Abstract
The lck gene encodes a lymphocyte-specific protein-tyrosine kinase that is implicated in T cell maturation and signaling. In mammals, the transcription of the lck gene is regulated by two independent promoters, the proximal promoter, which is active in thymocytes, and the distal promoter, which dominates in mature T cells. In the human and mouse lck gene loci, the two promoter elements are separated by at least 40 kb and 10 kb, respectively. In this study, we have cloned and sequenced 60 kb from the pufferfish (Fugu rubripes) lck locus. The promoter region of the Fugu lck spans only 4.2 kb and contains a proximal and a distal promoter in the 2.3-kb region adjacent to the coding sequence. By generating transgenic mice, we have demonstrated that the compact promoter of the Fugu lck contains regulatory elements that direct expression to lymphoid organs of mice. We were able to localize the regulatory elements to a short region of 830 bp without losing specificity to cultured human T cell line. These results show that the basic mechanisms that mediate lymphocyte-specific expression are conserved between teleosts and mammals. The short promoter of the Fugu lck isolated by us offers a powerful tool for labeling T cells, targeting expression, and manipulating T cell activity in fishes as well as in mammals.
The specificity of gene expression for cell types or developmental stages and the response of genes to physiological stimuli are mediated through a combinatorial interaction of promoter sequences, enhancers, and suppressors. These regulatory elements are composed of short stretches of DNA that are generally found in the promoter region of genes but also can be located in introns or dispersed over many kilobases upstream and downstream of genes (1). Finding these short elements in mammalian genomes is a formidable task because the intergenic regions in these genomes are large and complex. The genome of the pufferfish, Fugu rubripes, which is approximately eight times smaller than the human and mouse genomes, contains compact intergenic and intronic regions that are devoid of repetitive sequences (2, 3). This genome offers an attractive model for the rapid screening of noncoding sequences for conserved putative regulatory elements. Conserved regulatory elements driving cell- and stage-specific expression of developmental control genes such as the Hoxb-4, Otx2, Wnt-1, and Pax6 were identified by this strategy (4–7). Conserved regulatory elements were identified in the Fugu neurohypophyseal gene that express specifically in a subset of neurons in the hypothalamus of transgenic rats (8), and the Fugu tyrosinase gene was shown to contain conserved elements that directed melanocyte and retinal pigment epithelium-specific expression in transgenic mice (9). The hypothesis can be proposed that, where physiological systems show conservation over long stretches of evolutionary time, the genes specifying these processes are likely to be conserved not only in their coding sequences but also in their regulatory sequences. The immune systems of mammals and teleosts are similar, and we have now extended these transgenic experiments to genes of the immune system.
The major lymphoid organs of teleosts are the thymus, spleen, kidney, and the gut-associated lymphoid tissue (10). Teleosts lack bone marrow and lymph nodes. Although it has been shown that teleosts possess different populations of lymphocytes that are analogous to the mammalian T cells and B cells, it has not been possible to unequivocally identify T cells and B cells because of a lack of reagents for surface markers such as CD4, CD8, and CD19. Indirect evidence such as the expression of genes for RAG1 and RAG2, TdT, T cell receptors, and lck in the developing thymus of teleosts suggests that T cell maturation occurs in the thymus similar to that in mammals (11). We chose the gene encoding p56lck because this gene in mammals shows a tightly controlled lymphocyte-specific expression (12). Although the exact cellular role of p56lck is not known, it is associated with T cell maturation and signaling (13, 14). In humans and mice, the lck gene is differentially transcribed from two independent promoters, a proximal promoter, which lies within the intron in the 5′-untranslated region (UTR) and is active in thymocytes, and a distal promoter, located upstream of the 5′-UTR intron, which is active predominantly in mature T cells and to a lesser extent in thymocytes. There is a gradual shift of transcriptional activity from the proximal promoter to the distal promoter as the thymocytes mature to become T cells (15, 16). Consequently the transcripts generated from the proximal promoter are found only in the thymus whereas the distal promoter transcripts are found both in the thymus and peripheral T cells. The proximal and distal promoters of lck are separated by >40 kb and 10 kb in humans (UCSC Human Genome browser at http://genome.ucsc.edu) and mice (17), respectively.
Transgenic experiments in mice have shown that a 3.2-kb fragment of the mouse lck proximal promoter directs expression to the thymus (18). On the other hand, a 2.6-kb fragment of the human lck distal promoter and a 3-kb fragment of the mouse lck distal promoter were able to direct correct expression to the thymus, spleen, and peripheral T cells (15, 19). These studies indicate that the two promoters act independently. In subsequent studies to localize the proximal promoter elements in the mouse lck, sequences between −584 and +37 relative to the proximal transcription start site were found to direct tissue-specific and temporally correct expression (20). This region contains binding sites for five nuclear proteins, of which one protein was found in cell lines that do not express from the proximal promoter and another in cell lines in which the proximal promoter is active, indicating the presence of silencing as well as positive regulatory factors within the proximal promoter region (20). Recently, it was demonstrated that a Kruppel-like zinc finger-binding protein, mtβ, binds to the −365 to −328 region of the mouse proximal promoter (21). Mutation of the binding site or a reduction in the mtβ protein expression level significantly impaired the proximal-promoter activity, suggesting that mtβ protein is a critical transactivator for the proximal promoter (21).
