Abstract
Autism is a spectrum of neurodevelopmental disorders with a primarily genetic etiology exhibiting deficits in (1) development of language and (2) social relationships and (3) patterns of repetitive, restricted behaviors or interests and resistance to change. Elevated platelet serotonin (5-HT) in 20%–25% of cases and efficacy of selective 5-HT reuptake inhibitors (SSRIs) in treating anxiety, depression, and repetitive behaviors points to the 5-HT transporter (5-HTT; SERT) as a strong candidate gene. Association studies involving the functional insertion/deletion polymorphism in the promoter (5-HTTLPR) and a polymorphism in intron 2 are inconclusive, possibly because of phenotypic heterogeneity. Nonetheless, mounting evidence for genetic linkage of autism to the chromosome 17q11.2 region that harbors the SERT locus (SLC6A4) supports a genetic effect at or near this gene. We confirm recent reports of sex-biased genetic effects in 17q by showing highly significant linkage driven by families with only affected males. Association with common alleles fails to explain observed linkage; therefore, we hypothesized that preferential transmission of multiple alleles does explain it. From 120 families, most contributing to linkage at 17q11.2, we found four coding substitutions at highly conserved positions and 15 other variants in 5′ noncoding and other intronic regions transmitted in families exhibiting increased rigid-compulsive behaviors. In the aggregate, these variants show significant linkage to and association with autism. Our data provide strong support for a collection of multiple, often rare, alleles at SLC6A4 as imposing risk of autism.
Introduction
In 1943, Leo Kanner published a series of 11 case reports of children with a condition he termed “infantile autism” or “autistic disturbances of affective contact” (Kanner 1943). Autism (MIM 209850) is now recognized as a spectrum of phenotypes that spans a range of clinical severity but fundamentally represents deficits in three domains: (1) development and use of language, (2) development of social relationships and interactions with family and peers, and (3) patterns of repetitive behaviors, restricted interests and activities, and a strong desire to maintain “sameness” in environment and daily routines (reviewed by Folstein and Rosen-Sheidley [2001]). Pharmacotherapies include antipsychotic medications for disruptive and aggressive behaviors and selective serotonin-reuptake inhibitors (SSRIs) for treatment of anxiety, depression, and repetitive behaviors (Cook and Leventhal 1996; Hollander et al. 2005). Of interest regarding the partial efficacy of SSRIs are findings of elevated levels of platelet serotonin (5-hydroxytryptamine [5-HT]) in ∼20%–25% of affected individuals (Schain and Freedman 1961) and correlation with levels in first-degree relatives (Cook et al. 1993). Other findings supporting 5-HT involvement in autism are reviewed elsewhere (Cook and Leventhal 1996; Veenstra-VanderWeele et al. 2000).
Since the first twin study of autism by Folstein and Rutter in 1977 (Folstein and Rutter 1977), evidence has mounted to support a predominantly genetic etiology of autism (reviewed by Folstein and Rosen-Sheidley [2001]). The prevalence of narrowly defined autism is ∼1/1,000 (Chakrabarti and Fombonne 2001), and inclusion of the broader spectrum increases this rate to ∼1/300–1/500 (Fombonne 2003; Yeargin-Allsopp et al. 2003). Males are affected more often, with a male:female ratio of 4:1. Twin data show that MZ twins have an average concordance of ∼60%–70% for classic autism and up to 90% when milder language and social deficits seen in the broader phenotype are considered. This contrasts with DZ twin concordance rates, shown to be 0%–10%, depending on the study. Sibling-recurrence risk in narrowly defined autism is ∼6%–8% (Jones and Szatmari 1988; Ritvo et al. 1989). Modeling the above data has led to estimates of ∼5–15 genes that contribute to genetic risk, possibly involving epistasis (Pickles et al. 1995), and locus heterogeneity (Risch et al. 1999). The data may best be explained by oligogenic inheritance, with different families possessing varying constellations of risk alleles (Folstein and Rosen-Sheidley 2001). A genetic-heterogeneity framework has important implications for the clinical variability observed in autism, in that specific risk loci, or distinct alleles at a given locus, are likely to influence the phenotype differently.
Several groups have undertaken family-based genetic studies to (1) identify regions of the genome commonly inherited by affected family members in multiplex-family samples (i.e., genomic linkage screens) and/or (2) test-specific loci for evidence of common alleles that confer genetic risk on the basis of tests of allelic association (reviewed by Folstein and Rosen-Sheidley [2001] and Veenstra-VanderWeele et al. [2004]). Chromosomal intervals identified in linkage studies with use of either a categorical diagnosis or indexing on specific traits include 7q, 2q, and 17q (International Molecular Genetic Study of Autism Consortium [IMGSAC] 1998, 2001a, 2001b; Ashley-Koch et al. 1999; Collaborative Linkage Study of Autism (CLSA) et al. 1999; Philippe et al. 1999; Risch et al. 1999; Auranen et al. 2000; Buxbaum et al. 2001, 2004; CLSA 2001; Liu et al. 2001; Shao et al. 2002a, 2002b; Yonan et al. 2003; McCauley et al. 2004, 2005; Stone et al. 2004; Cantor et al. 2005). For 17q, second-stage genomic screens with use of different samples (IMGSAC 2001a; Yonan et al. 2003) detected highly suggestive or significant linkage at or near the serotonin transporter (SERT) locus (SLC6A4 [MIM 182138]) and evidence of a sex-restricted pattern of genetic effects (Stone et al. 2004). Our own reports of linkage to this region, with use of family data sets that contain partial overlap with the Autism Genetics Resource Exchange (AGRE) sample (Yonan et al. 2003), lend further support to involvement of this region (McCauley et al. 2004, 2005).
Studies of allelic association at SLC6A4 with autism have focused primarily on a functional, insertion-deletion polymorphism in the promoter (5-HTTLPR) or a variable number tandem repeat (VNTR) marker in intron 2. Results of these studies are inconsistent, with associations shown either to the short (S) or long (L) alleles or absent association (Cook et al. 1997; Klauck et al. 1997; Maestrini et al. 1999; Persico et al. 2000; Tordjman et al. 2001; Yirmiya et al. 2001). Four studies have reported more-comprehensive analyses of common alleles and haplotypes that span the locus, including multiple SNPs (Kim et al. 2002; Conroy et al. 2004; McCauley et al. 2004; Devlin et al., in press). These studies find at least nominal association of the S allele at 5-HTTLPR and other markers or haplotypes. Our study of multiplex families (McCauley et al. 2004) also showed very suggestive linkage that cannot be explained by the modest association of HTTLPR and rs140700 at SLC6A4. The two possible explanations for these results are (1) that SLC6A4 is not the risk locus accounting for linkage or (2) that multiple different alleles at SLC6A4 contribute to genetic risk independently. We report further investigation of chromosome 17 linkage in families with autism and an in-depth analysis of the SLC6A4 gene, testing the hypothesis that allelic heterogeneity may account for the genetic liability to autism. Using a large sample of multiplex families, we find that SLC6A4 exhibits strong evidence of linkage to autism, driven by allele-sharing in males. We find multiple coding and noncoding variants preferentially transmitted to affected individuals, and we identify significant correlations with increased rigid-compulsive behaviors, which indicates that SLC6A4 is a likely susceptibility locus for autism, where allelic heterogeneity supports disease risk.
Subjects and Methods
Sample and Genetic Analyses
The sample for the present study consisted of 73 families recruited from the Tufts-Vanderbilt Consortium and 267 from the AGRE Consortium (table 1). The demographics of these populations have been reported elsewhere (Yonan et al. 2003; McCauley et al. 2005) as have diagnostic inclusion and exclusion criteria for linkage analysis (McCauley et al. 2005). Families were selected on the basis of the presence of a single proband who met full Autism Diagnostic Interview (ADI) criteria for autism and a second sibling who received the diagnosis of autism or presented on the broader spectrum. In the current report, we further examined linkage in a larger sample of 341 families with at least two affected children; for association studies and screening for known variants (e.g., Gly56Ala), we included an additional 43 trios (proband and both parents). As reported elsewhere (McCauley et al. 2005), Tufts/Vanderbilt families were genotyped at deCODE by use of the deCODE 500 marker panel with an average intermarker spacing of ∼8 cM. Two-point and multipoint heterogeneity LOD (HLOD) scores for individual and combined samples, respectively, were calculated with Allegro, under both dominant and recessive models (Gudbjartsson et al. 2000). Disease-allele frequencies were estimated to be 0.01 and 0.1 for dominant and recessive models, respectively. Phenotypic status was considered only for affected individuals, and other family members were designated as having an unknown phenotypic status. Nonparametric allele-sharing LOD* values were calculated using affected relative pair data that was based on an exponential model with use of the Spairs scoring function, as recommended by McPeek (1999). Nonparametric linkage (NPL) scores and corresponding P values were also calculated with Allegro. The position of chromosome 17 markers is based on the deCODE genetic map (Kong et al. 2002).
