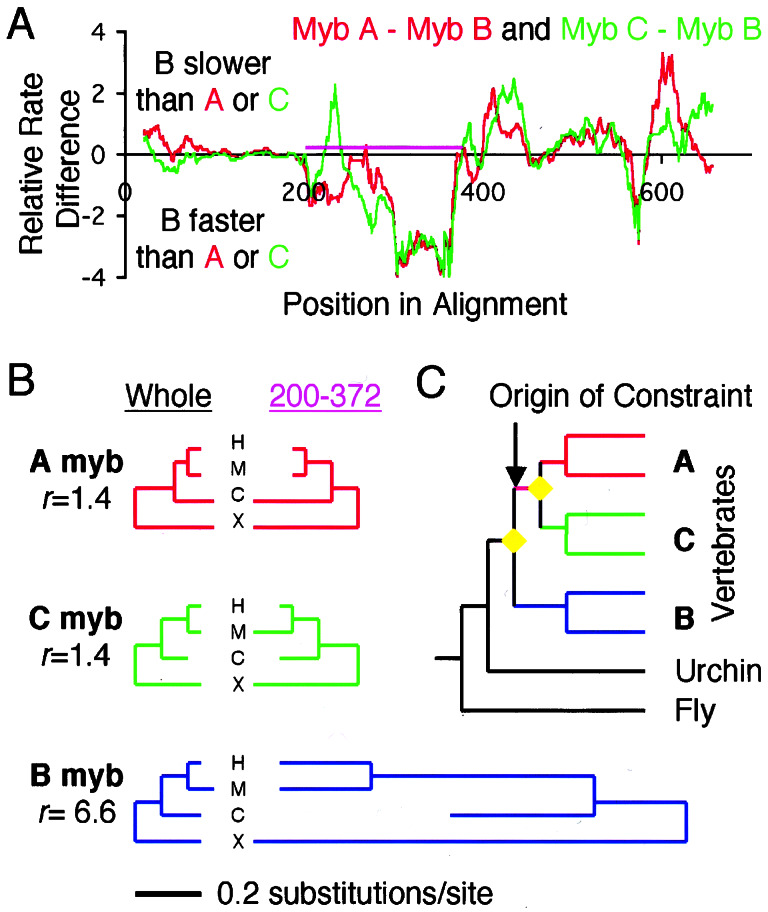
In these analyses, differences between aligned RPs of paralogs are first used to identify candidate regions where rates greatly differ. Then, the rate of evolution in the candidate region, not just in the windows, is calculated and compared with the average rate of the entire protein. In the alignment of the Mybs, the section from position 200 to 372 exhibits the greatest difference in relative rates between A-myb or C-myb and B-myb, with A and C having similar rates and B obviously evolving more quickly (Fig. 4A). In both A-myb and C-myb, this region contains a domain that has been shown to be capable of transcriptional activation (34, 35) and binding to the coactivator, CBP (36). Whether the corresponding section in B-myb is also capable of activating transcription is controversial (33). Our analyses show a lack of constraints in that section of B-myb, with a 6.6-fold faster rate than the average of the entire protein (Fig. 4B). In contrast, in both A-myb and C-myb, the corresponding section evolves only 1.4 times faster than the average, and the transcriptional activation domain is one of the most slowly evolving ECRs (data not shown). Given the evolutionary relationship of the Myb genes, we can map the origin of the constraint, and the likely origin of the transcriptional activation function, on the ancestral lineage of A- and C-myb, after the B-myb lineage diverged (Fig. 4C).
Figure 4.
The Myb case study for illustration of inference of evolved differences between paralogs. (A) Differences in relative rates between A myb and B myb (red), and C myb and B myb (green). Purple bar delineates the region C-terminal to the myb repeats that contains the acidic activator domain in A- and C-myb, comprising alignment positions 200–372. (B) Trees for each paralog relating the orthologs from human (H), mouse (M), chick (C), and Xenopus (X), with branch lengths proportional to the number of substitutions. Left set of trees are calculated from the entire alignment; right set of trees are calculated from just the positions corresponding to the region indicated in purple in A. This region is constrained equally in A and C, but in B it evolves much faster by comparison. (C) Most likely position of the origin of the constraint, which is shared between A and C, but not present in B and invertebrates. Yellow diamonds are gene duplications; vertebrate subtrees are simplified for display.
Discussion
We show that local rates of protein evolution are generally stable regardless of the strength of the constraint, a phenomenon that manifests itself as virtually superimposable RPs of independent sets of sequences for the same protein. This stability extends to more distant paralogs such as Wnt1 and Wnt5 despite differences in their average rates. This finding has the interesting implication that, even in the absence of a true molecular clock for the absolute rate of each protein's evolution, there is a relative clock for the local rate of evolution that is primarily governed by physicochemical constraints on protein structure and function. Our analyses uncover this clock with normalization of the windows' rates by the average rate of the protein, which factors out time and those contributions to the evolutionary rate that are due to organismal and population parameters.