In this study, we have sequenced 60 kb from the Fugu lck locus and shown that the Fugu lck-promoter region spans only 4.2 kb of which only 2.3 kb is adequate to direct expression to lymphoid organs in transgenic mice and in cell lines derived from human T cell lineage. The compact Fugu promoter offers a powerful tool for targeting gene expression and manipulating T cell activity.
Materials and Methods
Isolation and Sequencing of Fugu lck Cosmid.
A genomic fragment of the Fugu lck gene was amplified by PCR using degenerate primers that are complementary to the human lck exon 10 (5′-TGY AAR ATH GCN GAY TTY GG-3′) and exon 11 (5′-GCY TCN GGN GCN GTC CAY TT-3′), cloned into a T-vector and sequenced. This fragment was used as a probe to screen a gridded Fugu cosmid library (Greg Elgar, UK-Human Genome Mapping Project Resource Center) and two positive cosmids, 88A8 and 52C1, were isolated. Restriction enzyme analyses revealed that the two cosmids overlap by ≈20 kb. A contiguous sequence of ≈60 kb was determined from the two cosmids by a combination of “shotgun” sequencing and primer walking on an Applied Biosystems 377 automated DNA sequencer. Genes were identified by their homology to known genes by blast searching of public databases maintained at the National Center for Biotechnology Information. The exon–intron structure of the Fugu lck gene was confirmed by reverse transcription (RT)-PCR and the transcription start site was mapped by 5′-rapid amplification of cDNA ends (RACE) (SMART RACE, CLONTECH).
Generation of Transgenic Mice.
Transgenic mice were generated by using standard procedures. In brief, fertilized one-cell stage eggs were isolated from superovulated FVB/N mice and microinjected with linearized Fugu cosmid 52C1 or with Fugu lck promoter-driven enhanced green fluorescent protein (EGFP) constructs in which the plasmid backbone was removed by restriction digestion and gel purification. Cosmid 52C1 was linearized by digesting with SfiI that has a single restriction site at 9 kb upstream of the lck gene (Fig. 1a). Transgenic mice were identified by PCR analysis of genomic DNA isolated from tail biopsies. The founder mice were mated with wild type to produce independent transgenic lines.
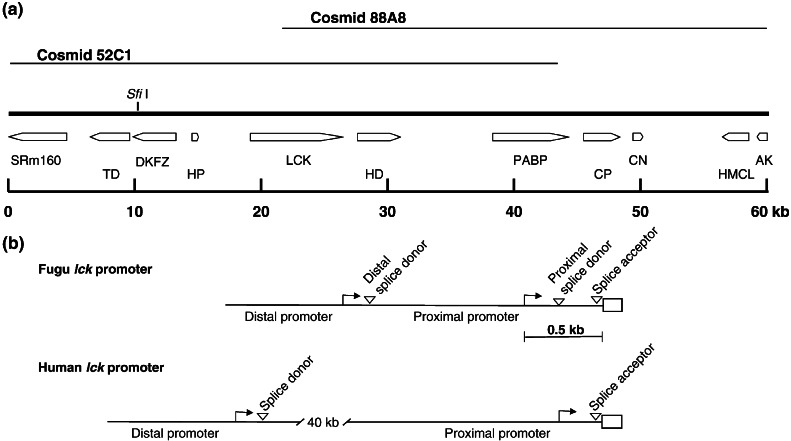
Figure 1.
(a) Schematic representation of the Fugu lck locus. Only the unique site for SfiI is shown. Arrows represent genes and indicate the direction of transcription. Only partial sequences for the genes SRm160 (SRm160/300 splicing coactivator) and AK (homolog of human AK010736 protein) are present in this sequence. Other genes are TD, threonine dehydrogenase; DKFZ, Fugu homolog of human DKFZP434K171 protein; HP, Fugu homolog of a human hypothetical protein; LCK, p65lck; HD, histone deacetylase; PABP, poly(A)-binding protein; CP, cyclophilin; CN, connexin; and HMCL, HMG coA lyase. (b) Schematic representation of the Fugu and human lck-promoter regions. The first coding exon of lck is shown as an open box. The transcription start sites are shown by arrows, and the splice donor and acceptor sites are marked by arrowheads. In the Fugu promoter, the 5′-UTR intron is spliced by using the proximal splice donor when the transcripts are initiated from the proximal promoter and the distal splice donor when the transcripts are generated from the distal promoter.
Expression Analysis.