Table 1.
Autism Family Samples for Linkage and Association Studies
|
No. of Families |
|||||||
| Multiplex |
Trio |
||||||
| Sample | MO | FC | Total | MO | FC | Total | Grand Total |
| AGRE | 156 | 111 | 267 | 11 | 5 | 16 | 283 |
| Tufts/Vanderbilt University | 46 |
28 |
74 |
22 |
5 |
27 |
101 |
| Total | 202 | 139 | 341 | 33 | 10 | 43 | 384 |
Association tests of 5-HTTLPR and rs140700 were performed using the pedigree disequilibrium test (PDT) statistic (Martin et al. 2000), a variant of the transmission/disequilibrium test (TDT), developed for use with general pedigrees. Genotype analysis of these markers has been reported elsewhere (McCauley et al. 2004). Other tests of association or comparison of allele frequencies involved generation of a χ2 statistic and corresponding P values with use of standard 2×2 contingency tables. Comparison of male:female affection for autism in the presence of Gly56Ala alleles was performed using Fisher’s exact test. Comparison of ADI-derived variable cluster scores was performed using the T test—with incorporation of subject numbers, means, and standard deviations for the overall data set and either (1) individuals within a given family or (2) family means across multiple families—to evaluate the significance of score differences. Significance is reported as two-tailed P values. Approval for these studies was granted by the respective institutional review boards at Tufts University School of Medicine/New England Medical Center and Vanderbilt University Medical Center. All studies were performed with the informed consent of the families participating in the research.
Variant Discovery
Known but rare SLC6A4 coding variants reported in previous studies (Cargill et al. 1999; Glatt et al. 2001; Hahn and Blakely 2002; Ozaki et al. 2003) were screened in 327 multiplex families and 57 parent-child trios by use of TaqMan allelic discrimination assays (table 2). When possible, variants were assayed within exons to permit inclusion of in vitro mutagenized plasmid cDNA samples either alone or mixed with wild-type cDNAs, to provide homozygous and heterozygous controls, respectively. ABI TaqMan (Holloway et al. 1999) reactions were performed in a 5-μl volume in accordance with manufacturer’s recommendations (Applied Biosystems). Cycling conditions included an initial denaturation at 95°C for 10 min, followed by 50 cycles of 92°C for 15 s and 60°C for 1 min. Samples were analyzed using an ABI 7900HT Sequence Detection System.
Table 2.
SLC6A4 SNP Marker Information
|
Primers |
Probes |
|||||||||||
| Marker Number | Marker Type | SLC6A4 Region | dbSNPa/Celera Number or Allele | Alleles | MAF | Intermarker Distance(bp) | Forward | Reverse | VIC | FAM | Product Size (bp) | Assay |
| 1 | SNP | Promoter 1A | rs1050565hCV7473213 | T/C | .32 | 10,035 | NA | NA | NA | NA | AoD | |
| 2 | SNP | Promoter 1A | rs2020930 hCV11424041 | G/A | .03 | 1,734 | 5′-GCTCAAGCAGGTGAACAAAGAAA-3′ | 5′-CTGGGCAGCTGGGAAGAG-3′ | 5′-VIC-AACTATTGCTATGCGGTGAT-MGB-NFQ-3′ | 5′-FAM-TTGCTGTGCGGTGAT-MGB-NFQ-3′ | 72 | AbD |
| 3 | Ins/Del | Promoter 1A | 5-HTTLPR | 528(L)/484(S) | .44 | 2,549 | 5′-GGCGTTGCCGCTCTGAATGC-3′ | 5′-GAGGGACTGAGCTGGACAACCAC-3′ | 484/528 | Gel | ||
| 4 | SNP | Intron 1A | rs2020933 hCV11424045 | A/T | .07 | 295 | 5′-TGTATGTATTTTTACCATCAGTTTTGTCCAGAA-3′ | 5′-GAGAGTTAGCTAGCAGGCTCATAAAT-3′ | 5′-VIC-CATTGACCAGGTTCAC-MGB-NFQ-3′ | 5′-FAM-CATTGACCTGGTTCAC-MGB-NFQ-3′ | 81 | AbD |
| 5 | SNP | Intron 1A | rs2020934 hCV7911197 | C/T | .47 | 5 | 5′-TTTTCCTGCCACGCACTCT-3′ | 5′-GCACAAACCTCATAAGAACCTGCTT-3′ | 5′-VIC-ACCGTTCCAATATGG-MGB-NFQ-3′ | 5′-FAM-CCGTTCCAACATGG-MBG-NFQ-3′ | 80 | AbD |
| 6 | SNP | Intron 1A | rs2020935 hCV11424046 | T/A | .07 | 11,477 | 5′-TGGCAGTGACCGTTCCAA-3′ | 5′-TTGCTCAATTTGCACAAACCTCAT-3′ | 5′-VIC-CTGCTTCTCACTCATCCA-MGB-NFQ-3′ | 5′-FAM-TGCTTCTCACTCAACCA-MGB-NFQ-3′ | 68 | AbD |
| 7 | SNP | Intron 1A | rs25528 hCV1841705 | A/C | .16 | 80 | 5′-CCCAGTGGAGGCACAGG-3′ | 5′-GAGTGTGCAGGTTACTGATGCT-3′ | 5′-VIC-TGGTTGGTGTCGCCG-MGB-NFQ-3′ | 5′-FAM-TGGTTGGTTTCGCCG-MGB-NFQ-3′ | 62 | AbD |
| 8 | SNP | Exon 1B | rs6354 hCV1841706 | A/C | .17 | 931 | 5′-GGAGGCAAGGCGACCTT-3′ | 5′-CTGTGGCTAAGCCCCTTGTTATT-3′ | 5′-VIC-CTTGCCCTCTATTGCAG-MGB-NFQ-3′ | 5′-FAM-TTGCCCTCTCTTGCAG-MGB-NFQ-3′ | 58 | AbD |
| 9 | SNP | Exon 2 | Thr4Ala | A/G | 157 | 5′-GTCATTTACTAACCAGCAGGATGGA-3′ | 5′-CGCTGATAGCTGCTTCTGAGA-3′ | 5′-VIC-ATTCAAGGGCGTCGTC-MGB-NFQ-3′ | 5′-FAM-CAAGGGCGCCGTC-MGB-NFQ-3′ | 62 | AbD | |
| 10 | SNP | Exon 2 | Gly56Ala | G/C | .01 | 310 | 5′-GGGTACTCAGCAGTTCCAAGTC-3′ | 5′-GGGATAGAGTGCCGTGTGT-3′ | 5′-VIC-CTGGTGCGGGAGAT-MGB-NFQ-3′ | 5′-FAM-CTGGTGCGGCAGAT-MGB-NFQ-3′ | 56 | AbD |
| 11 | VNTR | Intron 2 | VNTR | 9/10/12 | .02/.39/.59 | 3,269 | 5′-TGGATTTCCTTCTCTCAGTGATTGG-3′ | 5′-TCATGTTCCTAGTCTTACGCCAGTG-3′ | 345/360/390 | Gel | ||
| 12 | SNP | Exon 4 | Lys201Asn | G/C | 40 | 5′-GCTATACTACCTCATCTCCTCCTTCAC-3′ | 5′-TGGTGCAGTTGCCAGTGTT-3′ | 5′-VIC-CCAGGAGTTCTTGCAGC-MGB-NFQ-3′ | 5′-FAM-CAGGAGTTGTTGCAGC-MGB-NFQ-3′ | 83 | AbD | |
| 13 | SNP | Exon 4 | Glu215Lys | G/A | 1,802 | 5′-TCCTGGAACACTGGCAACTG-3′ | 5′-GAATGGAGGGTCCAGGTGATG-3′ | 5′-VIC-AATTACTTCTCCGAGGACAA-MGB-NFQ-3′ | 5′-FAM-AATTACTTCTCCAAGGACAA-MGB-NFQ-3′ | 65 | AbD | |
| 14 | SNP | Intron 5 | rs140700 hCV7473202 | G/A | .07 | 195 | 5′-ACTCCAAGGGTTGTGATCTTTCTG-3′ | 5′-GGGTGAATGGATGTCAGTGTCTTTT-3′ | 5′-VIC-ACCACCTCACCCTCCT-MGB-NFQ-3′ | 5′-FAM-CACCTCGCCCTCCT-MGB-NFQ-3′ | 89 | AbD |
| 15 | SNP | Exon 6 | Ser293Phe | C/T | 505 | 5′-GTGACAGCCACCTTCCCTTATATC-3′ | 5′-GTGGCACCCCTCACCAG-3′ | 5′-VIC-CAGGACAGAAAGGAT-MGB-NFQ-3′ | 5′-FAM-CAGGACAAAAAGGAT-MGB-NFQ-3′ | 56 | AbD | |
| 16 | SNP | Exon 7 | Pro339Leu | C/T | 2,811 | 5′-AGCCGCTCAGATCTTCTTCTCT-3′ | 5′-TTGAACTTGTTGTAGCTAGCAAAAGC-3′ | 5′-VIC-CAAAGCCCGGACCAA-MGB-NFQ-3′ | 5′-FAM-CAAAGCCCAGACCAA-MGB-NFQ-3′ | 75 | AbD | |
| 17 | SNP | Exon 8 | Leu362Met | C/A | 1,504 | 5′-CGGCCCCTTGGGTTTTC-3′ | 5′-GAAGCTCGTCATGCAGTTCAC-3′ | 5′-VIC-CCAGAGATGCCCTGGTGA-MGB-NFQ-3′ | 5′-FAM-CAGAGATGCCATGGTGA-MGB-NFQ-3′ | 68 | AbD | |
| 18 | SNP | Exon 9 | Ile425Val | A/G | 8,181 | 5′-GCAGAAGCGATAGCCAACATG-3′ | 5′-CAAGCCCAGCGTGATTAACATC-3′ | 5′-VIC-TTTCTTTGCCATCATCTT-MGB-NFQ-3′ | 5′-VIC-TCTTTGCCGTCATCTT-MGB-NFQ-3′ | 78 | AbD | |
| 19 | SNP | Exon 13 | Lys605Asn | A/C | 4,687 | 5′-TCCCCACATATATAGCTTATCGGTTGA-3′ | 5′-CAAAACAATTAGTAGTCTGAACACACACA-3′ | 5′-VIC-CACGTACCTCTTTAAAT-MGB-NFQ-3′ | 5′-FAM-ACGTACCTCGTTAAAT-MGB-NFQ-3′ | 105 | AbD | |
| 20 | SNP | Exon 14 | Pro621Ser | C/T | … | 5′-GCGTATTATTAAAAGTATTACCCCAGAAACAC-3′ | 5′-CACAGCATTCAAGCGGATGTC-3′ | 5′-VIC-CACAAGGAATTTCT-MGB-NFQ-3′ | 5′-FAM-CACAAGAAATTTCT-MGB-NFQ-3′ | 73 | AbD | |
From the Single Nucleotide Polymorphism Web site.