We term the methodology described here “Evolution–Structure–Function” (ESF) analyses. ESF analyses estimate rates within a window, normalize the local rates by the average rate, smooth the rates to detect ECRs, and then rank the ECRs by the rate of the most slowly evolving window. This combination of algorithms distinguishes the method from similar approaches that either require structures (7, 8), do not predict or rank ECRs, or separately estimate rates for each position (9). One key assumption we make is that of a strong correlation of rates in sites that are close in the alignment. We make this assumption to increase power (allowing analyses of rather closely related sequences) and to reduce the statistical uncertainty associated with estimates of rates in single positions. One potential drawback of this approach is a loss of sensitivity with respect to detection of small conserved regions. Another drawback, which may be counterbalanced by post-ESF scrutiny of the alignment for fully conserved positions, is that the resolution of the method is region-based and not focused on the individual amino acid.
ESF analyses provides specific, first-principle hypotheses as to where a protein's functional regions most likely reside. The dissociation of functional from structural constraints is perhaps the most important practical feature of ESF analyses. It is best illustrated by the EGF repeats in Notch and its ligands, which, despite very similar structures, evolve at vastly different rates. Depending on the resolution desired, ESF analyses alone, or in combination with other comparative analyses such as the one described above for the conserved fucosylation site in EGF repeat 26 of Notch, can generate precise, experimentally testable predictions of function.
Natural selection has generated an extraordinary trove of experimental data that is hidden in extant sequences by testing millions of point mutants and rejecting the vast majority with a probability proportional to their deleteriousness. Because average evolutionary rates of proteins vary greatly in the proteome (2), the species that will reveal these data with the best signal-to-noise ratio range from closely to distantly related. Thus, the information gained from comparative analyses such as the ones described here would be maximized if a diverse set of genomes were to be sequenced, not all of which need belong to experimental model organisms.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for constructive criticisms and Joseph Lipsick for discussions about mybs. This study was supported by a Terman Fellowship (to A.S.). E.A.S. and A.L.S. were supported by training grants from the National Human Genome Research Institute and National Cancer Institute, respectively.
Abbreviations
- ECR
evolutionarily constrained region
- RP
rate profile
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 2.Li W H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 3.Uzzell T, Corbin KW. Science. 1971;172:1089–1096. doi: 10.1126/science.172.3988.1089. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z. Mol Biol Evol. 1993;10:1396–1401. doi: 10.1093/oxfordjournals.molbev.a040082. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein J, Churchill G A. Mol Biol Evol. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Kumar S. Mol Biol Evol. 1996;13:650–659. doi: 10.1093/oxfordjournals.molbev.a025625. [DOI] [PubMed] [Google Scholar]
- 7.Lichtarge O, Bourne H R, Cohen F E. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 8.Dean A M, Golding G B. Pac Symp Biocomput. 2000;2000:6–17. doi: 10.1142/9789814447331_0002. [DOI] [PubMed] [Google Scholar]
- 9.Armon A, Graur D, Ben-Tal N. J Mol Biol. 2001;306:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi J, Hasegawa M. Comput. Sci. Monographs. Vol. 27. Tokyo: Institute of Statistical Mathematics; 1992. [Google Scholar]
- 13.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 14.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 15.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 16.Walker D R, Bond J P, Tarone R E, Harris C C, Makalowski W, Boguski M S, Greenblatt M S. Oncogene. 1999;19:211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 17.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 18.Sayle R A, Milner-White E J. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 19.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall T M T, Porter J A, Beachy P A, Leahy D J. Nature (London) 1995;378:212–215. doi: 10.1038/378212a0. [DOI] [PubMed] [Google Scholar]
- 21.Hall T M T, Porter J A, Young K E, Koonin E V, Beachy P A, Leahy D J. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- 22.Porter J A, von Kessler D P, Ekker S C, Young K E, Lee J J, Moses K, Beachy P A. Nature (London) 1995;374:363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- 23.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 24.Rebay I, Fleming R J, Fehon R G, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 26.Graham T A, Weaver C, Mao F, Kimelman D, Xu W. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 27.Huber A H, Weis W I. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 28.Jones, S. & Sgouros, J. (2001) Gen. Biol.2, RESEARCH0009. [DOI] [PMC free article] [PubMed]
- 29.Moloney D J, Panin V M, Johnston S H, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine K D, Haltiwanger R S, Vogt T F. Nature (London) 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 30.Kelley M R, Kidd S, Deutsch W A, Young M W. Cell. 1987;51:539–548. doi: 10.1016/0092-8674(87)90123-1. [DOI] [PubMed] [Google Scholar]
- 31.Bhanot P, Brink M, Samos C H, Hsieh J C, Wang Y, Macke J P, Andrew D, Nathans J, Nusse R. Nature (London) 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 32.Sidow A. Proc Natl Acad Sci USA. 1992;89:5098–5102. doi: 10.1073/pnas.89.11.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganter B, Lipsick J S. Adv Cancer Res. 1999;76:21–60. doi: 10.1016/s0065-230x(08)60773-3. [DOI] [PubMed] [Google Scholar]
- 34.Sakura H, Kanei-Ishii C, Nagase T, Nakagoshi H, Gonda T J, Ishii S. Proc Natl Acad Sci USA. 1989;86:5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golay J, Loffarelli L, Luppi M, Castellano M, Introna M. Oncogene. 1994;9:2469–2479. [PubMed] [Google Scholar]
- 36.Parker D, Rivera M, Zor T, Henrion-Caude A, Radhakrishnan I, Kumar A, Shapiro L H, Wright P E, Montminy M, Brindle P K. Mol Cell Biol. 1999;19:5601–5607. doi: 10.1128/mcb.19.8.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.