Total RNA was extracted from various tissues of Fugu or transgenic mice by using TRIzol reagent (GIBCO/BRL), and the expression pattern was analyzed by RT-PCR or Northern analysis. The 5′-RACE was carried out by using the SMART RACE kit (CLONTECH). Total RNA was pretreated with RNase-free DNase I (Boehringer Mannheim) before RT-PCR. First strand cDNA was synthesized from the total RNA by using the oligo dT primer and SUPERSCRIPT II reverse transcriptase (GIBCO/BRL). RT-PCR- and RACE-primer sequences are available on request. Representative RT-PCR products were cloned and sequenced to confirm their sequences. For Northern analysis, total RNA was fractionated on a 1.2% agarose gel containing formaldehyde, transferred to a Hybond-N nylon membrane (Amersham Pharmacia), and probed with [α-32P]-labeled-coding sequence for EGFP.
Preparation of lck Promoter-EGFP Constructs.
Different lengths of the Fugu lck promoter together with 84 bp of the first coding exon were amplified from the genomic DNA by PCR using specific primers that contained KpnI sites at the 5′-end. The PCR-generated promoter sequences were digested with KpnI and cloned into the KpnI site in the multiple cloning site of pEGFP-1 vector (CLONTECH) such that the initiation codons of the lck and EGFP are in the same ORF. The recombinant Fugu lck promoter-EGFP construct clones were sequenced to confirm the orientation of the promoter and EGFP-coding sequences. Deletions within the Fugu lck promoter-EGFP constructs were made by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene). The sequences of primers used for making these constructs are available on request.
Transfection into Cell Lines.
All cell lines except 293T cell lines were obtained from American Type Culture Collection. The 293T cells were obtained from Inder Verma's lab. Jurkat cells (human leukemic T cell line) and EL4 (murine lymphoma T cell line) were grown in RPMI 1640 medium supplemented with 10% FBS, sodium pyruvate, glutamine, penicillin, and streptomycin. The 293T (human kidney cells), HeLa (human cervix adenocarcinoma), Huh7 (human hepatoma), HT29 (human colon cancer), Daudi (human B lymphoma cells), Raji (human B lymphoma cells), and C2C12 (mouse myoblast) cells were grown in DMEM with the same supplements as for Jurkat cells.
The transfection reagent 1,2-dimyristyloxypropyl-N,N-dimethyl-N-hydroxyethylammonium bromide (DMRIE, GIBCO/BRL) was used for transfecting DNA into cell lines. Then 2 μg/ml of cosmid DNA or lck promoter-EGFP construct DNA was combined with 16 μg/ml of DMRIE, and the transfection and cell culture were carried out by using manufacturer's instructions. pEGFP-N1, a vector containing EGFP driven by the cytomegalovirus promoter (CLONTECH) was used as a control. EGFP expression in transfected cells was determined by using fluorescence visualization in a Leica DMIL-inverted microscope fitted with FITC filter or by Western analysis.
Western Blot Analysis.
Protein was extracted from transfected cells by using 1% Triton-X with 1 mM PMSF (Sigma) in PBS, and protein concentration was determined by Bradford protein assay (Bio-Rad). Western blots were performed by using the enhanced chemiluminescence (ECL) Western blotting analysis kit (Amersham Pharmacia). Mouse mAb against GFP (CLONTECH) was used to probe the expression of EGFP.
Results
Genomic Organization of the Fugu lck Locus.
We sequenced a total of 60 kb from two overlapping cosmids, 52C1 and 88A8. This locus contains 11 protein-coding genes (Fig. 1a). One of these genes was identified based on its high homology to an hypothetical human protein, which was predicted by using computational methods. The presence of the sequence for this hypothetical gene in the Fugu confirms that it is indeed a real gene. The Fugu lck gene is flanked by the hypothetical protein gene at 4.2 kb upstream and the histone deacetylase gene at 0.9 kb downstream. The human ortholog of the hypothetical gene present 5′ to the Fugu lck gene is found on human chromosome 8. The human gene for lck is present on chromosome 1 and is flanked upstream by another hypothetical protein (FLJ10547) at ≈40 kb and downstream by the histone deacetylase gene at 8 kb (UCSC Human Genome Browser). The differences in the gene order in the Fugu and human lck loci indicate that there have been rearrangements in this locus since the divergence of the human and Fugu lineages.
Structure of the Fugu lck Gene.
The Fugu lck gene consists of 12 coding exons similar to the human and mouse lck genes (17, 22). It codes for a protein of 502 aa, which is 71% identical to the human, mouse, and chicken p56lck (Fig. 2). The identity is higher (78%) in the carboxyl-terminal half, which contains the kinase domain. The residues that have been shown to be important for the function of lck in mammals, such as the two cysteines and a serine in the amino-terminal region, the lysine residue that catalyzes the interaction with ATP, and the two tyrosine phosphorylation sites in the carboxy region (23) are all conserved in the Fugu lck (Fig. 2), indicating that this is indeed the Fugu ortholog of the mammalian lck.