Screening for unknown variants involved arbitrary selection of one affected individual from each of 120 families, ranked by family-specific nonparametric LOD* scores from the overall 341-family data set. Screening of PCR products for all exons was performed on the first 24 samples, with temperature-gradient capillary electrophoresis (TGCE) (Li et al. 2002), on a 96-capillary Reveal system, in accordance with manufacturer’s recommendations (Spectrumedix). Putative variants detected by TGCE were confirmed by direct sequencing of PCR product by use of ABI dye terminators in the Center for Molecular Neuroscience Neurogenomics Core. The promoter for the initial 24 individuals and both exons and the promoter for samples from one proband from each of the remaining 96 (of 120) families were analyzed for variation by double-stranded direct sequencing of PCR products with use of ABI dye terminator chemistry. ABI electropherogram data obtained from Vanderbilt Cores or from Polymorphic DNA Technologies were imported and were analyzed for variation with the Phred/Phrap/Consed and PolyPhred suite of sequence analysis tools (Nickerson et al. 1997, 2001; Gordon et al. 2001). Variant confirmation and segregation of rare variants were determined by sequencing available family members and the original proband in the same manner. Location of variation within the gene was documented in table 3 with nomenclature described by den Dunnen and Antonarakis (2001).
Table 3.
TD of Multiple Coding and Noncoding Alleles at SLC6A4[Note]
|
No. of Transmissions to |
No. of Autistic |
|||||||
| Location anddbSNP Number | Positiona | Protein | No. of Families | All Affected Individualsb | All but Probandc | No. of NT | Males | Females |
| Exon 1b: | ||||||||
| ss38318598 | c.147C→A | 5′ UTR | 2 | 4 | 2 | 1 | 3 | 1 |
| Exon 2: | ||||||||
| rs6355 | c.462G→C | Gly56Alad | 7 | 15 | 8 | 5 | 11 | 4 |
| Exon 9: | ||||||||
| ss38318599 | c.1568A→C | Ile425Leu | 1 | 2 | 1 | 0 | 2 | 0 |
| Exon 10: | ||||||||
| ss38318600 | c.1688T→C | Phe465Leu | 1 | 2 | 1 | 1 | 1 | 1 |
| Exon 12: | ||||||||
| ss38318601 | c.1943G→C | Leu550Val | 1 | 2 | 1 | 0 | 2 | 0 |
| Exon 13: | ||||||||
| rs6352 | c.2110A→C | Lys605Asn | 1 | 0 | 0 | 2 | 0 | 0 |
| Exon 14: | ||||||||
| rs13306796 | c.2516A→G | 3′ UTR | 1 | 2 | 1 | 0 | 2 | 0 |
| Promoter: | ||||||||
| ss38318589 | g.15622G→A | NA | 1 | 1 | 0 | 1 | 1 | 0 |
| rs2020932 | g.14519A→T | NA | 3 | 8 | 5 | 0 | 6 | 0 |
| ss38318590 | g.14289A→C | NA | 1e | 1 | 0 | 1 | 1 | 0 |
| rs25533 | g.13912T→C | NA | 1 | 4 | 3 | 0 | 2 | 0 |
| ss38318591 | g.13754C→T | NA | (1)f | (1) | (0) | (1) | (1) | (0) |
| Intron 1a: | ||||||||
| ss38318592 | IVS1a+20C→T | NA | 1 | 1 | 0 | 0 | 1 | 0 |
| ss38318593 | IVS1a+133G→A | NA | 1 | 1 | 0 | 0 | 1 | 0 |
| hcv11414117 | IVS1a−47G→C | NA | 5 | 10 | 5 | 2 | 8 | 2 |
| hcv11414114 | IVS1a−25G→A | NA | 4 | 9 | 5 | 0 | 7 | 2 |
| Intron 1b: | ||||||||
| ss38318594 | IVS1b+28G→A | Gly56Alag | 3 | NA | NA | NA | NA | NA |
| Intron 6: | ||||||||
| ss38318595 | IVS6−44G→C | NA | 1 | 2 | 1 | 0 | 2 | 0 |
| Intron 7: | ||||||||
| ss38318596 | IVS7+83C→T | NA | 4 | 9 | 5 | 0 | 9 | 2 |
| Intron 8: | ||||||||
| ss38318597 | IVS8−33C→T | NA | 2 | 4 | 2 | 0 | 4 | 0 |
| Total | 31 | 76 | 40 | 12 | 63 | 12 | ||
Note.— NA=not applicable.
Changes in the cDNA are indicated relative to the GenBank SLC6A4 reference sequence (accession number NM_001045); genomic variants are designated by +1 corresponding to the initiating ATG or position within an intron.
Transmissions to all affected individuals, including the proband in whom the variant was first identified (T:NT=76:12; χ2=26.82; 1 df; P=2.2×10-7).
Transmissions excluding the screened proband in whom the variant was initially identified (T:NT=39:12; χ2=8.13; 1 df; P=.0042).
Four 56Ala NTs derive from two heterozygous couples transmitting only one 56Ala allele to affected children.
Redundant transmissions are not counted toward total transmissions.
Family does not contribute to linkage; corresponding counts are not included in the totals.
IVS1b+28G→A lies on the Gly56Ala allele; therefore, to avoid redundancy, transmission was not considered.
Functional Analysis of Gly56Ala Activity and Regulation
Epstein-Barr virus (EBV)–transformed lymphocyte cell lines from AGRE or Tufts/Vanderbilt families with autism who carried either the Gly56 or 56Ala allele were obtained from the National Institute of Mental Health (NIMH) Center for Collaborative Genetic Studies on Mental Disorders repository. Lymphocytes were cultured in suspension in RPMI 1640 medium (supplemented with 15% fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin) at 37°C in a humidified incubator at 5% CO2 prior to assay. Lymphocytes were pelleted at 1,500 rpm for 5 min and were washed with Krebs-Ringers-HEPES (KRH) assay buffer. A total of 1×106 cells in triplicate were prewarmed (37°C) in a shaking water bath (10 min) in 12×75 polypropylene tubes in KRH buffer that contained 100 μM pargyline and 100 μM ascorbic acid. After a 5-min incubation with [3H]5-HT (20 nM) at 37°C, uptake assays were terminated by immersion on ice, and uptake in pelleted, 1% SDS-extracted cells was quantitated by scintillation spectrometry. Specific 5-HT uptake was determined by subtracting the amount of [3H]5-HT accumulated in the presence of 10 μM paroxetine (SmithKline Beecham). [3H]L-glutamate transport assays were conducted as described for [3H]5-HT, except with use of 100 nM substrate and definition of nonspecific uptake, with parallel accumulation acquired at 4°C.
Results
Linkage
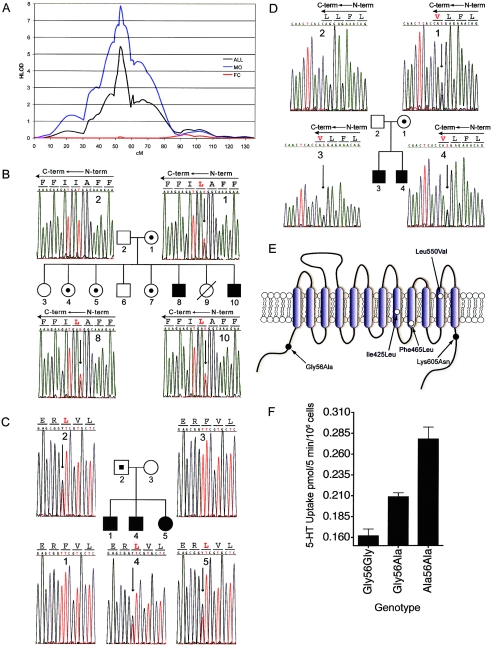
On the basis of our initial linkage results (McCauley et al. 2004) and other reports of linkage to 17q (IMGSAC 2001a; Yonan et al. 2003), we substantially increased our sample of 73 Tufts/Vanderbilt and 85 AGRE families through acquisition from the NIMH repository; we reassessed evidence of linkage in the larger sample that contained 182 additional AGRE families. We found striking linkage in an overall data set of 341 multiplex families, with a peak recessive HLOD (HLODREC) score of 5.8 (fig. 1A) at two adjacent markers at ∼53 cM (D17S1800 [∼1.4 Mb from SLC6A4] and D17S1294 [∼150 kb from SLC6A4]). Results from all parametric and NPL analyses are detailed in table 4. Influenced by two recent reports (Stone et al. 2004; Weiss et al. 2005), we incorporated a sex-specific approach in our linkage analysis of chromosome 17. We queried for male-specific effects by splitting the data set into (1) families containing only affected males (MO) and (2) the remaining female-containing (FC) families, similar to the approach of Stone and colleagues (2004). Linkage increased substantially in the MO sample (HLODREC=8.0; n=202), with negligible contribution from the 138 remaining FC families. The magnitude of linkage overall and in the MO families, extraordinary for a complex behavioral disorder, encouraged us to pursue more-detailed studies to identify autism-associated alleles within SLC6A4.
Figure 1.
Male-biased linkage of autism and novel coding variants at the 17q11.2 SLC6A4 locus. A, Male-biased linkage of autism to 17q11.2. Multipoint linkage analysis on chromosome 17 is shown for the overall 341-family data set (black line), 202 MO families (blue line), or the remaining 138 FC families (red line). HLOD scores were calculated under a recessive model and were plotted as a function of marker position in centimorgans (cM) along chromosome 17. B–D, Sequence detection of novel nonsynonymous SLC6A4 variants in families with autism. Sequence-based detection is shown for each of the three novel coding variants, with corresponding pedigrees. B, Ile425Leu. C, Phe465Leu. D, Leu550Val. Blackened circles or squares reflect individuals with an autism diagnosis, unblackened circles or squares reflect individuals without autism, and allele carriers without autism are indicated by small blackened circles or squares within the larger pedigree symbol. Electropherogram data is shown in either sense (B) or antisense (A and C) orientations, with corresponding coding sequence. Antisense sequences (A and C) indicate the reversed orientation of amino acid codons, represented by lines across each three-base sequence. Variant amino acids are shown in red, and corresponding heterozygous sequence changes are indicated by an arrow. Individual numbers in the respective pedigrees correspond to numbers within each of the sequence frames. E, Schematic representation of the 5-HT transporter. Amino acid substitutions are indicated by location within transmembrane or cytoplasmic domains. F, Dosage-dependent elevated 5-HT transporter activity of Ala56-encoded hSERT in native lymphoblastoid cells. Lymphocytes genotype-matched at 5-HTTLPR (L/L) and the intron 2 VNTR (10/10) and bearing Gly/Gly–, Gly/Ala–, or Ala/Ala–encoding genotypes at residue 56 were assayed for [3H]5-HT transport activity, as described (see the “Subjects and Methods” section). Three independent experiments were performed in triplicate for each line, and the combined basal uptake data were plotted.
Table 4.
Autism Linkage Data for Chromosome 17
| HLOD |
||||||||
| Family Sample | Marker | Positiona(cM) | Dominant | Recessive | LOD* | LOD* Pb | NPL | NPL Pb |
| Overall (n=340) | D17S1800 | 53 | 4.98 | 5.44 | 5.82 | 1.59×10-7 | 4.88 | 6.63×10-8 |
| MO (n=189) | D17S1800 | 53 | 5.35 | 7.86 | 6.65 | 4.85×10-8 | 5.18 | 1.64×10-8 |
| FC (n=138) | D17S1800 | 53 | .4 | .06 | .46 | .085 | 2.21 | .073 |
Position on the deCODE chromosome 17 genetic map.
P values are nominal and are not corrected.
Association
Initially, we tested for evidence of allelic association using the two markers (5-HTTLPR and the intron 5 SNP rs140700) that previously demonstrated modest association in a smaller sample (McCauley et al. 2004). In a sample of 384 combined multiplex and trio families, we detected evidence of allelic association with autism (i.e., transmissions [T]>nontransmissions [NT]) only at the intron 5 marker rs140700 (minor allele T:NT = 93:124; χ2=4.47; P=.03). However, when MO and FC subsets were examined separately, modest association of the S allele of 5-HTTLPR was detected in the MO data set (n=235 families; T:NT=362:314; χ2=4.90; P=.03) as well as persistent undertransmission of the minor allele at rs140700 (T:NT=47:76; χ2=6.84; P=.009), whereas no association or trend toward association was seen in the 149 FC families (HTTLPR-S: T:NT = 223:231, χ2=0.07, P=.79; rs140700: T:NT=46:48, χ2=0.021, P=.88). The association in MO families is consistent with previous results (McCauley et al. 2004) and the male bias in linkage (Stone et al. 2004), but it fails to explain the highly significant linkage in MO families shown above.
Allelic Heterogeneity at SLC6A4
Since common alleles across the SLC6A4 locus do not explain the observed linkage, we considered the hypothesis that multiple, possibly rare, SLC6A4 risk alleles exist and confer risk of autism. We tested our hypothesis using two parallel strategies. We first screened all families for known rare coding variants to determine if one or more is present at elevated frequency in our autism sample and, by inference, is potentially related to disease risk. Our second approach involved (1) selecting unrelated probands from the multiplex families that contribute most to linkage and, at minimum, have a positive LOD score at the linkage peak and (2) screening their SLC6A4 exons and promoter sequences for novel variants. In our combined sample of 384 multiplex families and parent-child trios, we screened for known nonsynonymous variants using TaqMan allelic discrimination assays. We detected multiple individuals who carried one or two copies of the 56Ala-encoding allele and a single subject heterozygous for an allele encoding the nonsynonymous variant Lys605Asn.
Detailed analysis of families with the Gly56Ala substitution reveal its presence in both “linked” (positive LOD scores) and “unlinked” multiplex families and trios. Within the 120 families with the highest family-specific LOD scores, we found the 56Ala allele present on 11 (2.3%) of 480 independent chromosomes. Three homozygous 56Ala subjects were identified in these families. In contrast, Glatt et al. (2001) reported the 56Ala variant occurred on only 4 of 900 chromosomes in the only large nonclinical comparison sample described, with a minor-allele frequency (MAF) of 0.44% and no homozygous subjects. In our remaining families with autism, we found a lower frequency of the 56Ala allele, with 12 (1.1%) of 1,056 chromosomes harboring the variant; no homozygotes were identified for this group. An alternative comparison comes from our screening of a predominantly white population ascertained for Axis I Mood Disorders. Here, we identified 3 (1.1%) of 272 chromosomes with 56Ala alleles (J. R. Field, H.C.P., R.D.B., E. Sanders-Bush, and R. C. Shelton, unpublished findings).