Figure 2.
Comparison of the conceptual translation of the Fugu lck to the human and chicken p56lck amino acid sequences. The residues that have been shown to be essential for the function of lck are boxed.
The human and mouse lck genes contain an untranslated exon in the 5′-region. The distal promoter lies upstream of this untranslated exon, and the proximal promoter is present within the 5′-UTR intron. This intron is >40 kb in humans (22; UCSC Human Genome Browser) and >10 kb in mouse (17). The 5′-RACE analysis of the Fugu lck cDNA from the kidney, gills, and spleen revealed the presence of a 1.4-kb intron in the 5′-UTR located 5 bp upstream of the lck initiation codon and mapped the transcription start site to 154 bp upstream of this intron (Fig. 1b). This promoter is analogous to the distal promoter of the human lck. Because we did not have RNA from the Fugu thymus, we could not ascertain whether the Fugu lck gene contains a proximal promoter that is active in thymocytes. The overall size of the Fugu lck gene, from the transcription start site to the poly(A) signal, is 9 kb. The human lck gene is approximately sixfold larger than the Fugu gene mainly because of the large intron (>40 kb) in the 5′-UTR.
Expression Pattern of Fugu lck Gene.
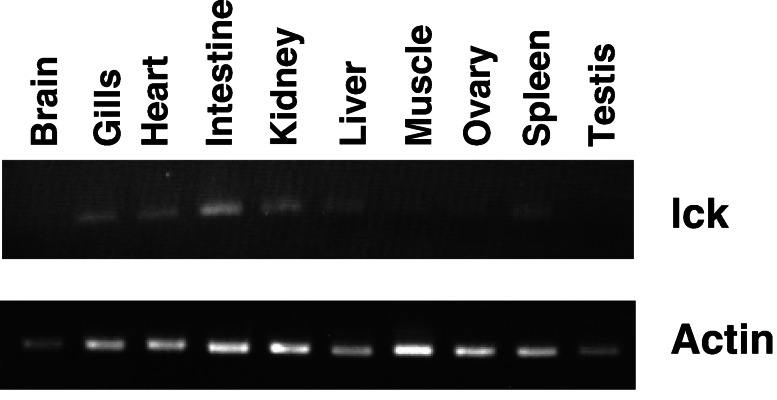
RT-PCR analysis showed that the Fugu lck expresses in the gills, heart, intestine, kidney, and spleen (Fig. 3). Weaker RT-PCR bands were detected in the liver, muscle, and ovary. The expression of Fugu lck in the kidney, spleen, and intestine is consistent with the lymphocyte-specific expression of lck in mammals. The expression seen in other Fugu tissues such as the gills and heart is presumably because of the high levels of circulating lymphocytes. The mouse lck gene also expresses in the peripheral tissues (Fig. 4).
Figure 3.
Expression pattern of lck in Fugu. Fugu lck sequence was amplified by RT-PCR using primers complementary to exons 1 and 3. Cytoplasmic actin (coding exons 2 and 3) was amplified as a control for the quality of cDNA.
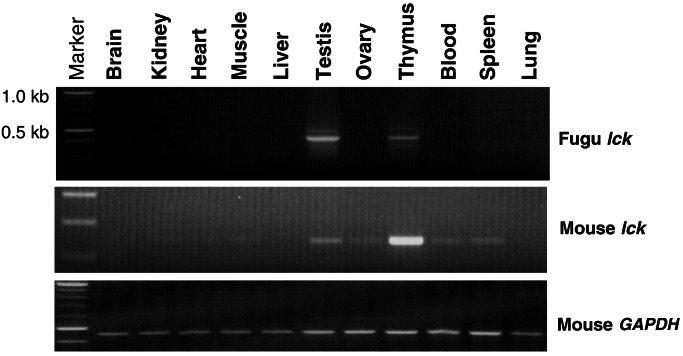
Figure 4.
Expression profile of the Fugu and mouse lck genes in transgenic mice bearing the Fugu lck cosmid 52C1. The Fugu and mouse lck gene fragments were amplified by RT-PCR. The mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was amplified as a control for the quality of cDNA.
Fugu lck-Promoter Elements.
The Fugu lck promoter lacks typical TATA and CAAT box sequences similar to the mammalian lck promoter. We scanned the Fugu-promoter region by using the Transcription Element Search System on http://www.cbil.upenn.edu/tess and identified several consensus sites for the T cell-specific nuclear factor, TCF1-α/LEF1 (5′-CANAG-3′) and a single site for the lymphocyte-specific factor LYF-1 (5′-PyPyTGGGAGPu-3′). Sites for these factors also are found in the proximal promoters of the human and mouse lck (20). Further analysis of the Fugu promoter also identified a G-rich stretch (TGAGGGGGTCACGGTTG) that is similar to the mtβ-binding site present in the proximal promoter of the mouse lck (21). The G-rich stretch in the Fugu lck is present 507 bp upstream of the translation initiation site, a region analogous to the proximal promoter of the mouse lck gene.