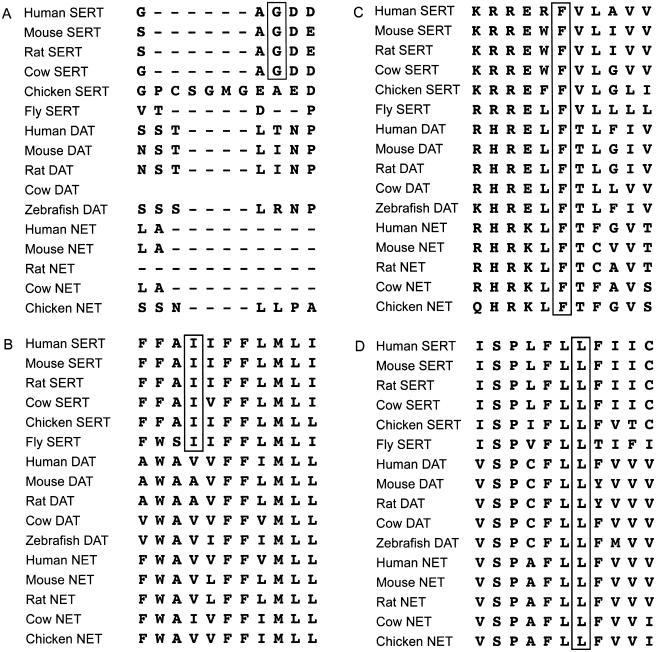
A parallel strategy involved screening the promoter, exons, and flanking sequence of unrelated probands from, initially, 24 families who most contributed to linkage in this region, on the basis of ranking for family-specific LOD scores. TGCE followed by direct sequencing of PCR products comprised the initial effort, and one proband sample from each of the remaining 96 (of 120) families was subjected to direct sequencing of PCR products. In addition to the independent identification of one 56Ala homozygote, two novel coding variants (Ile425Leu and Leu550Val) (fig. 1B and 1D) were detected in the 24 most-linked families (4/48=8.3% allele frequency). In the remaining 96 families, several 56Ala alleles and a Phe465Leu nonsynonymous substitution (fig. 1C) were detected (table 3). All three coding substitutions occur within transmembrane domains (fig. 1E) and are highly conserved (fig. 2). The least conserved of the novel variants corresponds to residue Ile425, which is present within all SERT proteins from human to Drosophila. The other two nonsynonymous variants occur at residues conserved from human to Drosophila in all monoamine transporters—including SERT, the norepinephrine transporter (NET), and the dopamine transporter (DAT)—for which sequence was available.
Figure 2.
ClustalW alignment of hSERT amino acid sequence versus other species of SERT and other biogenic amine transporters. Gly56 is conserved in mammalian species of SERT (A). Ile425 is conserved in all SERT proteins from human to Drosophila (B). Phe465 is conserved in all species shown for all monoamine transporters and is also conserved in glycine, γ-aminobutyric acid, and creatine transporters (C). Leu550 is conserved in all monoamine transporters from human to Drosophila (D).
To consider the genetic relevance of the coding variants to autism risk, we asked if these variants segregated with disease in each family. Two of the three novel coding variants were transmitted to all affected individuals (5 males and 1 female) in the three families (fig. 1B–1D). One affected male did not inherit the paternally transmitted Phe465Leu variant allele. The 425Leu allele exhibited a particularly intriguing overall segregation pattern (fig. 1B). This variant was maternally transmitted to both affected sons. Three unaffected daughters also inherited the allele, but two other unaffected siblings, one male and one female, did not. When the clear sex bias in linkage is considered, segregation of the 425Leu allele in this family is consistent with but not proof of male-biased genetic risk or elevated penetrance associated with the allele. No unaffected siblings were present in the other two families. The 56Ala allele was detected in seven linked families, in which the T:NT was 15:5 (table 3). Of the five nontransmissions, four correspond to two distinct instances in which both parents were carriers (expected to occur in ∼1 of every 2,000 couples, under the assumption of Hardy-Weinberg equilibrium [HWE]), and offspring received only one Ala56 allele. The other nontransmitted 56Ala allele was present in the mother of the family that paternally transmitted the Phe465Leu variant. Of the three homozygous individuals in the 120 linked/screened families, two were affected male offspring and one was a mother for whom medical history information was unavailable but who transmitted the allele to two affected male offspring. Analysis of the unlinked multiplex and simplex families with a 1.1% (12/1056) 56Ala frequency did not show bias in transmission (data not shown). There was a significant effect in a larger 643 multiplex/trio data set for autism in males carrying one or two 56Ala alleles compared with females (23:14 affected:unaffected males vs. 7:16 affected:unaffected females; P=.016).
Clinical Correlations
To explore phenotypic correlates with the coding variants, we compared scores for trait subsets of autism that were based on ADI-Revised (ADI-R)–derived variable clusters (table 5) identified elsewhere from a principal-components analysis of ADI-R items (Tadevosyan-Leyfer et al. 2003). These clusters reflect (1) language, (2) social intent, (3) developmental milestones, (4) savant skills, (5) rigid-compulsive, and (6) sensory-aversion aspects of the autism phenotype. There was a significant increase in rigid-compulsive behaviors associated with the novel variants (P=.0003) (table 5), and the effect was most pronounced for the Ile425Leu and Leu550Val families. The Ile425Leu substitution tracked with more-severe language deficits (P=.0031), although the brothers harboring the allele were generally more affected across most factor domains, with the exception of sensory aversions. The Leu550Val and Phe465Leu variants were associated with lesser impairment in language and social-intent domains, significantly so for the Phe465Leu variant (P<.0001 and P=.0021, respectively). The Gly56Ala variant (heterozygous or homozygous) in the linked families similarly demonstrated a significant association with more severe rigid-compulsive behaviors (P=.0085). When all linked families with individuals carrying a coding variant were considered together, the significance in elevated severity of rigid-compulsive behaviors increased (P=.0002). No consistent pattern was observed across all Gly56Ala families, regardless of sex, for other ADI clusters; however, there appeared to be a trend toward two subgroups. In one group, patients had greater severity for rigid-compulsive and sensory-aversion behaviors, with fewer impairments in language and social domains. The second group did not show the sensory-aversion finding but was more impaired in language (table 5).
Table 5.
ADI Cluster Scores for Families with SLC6A4 Coding Variant[Note]
|
Mean for Sample |
All Novel Variants |
Mean for Carriers with |
All G56A Carriers |
Male Gly56Ala Carriers |
All Coding |
||||||||||||||||
| ADI Cluster | No. of Subjects | Total | Ile425Leu | Leu550Val | Phe465Leu | Mean | P | No. ofSubjectswithG56A | G56A-1 | G56A-2 | G56A-3 | G56A-4 | G56A-5 | G56A-6 | Mean | P | No. ofSubjects | Mean | P | Mean | P |
| Language | 770 | .457 ± .268 | .838a | .314 | .231b | .461 ± .329 | NS | 13 | .647 | .378 | .552 | .207 | .378 | .436 | .462 ± .156 | NS | 9 | .541 ± .297 | NS | .442 ± .205 | NS |
| Social Intent | 770 | .439 ± .228 | .605c | .325 | .200d | .377 ± .207 | NS | 13 | .564 | .164 | .314 | .454 | .165 | .344 | .334 ± .158 | .075 | 9 | .444 ± .203 | NS | .370 ± .169 | .095 |
| Milestones | 758 | .738 ± .068 | .809 | .740 | .763 | .771 ± .035 | .052 | 13 | .723 | .734 | .750 | .780 | .757 | .759 | .742 ± .026 | NS | 9 | .754 ± .056 | NS | .757 ± .026 | .0069 |
| Savant Skills | 730 | .124 ± .150 | .000 | .083 | .028 | .037 ± .042 | .0031 | 13 | .088 | .671 | .111 | .033 | .671 | .109 | .215 ± .238 | NS | 9 | .150 ± .157 | NS | .199 ± .270 | NS |
| Rigid Compulsive | 747 | .288 ± .209 | .402 | .428 | .351 | .394 ± .039 | .0003 | 13 | .317 | .412 | .210 | .497 | .413 | .606 | .409 ± .138 | .0085 | 9 | .343 ± .138 | NS | .404 ± .11 | .0002 |
| Sensory Aversion | 708 | .331 ± .269 | .060 | .245 | .340 | .215 ± .142 | .08 | 11 | .305 | .667 | .319 | .667 | .667 | NA | .525 ± .195 | .0005 | 7 | .353 ± .272 | NS | .409 ± .231 | NS |
Note.— Values shown in bold italics are statistically significant (P<.05) in comparison with overall means. NS=not significant.
Ile425Leu-Language mean=.838±.34; P=.0031.
Social Intent mean=.605±.026; P=.011.
Phe465Leu-Language mean=.231±.0035; P<.0001.
Social Intent mean=.200±.06; P=.0021.
Functional Properties of Gly56Ala SERT
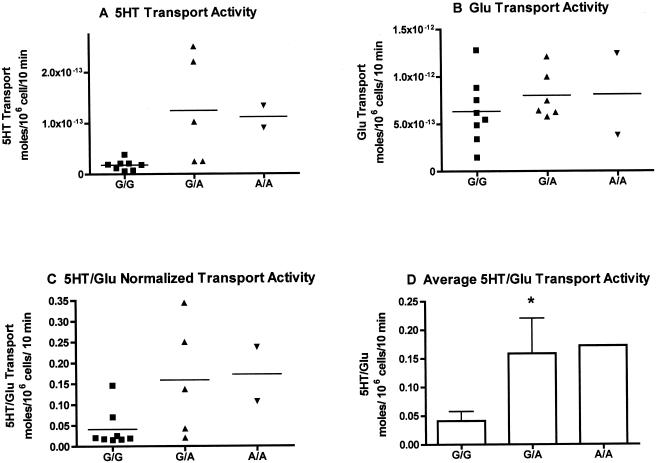
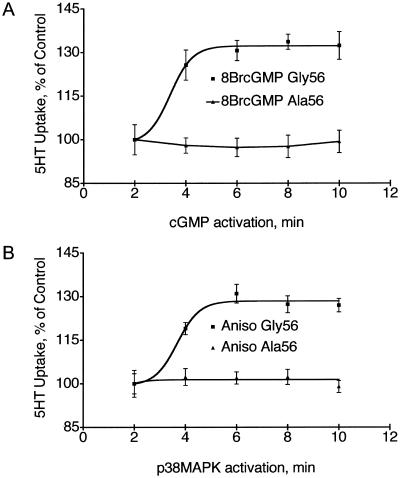
The presence of multiple homozygous 56Ala subjects in our multiplex autism sample allowed evaluation of the functional impact of a 56Ala substitution on SERT activity and regulation by use of genotyped, EBV-transformed lymphocytes, since SLC6A4 is natively expressed in those cells (Khan et al. 1996; Lesch et al. 1996; Faraj et al. 1997). Basal 5-HT transport activity was elevated in Ala56-expressing cells (T test, P<.05) compared with Gly56 homozygous, Gly56Ala heterozygous, or combined Gly56 genotypes; significance remains after normalization to L-glutamate transport activity and does not segregate with the SLC6A4 genotype (fig. 3). To control for potential confounding effects of variation in SLC6A4 gene expression and downstream basal 5-HT uptake associated with 5-HTTLPR and intron 2 VNTR genotypes (Lesch et al. 1996; Ogilvie et al. 1996; MacKenzie and Quinn 1999), we repeated studies with two cell lines of each Gly56Ala genotype that carried identical 5-HTTLPR (L/L) and intron 2 VNTR (10/10) genotypes. Figure 1F demonstrates a 56Ala dosage-dependent effect on basal 5-HT transport activity, with ∼75% increase evident for the 56Ala homozygous lines as compared with lines homozygous for 56Gly. In these cells, we also demonstrated that the 56Ala allele is refractory to regulation by acute application of activators of protein kinase G (PKG) or p38 mitogen-activated protein kinase (MAPK) (Miller and Hoffman 1994; Qian et al. 1997; Ramamoorthy and Blakely 1999; Zhu et al. 2004, 2005; Samuvel et al. 2005) (fig. 4A and 4B), which suggests that intrinsic features of function unlikely to be attributable to a linked, noncoding variant are evident. We recently described a similar loss of regulation despite normal surface density for 56Ala cDNA transfected into HeLa cells (Prasad et al., in press).
Figure 3.
Altered basal 5-HT transport activity and regulation associated with the Gly56Ala variant. [3H]5-HT (5HT) and [3H]L-glutamate (Glu) transport activities were defined as described in the “Subjects and Methods” section. A, 5-HT transport activity of EBV-transformed lymphocyte cell lines by genotype (G=Gly56; A=Alanine56). B, L-glutamate transport activity of the lines tested in panel A. C, Normalized 5-HT transport activity, with an individual cell lines 5-HT transport activity divided by its L-glutamate transport activity. D, Average by genotype of the values shown in panel C. An asterisk (*) denotes P<.05 (student’s t test) compared with Gly56 cell lines.
Figure 4.
SERT is refractory to regulation through PKG and p38 MAP kinase signaling pathways. A, SERT Ala56 lacks sensitivity to 8BrcGMP. Homozygous Ala56 cells (106/tube) were preincubated for various lengths of time at 37°C, with 10 μM 8BrcGMP prior to [3H]5-HT transport assays. B, SERT Ala56 lacks sensitivity to the p38 MAPK activator anisomycin; 8BrcGMP effects on Gly56 SERT are completely blocked by coincubation with the PKG inhibitor H8 (10 μM), whereas the p38 MAPK inhibitor SB203580 (1 μM) prevented anisomycin stimulation (data not shown). Data plotted represent mean data ± SD (n=3) for a single cell line of each genotype assayed in parallel. Findings were replicated with identical results in an additional line for each genotype.
Association of Heterogeneous Alleles with Autism
To more completely evaluate the SLC6A4 locus for novel alleles, we expanded our screen to cover proximal promoter sequences and noncoding 5′ and 3′ exons. We subsequently analyzed 31 families for allelic segregation patterns for rare, noncoding variants discovered across the locus. Variants were identified in the promoter, exon 1b, exon 14, and intronic sequences in the linked families (table 3). Several variants are known polymorphisms with existing database identifiers, although many are novel. Other polymorphisms with modest-to-high MAF were tested elsewhere for association and are not included in table 3. Allelic segregation patterns reveal a stark pattern of transmission disequilibrium (TD) when all variants are combined for purposes of considering segregation. Redundant allelic transmissions represented by two or more SNPs were not counted more than once. TD was evident from a T:NT count of 76:12, which represents a highly significant deviation from the null hypothesis of no association (χ2=26.82; 1 df; P=2.2×10-7). There is an a priori expectation of transmission (or de novo sequence change) to index cases in whom variants were originally detected. To reduce this bias, transmissions to these individuals were excluded, yet segregation remained significant (T:NT=40:12; χ2=8.13; 1 df; P=.0042). Therefore, these multiple coding and noncoding alleles persist in demonstrating a collective linkage to and allelic segregation with autism, despite failure to identify association of common alleles (apart from HTTLPR and rs140700) at SLC6A4 in these families.
Discussion
Two recent reports are notable for leveraging the sex bias of disease affection in autism to define an analytical framework that identified significant male-biased genetic effects at 17q. The AGRE Consortium reported a genomewide analysis with use of sex (as presented here) to stratify a 257-family data set (Stone et al. 2004), and they have recently replicated evidence for male-biased linkage to 17q (Cantor et al. 2005). They compared overall linkage with that of MO families and of FO families. They found a significant effect at 17q11.2 near the SLC6A4 locus (∼53 cM), with MO families showing an empirically significant increase of linkage from a maximum LOD score (MLS) of 3.2 in the overall data set to an MLS of 4.3 in the 148-family MO data set. Our analyses, which include AGRE and Tufts/Vanderbilt families, confirm these previous observations by the AGRE Consortium yet show more-pronounced overall and male-biased genetic effects at this locus. In the second report, Weiss et al. (2005) showed significant association of marker alleles at SLC6A4 and the 17q31 integrin β3 (ITGB3 [MIM 173470]) locus, with male-specific genetic influences on 5-HT levels. ITGB3 was detected earlier by Weiss et al. (2005) as a QTL for circulating serotonin levels. Our own recently published genomic linkage screen showed two adjacent linkage peaks on proximal 17q (McCauley et al. 2005). Conditional linkage analyses revealed locus- or peak-specific effects, which supports the premise that both SLC6A4 at 53 cM and another more telomeric locus (possibly ITGB3) contribute to genetic effects in this region. We speculate on the basis of published linkage patterns—including the AGRE replication study that showed linkage (MLS=3.6) to a more distal site in 17q21 (Cantor et al. 2005)—that overall linkage in this region represents two loci, with the likely stronger effect at SLC6A4.
We were struck by the extraordinary evidence of linkage, given that common alleles and haplotypes fully representing LD across the locus failed to explain the linkage, as would be predicted under the “common disease-common allele” hypothesis. Given the above observations, we considered the alternative hypothesis that allelic heterogeneity explained risk at SLC6A4. We have discovered three novel highly conserved coding variants in families that strongly contribute to linkage in this region. Two of the three novel coding variants (Ile425Leu and Leu550Val), a 56Ala homozygous proband, and a novel intron 6 SNP (IVS6-44G→C) were identified among 24 probands (8.3% allele frequency) from families with the highest family-specific LOD scores. These two novel alleles segregate to all affected individuals in their respective families and thus are associated with disease. These findings spawned further discovery efforts that led to the identification of 13 novel SNPs, including the Phe465Leu nonsynonymous variant. The family with this latter variant is of interest because the mother carried but did not transmit the 56Ala allele. She did transmit a rare genomic variant (hCV11414114) (table 3) to all affected children. The study by Glatt and colleagues (2001) did not detect the three novel coding alleles in the 450 individuals (900 chromosomes) sequenced; this indicates that these alleles are rare and have a frequency of <1/900, or <0.11%.