To identify conserved potential regulatory elements, we compared 2.3 kb of the Fugu promoter to ≈1 kb each of human proximal and distal promoter; 780 bp of mouse proximal promoter and 4 kb of mouse distal promoter that were available in the public database with a program, comseq.tr. This program is written in a string processing language, TRAC, which has an interpreter written in C. This program allows different length substrings of one string to be searched against the other with variable tolerance for mismatches. In general, only completely matching strings are searched for and additional similarity, if any, discovered by inspection. Our analysis identified an 11-bp element, GGAGGCAGGAA, (376–366 bp upstream of transcription start site) that is conserved in the Fugu promoter, and the distal as well as proximal promoters of both human and mouse. This element is 100% conserved in the human distal and mouse proximal promoters but has a single base mutation in the human proximal (AGAGGCAGGAA) and mouse distal (GGAGACAGGAA) promoters. Because this element is found in all of the vertebrate lck promoters, it is likely to be involved in the regulation of the lck gene. We designate it as putative regulatory element (PRE).
Expression of Fugu lck in Transgenic Mice Bearing Fugu lck Cosmid.
We generated seven independent founder transgenic mice bearing the Fugu cosmid, 52C1. Five of these founders transmitted the transgene to F1 generation. RT-PCR analysis detected expression of the Fugu lck in the thymus and testis from all of the five lines of transgenic mice (Fig. 4). Lower levels of expression were detected in the spleen, blood, heart, or skeletal muscle in some of the lines (data not shown). These results demonstrate that the Fugu lck locus contains conserved elements that direct expression to the lymphoid organs of mice.
Expression of Fugu lck-Promoter Driven EGFP Gene in Transgenic Mice.
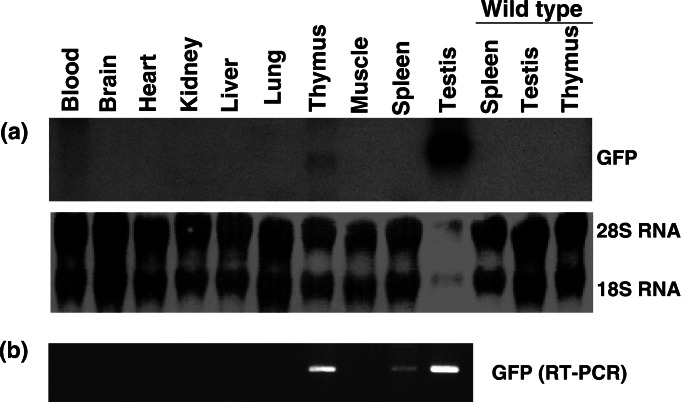
To define the boundaries of the Fugu lck promoter that directs expression to the mouse lymphoid organs, we generated transgenic mice bearing EGFP-constructs linked to 4 kb (Flck4) or 2.3 kb (Flck2) of the Fugu lck promoter. Construct Flck2 was generated by deleting 1.7 kb from the 5′-end of Flck4. Altogether, six founders bearing Flck4 and two founders bearing Flck2 were generated. Three of the Flck4 founders and both the Flck2 founders showed germline transmission. Northern analysis detected EGFP expression in the thymus and testis in the Flck4 lines and in testis in the Flck2 lines whereas RT-PCR showed expression in the thymus, spleen, and testis from both the lines (Fig. 5). These studies show that the full complement of regulatory elements that mediate lymphoid-specific expression in mice are present within 2.3 kb of the Fugu lck promoter.
Figure 5.
Expression of EGFP-driven Fugu lck promoter in transgenic mice. Representative Northern and RT-PCR analyses of EGFP expression driven by a 4-kb Fugu lck promoter are shown. (a) Northern analysis. Each lane contains ≈25 μg of total RNA with the exception of transgenic testis, which contains only 5 μg of total RNA. The Northern membrane was stained with methylene blue to visualize the 18S and 28S RNA. (b) RT-PCR analysis. The lanes correspond to the tissues shown in a.
Cloning the 5′-ends of the cDNA for the EGFP-Fugu lck promoter constructs revealed that the transcripts in the thymus and spleen from the transgenic mice were initiated from the Fugu distal promoter. In the testis, in addition to the transcripts from the distal promoter, an alternative transcript generated from a proximal promoter located 525 bp upstream of the Fugu lck initiation codon was detected. Interestingly, this transcript includes a 270-bp intron that uses an alternative proximal splice donor site but the same acceptor site as the large 1.4-kb intron (Fig. 1b). These results show that the Fugu lck contains a proximal and a distal promoter similar to the mammalian lck. We failed to detect the alternative transcript initiated from the proximal promoter in the thymus of transgenic mice presumably because of its low abundance in the thymus.