Cross-species conservation of novel coding-variant residues and their identification in linked families suggests an increased likelihood of altered functionality for the variant transporter, although future study will be required to fully elaborate this premise. All three novel substitutions occur within transmembrane domains (fig. 1E), and the 425Leu allele affects the identical residue and nucleotide—as does the Ile425Val mutant in the pedigrees described by Ozaki and colleagues (2003)—that segregates Asperger syndrome (MIM 608638), obsessive-compulsive disorder (OCD [MIM 164230]), and other psychiatric phenotypes. The Ile425Leu variant does, therefore, have an a priori increased likelihood of functional effect that is based on prior precedent from disease association and subsequent functional characterization of the Ile425Val substitution showing ∼2-fold elevated basal activity in transfection studies (Kilic et al. 2003; Prasad et al., in press).
The Gly56Ala substitution shows a very suggestive increase of MAF to 2.3% in the 120 linked families, compared with the 1.1% seen in the remaining families. A 2.3% allele frequency represents a noticeable increase over the single nonclinical reference study (Glatt et al. 2001), which shows 4 of 900 (0.44%) chromosomes (and no homozygotes) carrying a 56Ala allele. HWE would dictate a frequency of homozygous individuals to be ∼1/2,000 in unrelated individuals (or ∼1/5,000 on the basis of a 0.44% frequency in the Glatt et al. [2001] comparison sample). Therefore, our finding of three homozygotes and two additional (unrelated) instances of dual heterozygous couples is highly unlikely by chance (P≈0). The findings of (1) an apparent fivefold increase in 56Ala allele frequency in the linked families, with a trend toward overtransmission, (2) a substantial deviation from HWE, and (3) a male-biased trend toward autism affection in the presence of this allele compared with females indicates a role for the 56Ala allele as a genetic risk factor in autism. The infrequency of the allele, however, makes this more difficult to quantitate. Larger autism population studies are indicated.
Equally important are functional data that demonstrate that the 56Ala SERT protein displays an elevated basal activity and insensitivity to regulation through PKG and p38 MAPK signaling pathways in a native cell system. Importantly, down-regulation in response to protein kinase C–activating phorbol esters was equivalent between Gly56 and Ala56 lines (data not shown). We do not believe that PKG/p38 MAPK regulatory insensitivity derives directly from altered basal activities, since we have demonstrated transport regulation through these pathways with other variants that differed up to 10-fold in activity (Prasad et al., in press), although over-expression in transfected cells can blunt regulation (C.-B. Zhu and R.D.B., unpublished findings). Elevated basal 5-HT uptake is also not due to enhanced transcription, since RealTime PCR analysis revealed equal SERT mRNA levels in the Gly56 and Ala56 lines (data not shown). Elevated basal activity of the Ala56-encoded transporter is intriguing in light of a similar effect with 425Val-encoded SERT in patients with Asperger syndrome and OCD. Although altered regulation was not seen with the Ile425Val mutation, the elevated basal transporter activity may suggest a common functional mechanism important in autism, OCD, and other phenotypes. It is possible that prolonged expression of a variant lacking regulation induces other compensatory changes that influence basal transporter activity. Regardless, these findings suggest that Gly56Ala carriers may possess an inflexibility to regulatory stimuli that could thereby compromise appropriate demand-dependent modulation of SERT surface expression and/or catalytic activity.
Phenotypically, the three novel variants and Gly56Ala are significantly associated with increased rigid-compulsive behaviors. These include (1) stereotyped utterances, (2) unusual preoccupations, (3) compulsions/rituals, (4) resistance to trivial changes in the environment, and (5) unusual attachment to objects (Tadevosyan-Leyfer et al. 2003). This is important for several reasons. Our previous study of linkage in a smaller sample demonstrated a significant increase in linkage at ∼53 cM when the same rigid-compulsive trait was used to stratify the data set (McCauley et al. 2004). It is consistent with the finding of Asperger syndrome, OCD, and other traits in families segregating the functionally abnormal (Prasad et al., in press) Ile425Val substitution (Ozaki et al. 2003). Obsessive-compulsive–type traits and clinical OCD are seen more frequently in families with autism than in the general population (Bolton et al. 1998). Repetitive behaviors and associated anxiety in autism and OCD are often effectively treated with SSRIs, targeting the SERT protein (Hollander et al. 2005). Whereas other phenotypic findings associated with specific variants were variable across families with all four variants, increased severity for the rigid-compulsive domain was a consistent finding for these variants. Given the magnitude of the observed genetic effect, the possibility exists that our sample may be enriched by chance or selection methods for phenotypic traits (or regional alleles) that bias in favor of an effect at this locus.
The functionality of promoter and other noncoding variants is unknown; however, the segregation of these multiple variants in aggregate, in addition to the coding variants, provides additional genetic evidence of an allelic heterogeneity framework for disease risk involving SLC6A4. Several previously documented variants (rs2020932, hCV11414117, and hCV11414114) were each found in multiple families. These heterogeneous variants arose largely on independent haplotypic backgrounds, which indicates that they are rare independent events and are not the result of an effect of some common genetic background. The clustering of collectively associated rare genomic variants in the promoter and near the 5′ end of the gene raises the possibility of transcriptional effects at SLC6A4. Intronic or noncoding transcribed variants may, if they are risk factors with biological relevance, affect transcription, transcript stability, or RNA splicing (Pagani and Baralle 2004). An expression-based mechanism for potentially disease-related 5′ variants is consistent with association of the HTTLPR marker, since the insertion/deletion variant exhibits differential transcription. Even with a relatively conservative analysis of transmission by eliminating the subject from each family in whom the variant was discovered, association data in the presence of linkage must be interpreted cautiously. The analysis of transmissions in families with an a priori expectation of variable allele-sharing would inflate any measure of allelic association, and the TDT analysis of multiplex families in this unusual context would yield a measure of linkage (Spielman and Ewens 1996). Nevertheless, the context in which these observations are made supports the significance of association in the aggregate of these multiple variants. The modest association at 5-HTTLPR and the intron 5 SNP rs140700 in the current MO sample is not only consistent with but is also supportive of this proposed risk framework. Other common alleles are not associated with autism, whereas the heterogeneous rare alleles described in this report are. Thus, the most parsimonious model involves multiple different risk alleles (including 5-HTTLPR) that act in different families to collectively account for the observed linkage.
The import of our data is summarized by the constellation of findings, including (1) three novel highly conserved coding variants, one of which affects a residue (Ile425) with known phenotypic and functional relevance; (2) the dysfunctional properties of SERT encoded with the Gly56Ala substitution; (3) the stark deviation from HWE and increased frequency in linked families; (4) a phenotypic correlation between coding variants and increased rigid-compulsive behaviors; and (5) the aggregate association shown by heterogeneous promoter 5′ and intragenic noncoding variants. These data collectively support the premise that SLC6A4 represents a susceptibility locus for autism-spectrum disorders. Our findings must be examined in larger independent autism family populations with similar phenotypic representation to determine their ultimate significance. Given the linkage reported at SLC6A4 by IMGSAC (as well as AGRE) and incomplete screening in the current sample (only one affected individual per family was screened for exon and promoter variants), we suspect that additional coding and noncoding variants will be discovered at this locus. As they stand now, our findings provide compelling evidence that the SLC6A4 locus is a bona fide autism-susceptibility gene, with variants predisposing to rigid-compulsive traits.
Acknowledgments
This work was supported, in part, by National Institutes of Health (NIH) grants MH61009 (to J.S.S.), MH55135 (to S.E.F.), and DA07390 (to R.D.B.); Vanderbilt Kennedy Center Hobbs Research Awards (to J.S.S. and R.D.B.); a Family Research Award (to J.S.S.); generous support from the National Alliance for Autism Research (to J.S.S.); and a Pre-Doctoral fellowship (J.L.M.). We thank Jonathan L. Haines for helpful discussions and evaluation of the manuscript. We acknowledge the important contributions to this work by the staff at the Vanderbilt Center for Human Genetics Research, including the DNA Resources, Data Analysis, Family Ascertainment, and Bioinformatics Cores, and the Center for Molecular Neuroscience Neurogenomics Core. Some of this work was also supported by Vanderbilt General Clinical Research Center grant M01 RR-00095, NIH Center for Research Resources. We especially acknowledge the families that have participated in this research and resources provided by the NIMH Center for Collaborative Genetic Studies on Mental Disorders repository, without which this work would not have been possible.