The physiological significance of the high levels of expression of Fugu lck as well as EGFP transcripts directed by Fugu lck promoter in the testis of transgenic mice is not clear (Figs. 4 and 5). We have observed similar high levels of expression of Fugu hypothalamic genes in the testis of transgenic rats (8). The rodent testis appears to be permissive for fish transgenes.
Expression of Fugu lck-Promoter Driven EGFP Gene in Mammalian Cell Lines.
To characterize the Fugu lck-promoter region that is essential for the lymphocyte-specific expression, we analyzed the expression of Fugu-lck promoter–EGFP constructs in various mammalian lymphocyte and nonlymphocyte cell lines. Both Flck4 and Flck2 constructs showed expression only in Jurkat and EL4 cell lines that are derived from T cell lineages (Table 1). RT-PCR analysis showed that the distal promoter was active in Jurkat cells whereas the proximal promoter (found active in the testis of transgenic mice) was active in EL4 cells. To further localize the promoter region, we made shorter constructs by deleting (i) 1.3 kb from the middle of the 5′-UTR intron leaving behind 50 bp each at the 5′- and 3′-ends (Flck2ΔI), and (ii) 200 bp at the 5′-end of Flck2ΔI (Flck2ΔI200). These two constructs showed expression in Jurkat cells but not in 293T cells (Fig. 6). We then transfected construct Flck2ΔI200 into various lymphocyte and nonlymphocyte derived cell lines and confirmed that it expresses specifically in the Jurkat cell line (Table 1).
Table 1.
Expression of EGFP driven by Fugu lck promoter (Flck4, 4 kb; Flck2, 2.3 kb) in various cell lines
| Cell lines | C2C12 | Hela | Huh7 | HT29 | 293T | Jurkat | EL4 | Daudi | Raji |
|---|---|---|---|---|---|---|---|---|---|
| Constructs | |||||||||
| Flck4 | — | — | — | — | — | + | + | — | — |
| Flck2 | — | — | — | — | — | + | + | — | — |
| Flck2ΔI200 | — | — | — | — | — | + | — | — | — |
EGFP under the control of a CMV promoter was used as a positive control. +, expression of EGFP; —, no expression.
Figure 6.
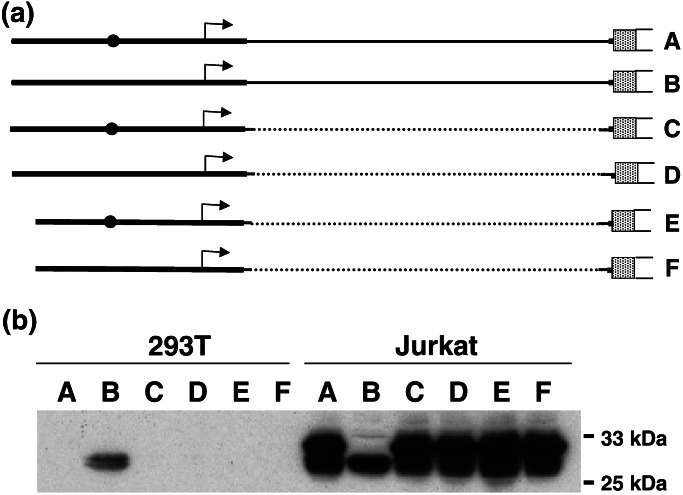
Expression of EGFP driven by Fugu lck promoter. (a) Various constructs tested in Jurkat and 293T cells. The stippled box represents the coding exon (84 bp) of the Fugu lck, and the open box represents the EGFP gene. EGFP is cloned in-frame with the lck exon. The thin line represents the intron in the 5′-UTR. The transcription start site is marked by an arrow, and the conserved 11-bp PRE is shown as a filled circle. A, Flck2; B, Flck2ΔPRE; C, Flck2ΔI (1.3 kb deleted from the 5′-UTR intron); D, Flck2ΔIPRE; E, Flck2ΔI200, and F, Flck2ΔI200PRE. (b) Western blot analysis of EGFP expression. The largest band corresponds to the lck-EGFP fusion protein (31.4 kDa), and the smallest band corresponds to the native EGFP (26.9 kDa). Intermediate size bands are proteins generated from two more ATGs present in-frame between the lck and the EGFP initiation codons. The lck-EGFP fusion protein was generated by transcripts initiated upstream of the intron (shown by arrow), whereas the native EGFP is from the transcripts initiated from a cryptic start site within the multiple cloning site of the EGFP vector. EGFP driven by a cytomegalovirus promoter was used as a positive control.
To determine the role of the 11-bp-conserved element PRE, we deleted this element from constructs Flck2, Flck2ΔI, and Flck2ΔI200 and analyzed their expression in Jurkat and 293T cells. Western analysis revealed that two major proteins, a 31.4-kDa lck-EGFP fusion protein and a 26.9-kDa native EGFP, were expressed from the Flck2 construct in Jurkat cells (Fig. 6). The 5′-RACE analysis showed that whereas the lck-EGFP fusion protein was generated from transcripts initiated from the lck distal promoter, the native EGFP was generated from a cryptic promoter that initiated transcription from the multiple cloning site of EGFP. Deletion of the PRE from the Flck2 construct abolished the expression of the lck-EGFP fusion protein by suppressing transcription but had no effect on the expression of native EGFP initiated from the cryptic promoter. However, deletion of the PRE from shorter constructs that lacked the 1.3-kb intronic sequence had no effect on the expression of either the lck-EGFP fusion protein or the native EGFP (Fig. 6). From these results it appears that, with regard to the lck transcription initiated from the distal lck promoter, PRE antagonizes a suppressor located in the 5′-UTR intron. Intriguingly, deletion of the PRE from Flck2 construct activated the transcription of EGFP transcripts initiated from the cryptic promoter in 293T cells (Fig. 6). The significance of the activation of the cryptic promoter in this kidney cell line is not clear.
Discussion
We have sequenced 60 kb from the Fugu lck locus and shown that it contains 11 protein-coding genes (on average one gene per 6 kb) with compact intergenic regions. A comparison with the human lck locus shows that there has been a genomic rearrangement in this locus after the divergence of the teleost and mammalian lineages. As a result, the Fugu and human lck genes are flanked by different genes in the immediate upstream region. The intergenic distance between the Fugu lck-coding sequence and its immediate upstream gene is only 4.2 kb compared to >80 kb between the human lck-coding sequence and its immediate upstream gene. By transgenic experiments, we have demonstrated that the compact promoter of the Fugu lck contains conserved regulatory regions that mediate lymphoid-specific expression in mice. Indeed, only 2.3 kb of the Fugu promoter is able to direct lymphoid-specific expression in transgenic mice. Furthermore, we also have demonstrated that only 830 bp of the Fugu promoter is required to confer specific expression in cell lines derived from human T cells. These cross-species experiments demonstrate that the molecular mechanisms that mediate T cell-specific transcription are conserved between the teleosts and mammals. This implies that the cis- and trans-elements that are involved in the lymphocyte specific transcription had fully evolved in the common ancestor of teleosts and mammals and have been conserved during 400 million years of evolution.
The present study shows that the Fugu lck promoter is as complex as that of the mammalian lck. Like the mammalian lck gene, Fugu lck contains a proximal and a distal promoter. The Fugu distal promoter is active in the Fugu spleen, kidney, and gills. We also found it to be active in the thymus and spleen from transgenic mice, and in Jurkat cell lines (derived from T cells). We could not ascertain whether the Fugu proximal promoter is active in the thymus (thymocytes) because of the lack of thymus tissue from Fugu. However, we found the Fugu proximal promoter to be active in EL4 cells that are derived from thymocytes. Furthermore, deletion of proximal promoter from the Fugu lck promoter-EGFP construct had no effect on the transcription initiated from the distal promoter in Jurkat cells. These results suggest that the Fugu proximal and distal promoters act independently and are activated at different stages of T cell development similar to the mammalian lck promoters (15, 16). We note that in both Fugu and the mammalian promoters, the proximal promoter is included within an intron that is spliced out in transcripts beginning at the distal promoter, although the topology is slightly different in the two cases. The same structure seems to be present in other genes with dual promoters and with two different 5′-sequences such as the human ST3Gal VI and protein phosphatase 2Cb genes (23, 24). Because there is an intron in the 5′-UTR, there is nothing to prevent its expansion and the distal promoter could be displaced by very large distances from the coding sequence. In the human lck gene, this distance is at least 40 kb. It is possible that some of the distant control elements in mammalian genomes can be accounted for in this way or evolved from such configurations. We also note that in principle, multiple promoter genes can create retropseudogenes that would remain functional, i.e., expressed from a proximal promoter, which would have to be retained in the transcript.
The promoter of the Fugu lck isolated by us is a powerful molecular marker for T cells and hence will be useful for identifying and labeling T cells in fishes. It also is a useful tool in studying the ontogeny, differentiation, and activation of lymphocytes in model fishes such as the zebrafish and medaka. Because the Fugu lck-promoter elements are recognized by the mammalian trans-acting factors, the Fugu promoter also can be used for targeting expression and manipulating T cell activity in mammals. The compact size of the Fugu lck promoter makes it a potential tool in gene therapy for targeting expression of therapeutic agents specifically to T cells. We predict that the compact promoters of the Fugu, which contain the full complement of regulatory elements within a short region, will be more efficient in applications in which a sustained and higher level of expression is preferred.
Acknowledgments
We thank the U.K.-Human Genome Mapping Project Resource Center for providing the Fugu cosmids; Inder Verma for 293T cells; B. H. Tay, S. Tohari, K. C. Lim and Y. Y. Yeoh for technical assistance; Esther Wong and the staff of Institute of Molecular and Cell Biology In Vivo Model System Unit for creating and maintaining transgenic mice colony. B.V. is an adjunct staff member of the Department of Paediatrics, National University of Singapore. This work was supported by the National Science and Technology Board of Singapore.
Abbreviations
- EGFP
enhanced green fluorescent protein
- lck
lymphocyte-specific c-src family protein tyrosine kinase
- RACE
rapid amplification of cDNA ends
- RT
reverse transcription
- UTR
untranslated region
- PRE
putative regulatory element
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF411956).
References
- 1.Bonifer C. Trends Genet. 2000;16:310–315. doi: 10.1016/s0168-9525(00)02029-1. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Nature (London) 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- 3.Venkatesh B, Gilligan P, Brenner S. FEBS Lett. 2000;476:3–7. doi: 10.1016/s0014-5793(00)01659-8. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Proc Natl Acad Sci USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 6.Kimura C, Takeda N, Suzuki M, Oshimura M, Aizawa S, Matsuo I. Development (Cambridge, UK) 1997;124:3929–3941. doi: 10.1242/dev.124.20.3929. [DOI] [PubMed] [Google Scholar]
- 7.Rowitch D H, Echelard Y, Danielian P S, Gellner K, Brenner S, McMahon A P. Development (Cambridge, UK) 1998;125:2735–2746. doi: 10.1242/dev.125.14.2735. [DOI] [PubMed] [Google Scholar]
- 8.Venkatesh B, Si-Hoe S L, Murphy D, Brenner S. Proc Natl Acad Sci USA. 1997;94:12462–12466. doi: 10.1073/pnas.94.23.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camacho-Hubner A, Rossier A, Beermann F. Genesis. 2000;28:99–105. doi: 10.1002/1526-968x(200011/12)28:3/4<99::aid-gene20>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 10.Zapata A G, Chiba A, Varas A. In: Fish Physiology. Iwama G, Nakanishi T, editors. Vol. 15. New York: Academic; 1996. pp. 1–62. [Google Scholar]
- 11.Hansen J D, Zapata A G. Immunol Rev. 1998;166:199–220. doi: 10.1111/j.1600-065x.1998.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 12.Marth J D, Peet R, Krebs E G, Perlmutter R M. Cell. 1985;43:393–404. doi: 10.1016/0092-8674(85)90169-2. [DOI] [PubMed] [Google Scholar]
- 13.Perlmutter R M. Science. 1989;245:344. doi: 10.1126/science.2787933. [DOI] [PubMed] [Google Scholar]
- 14.Veillette A, Bookman M A, Horak E M, Samelson L E, Bolen J B. Nature (London) 1989;338:257–259. doi: 10.1038/338257a0. [DOI] [PubMed] [Google Scholar]
- 15.Wildin R S, Garvin A M, Pawar S, Lewis D B, Abraham K M, Forbush K A, Ziegler S F, Allen J M, Perlmutter R M. J Exp Med. 1991;173:383–393. doi: 10.1084/jem.173.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adler H T, Reynold P J, Kelly C M, Sefton B M. J Virol. 1988;62:4113–4122. doi: 10.1128/jvi.62.11.4113-4122.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garvin A M, Pawar S, Marth J D, Perlmutter R M. Mol Cell Biol. 1988;8:3058–3064. doi: 10.1128/mcb.8.8.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaffin E, Beals C R, Wilkie T M, Forbush K A, Simon M I, Perlmutter R M. EMBO J. 1990;9:3821–3829. doi: 10.1002/j.1460-2075.1990.tb07600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wildin R S, Wang H U, Forbush K A, Perlmutter R M. J Immunol. 1995;155:1286–1295. [PubMed] [Google Scholar]
- 20.Allen J M, Forbush K A, Perlmutter R M. Mol Cell Biol. 1992;12:2758–2768. doi: 10.1128/mcb.12.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada A, Takaki S, Hayashi F, Georgopoulous K, Perlmutter R M, Takatsu K. J Biol Chem. 2001;276:18082–18089. doi: 10.1074/jbc.M008387200. [DOI] [PubMed] [Google Scholar]
- 22.Rouer E, Huynh T V, de Souza S L, Lang M, Fischer S, Benarous R. Gene. 1989;84:105–113. doi: 10.1016/0378-1119(89)90144-3. [DOI] [PubMed] [Google Scholar]
- 23.Anderson S J, Levin S D, Perlmutter R M. Adv Immunol. 1994;56:151–178. doi: 10.1016/s0065-2776(08)60451-4. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi A, Kaneta R, Morishita K, Matsumoto K. Biochem Biophys Res Commun. 2001;287:1148–1156. doi: 10.1006/bbrc.2001.5709. [DOI] [PubMed] [Google Scholar]
- 25.Seroussi E, Shani N, Ben-Meir D, Chajut A, Divinski I, Faier S, Gery S, Karby S, Kariv-Inbal Z, Sella O, et al. J Mol Biol. 2001;312:439–451. doi: 10.1006/jmbi.2001.4967. [DOI] [PubMed] [Google Scholar]