Biomaterials and phenotypic data provided through the NIMH Autism Genetics Initiative came from three projects: (1) Data and biomaterial collection was supported by NIH grant MH55135 (“Collaborative Linkage Study of Autism”) to S.E.F., and her key Clinical and Phenotypic Coordinators were Brian Winklosky and Beth Rosen-Sheidley, M.S., C.G.C. Coinvestigators included J.S.S. and Jonathan L. Haines. (2) Data and biomaterial collection was also supported by a supplement to NIH grant MH61009 (“Molecular Genetics of 15q11-q13 Defects in Autism”) and by Development Funds from the Vanderbilt Centers for Human Genetics Research and the Kennedy Center for Research on Human Development (to J.S.S.). The clinical and phenotypic coordinator for this project was Genea Crocket, M.S. (3) Data and biomaterial collection from the AGRE collection was supported by NIH grant MH64547 to Daniel H. Geschwind, M.D., Ph.D. (University of California at Los Angeles [UCLA]) and the Cure Autism Now Foundation. Coprincipal investigators include Stanley F. Nelson, M.D., and Rita Cantor, Ph.D. (UCLA) and Christa Lese Martin, Ph.D., and T. Conrad Gilliam, Ph.D. (University of Chicago). Coinvestigators include Maricela Alarcón, Ph.D., Kenneth Lange, Ph.D., and Sarah J. Spence M.D., Ph.D. (UCLA), David H. Ledbetter Ph.D. (Emory), and Hank Juo, M.D., Ph.D. (Columbia). Scientific oversight of AGRE is provided by the AGRE steering committee (chair: Daniel H. Geschwind, M.D., Ph.D.; members: W. Ted Brown, M.D., Ph.D., Maja Bucan, Ph.D., Joseph Buxbaum, Ph.D., T. Conrad Gilliam, Ph.D., David Greenberg, Ph.D., David Ledbetter, Ph.D., Bruce Miller, M.D., Stanley F. Nelson, M.D., Jonathan Pevsner, Ph.D., Carol Sprouse, Ed.D., Gerard Schellenberg, Ph.D., and Rudolph Tanzi, Ph.D.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SLC6A4 [accession number NM_001045])
- NIMH Center for Collaborative Genetic Studies on Mental Disorders, http://www.nimhgenetics.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autism, SLC6A4, ITGB3, Asperger syndrome, and OCD)
- Single Nucleotide Polymorphism, http://www.ncbi.nlm.nih.gov/SNP/ (for dbSNP numbers ss38318598, ss38318599, ss38318600, ss38318601, ss38318589, ss38318590, ss38318591, ss38318592, ss38318593, ss38318594, ss38318595, ss38318596, and ss38318597)
References
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 [DOI] [PubMed] [Google Scholar]
- Auranen M, Nieminen T, Majuri S, Vanhala R, Peltonen L, Jarvela I (2000) Analysis of autism susceptibility gene loci on chromosomes 1p, 4p, 6q, 7q, 13q, 15q, 16p, 17q, 19q and 22q in Finnish multiplex families. Mol Psychiatry 5:320–322 [DOI] [PubMed] [Google Scholar]
- Bolton PF, Pickles A, Murphy M, Rutter M (1998) Autism, affective and other psychiatric disorders: patterns of familial aggregation. Psychol Med 28:385–395 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman J, Keddache M, Smith CJ, Hollander E, Ramoz N, Reichert JG (2004) Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol Psychiatry 9:144–150 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor RM, Kono N, Duvall JA, Alvarez-Retuerto A, Stone JL, Alarcón M, Nelson SF, Geschwind DH (2005) Replication of autism linkage: fine-mapping peak at 17q21. Am J Hum Genet 76:1050–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 22:231–238 [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Fombonne E (2001) Pervasive developmental disorders in preschool children. JAMA 285:3093–3099 [DOI] [PubMed] [Google Scholar]
- Collaborative Linkage Study of Autism (2001) Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet 105:539–547 [PubMed] [Google Scholar]
- Collaborative Linkage Study of Autism, Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, et al (1999) An autosomal genomic screen for autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Conroy J, Meally E, Kearney G, Fitzgerald M, Gill M, Gallagher L (2004) Serotonin transporter gene and autism: a haplotype analysis in an Irish autistic population. Mol Psychiatry 9:587–593 [DOI] [PubMed] [Google Scholar]
- Cook EH, Leventhal BL (1996) The serotonin system in autism. Curr Opin Pediatr 8:348–354 [DOI] [PubMed] [Google Scholar]
- Cook EH Jr, Arora RC, Anderson GM, Berry-Kravis EM, Yan SY, Yeoh HC, Sklena PJ, Charak DA, Leventhal BL (1993) Platelet serotonin studies in hyperserotonemic relatives of children with autistic disorder. Life Sciences 52:2005–2015 [DOI] [PubMed] [Google Scholar]
- Cook EH Jr, Courchesne R, Lord C, Cox NJ, Yan S, Lincoln A, Haas R, Courchesne E, Leventhal BL (1997) Evidence of linkage between the serotonin transporter and autistic disorder. Mol Psychiatry 2:247–250 [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE (2001) Nomenclature for the description of human sequence variations. Hum Genet 109:121–124 [DOI] [PubMed] [Google Scholar]
- Devlin B, Cook Jr EH, Coon H, Dawson G, Grigorenko EL, McMahon W, Minshew N, Pauls D, Smith M, Spence MA, Rodier PM, Stodgell C, Network CG, Schellenberg GD. Autism and the serotonin transporter: the long and short of it. Mol Psychiatry (in press) [DOI] [PubMed] [Google Scholar]
- Faraj BA, Olkowski ZL, Jackson RT (1997) Prevalence of high serotonin uptake in lymphocytes of abstinent alcoholics. Biochem Pharmacol 53:53–57 [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M (1977) Genetic influences and infantile autism. Nature 265:726–728 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B (2001) Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet 2:943–955 [DOI] [PubMed] [Google Scholar]
- Fombonne E (2003) The prevalence of autism. JAMA 289:87–89 [DOI] [PubMed] [Google Scholar]
- Glatt CE, DeYoung JA, Delgado S, Service SK, Giacomini KM, Edwards RH, Risch N, Freimer NB (2001) Screening a large reference sample to identify very low frequency sequence variants: comparisons between two genes. Nat Genet 27:435–438 [DOI] [PubMed] [Google Scholar]
- Gordon D, Desmarais C, Green P (2001) Automated finishing with autofinish. Genome Res 11:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Hahn MK, Blakely RD (2002) Monoamine transporter gene structure and polymorphisms in relation to psychiatric and other complex disorders. Pharmacogenomics J 2:217–235 [DOI] [PubMed] [Google Scholar]
- Hollander E, Phillips A, Chaplin W, Zagursky K, Novotny S, Wasserman S, Iyengar R (2005) A placebo controlled crossover trial of liquid fluoxetine on repetitive behaviors in childhood and adolescent autism. Neuropsychopharmacology 30:582–589 [DOI] [PubMed] [Google Scholar]
- Holloway JW, Beghe B, Turner S, Hinks LJ, Day IN, Howell WM (1999) Comparison of three methods for single nucleotide polymorphism typing for DNA bank studies: sequence-specific oligonucleotide probe hybridisation, TaqMan liquid phase hybridisation, and microplate array diagonal gel electrophoresis (MADGE). Hum Mutat 14:340–347 [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- ——— (2001a) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2001b) Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet 10:973–982 [DOI] [PubMed] [Google Scholar]
- Jones MB, Szatmari P (1988) Stoppage rules and genetic studies of autism. J Autism Dev Disord 18:31–40 [DOI] [PubMed] [Google Scholar]
- Kanner L (1943) Autistic disturbance of affective contact. Nervous Child 2:217–250 [Google Scholar]
- Khan NA, Meyneil JP, Deschaux P (1996) Ca2+/calmodulin and protein kinase C regulation of serotonin transport in human K562 lymphocytes. Cell Immunol 172:269–274 [DOI] [PubMed] [Google Scholar]
- Kilic F, Murphy DL, Rudnick G (2003) A human serotonin transporter mutation causes constitutive activation of transport activity. Mol Pharmacol 64:440–446 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Cox N, Courchesne R, Lord C, Corsello C, Akshoomoff N, Guter S, Leventhal BL, Courchesne E, Cook Jr EH (2002) Transmission disequilibrium mapping at the serotonin transporter gene (SLC6A4) region in autistic disorder. Mol Psychiatry 7:278–288 [DOI] [PubMed] [Google Scholar]
- Klauck SM, Poustka F, Benner A, Lesch KP, Poustka A (1997) Serotonin transporter (5-HTT) gene variants associated with autism. Hum Mol Genet 6:2233–2238 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL (1996) Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274:1527–1531 [DOI] [PubMed] [Google Scholar]
- Li Q, Liu Z, Monroe H, Culiat CT (2002) Integrated platform for detection of DNA sequence variants using capillary array electrophoresis. Electrophoresis 23:1499–1511 [DOI] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, the Autism Genetic Resource Exchange, Ott J, Gilliam TC (2001) A genomewide screen for autism susceptibility loci. Am J Hum Genet 69:327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie A, Quinn J (1999) A serotonin transporter gene intron 2 polymorphic region, correlated with affective disorders, has allele-dependent differential enhancer-like properties in the mouse embryo. Proc Natl Acad Sci USA 96:15251–15255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrini E, Lai C, Marlow A, Matthews N, Wallace S, Bailey A, Cook EH, Weeks DE, Monaco AP, the International Molecular Genetic Study of Autism Consortium (1999) Serotonin transporter (5-HTT) and γ-aminobutyric acid receptor subunit β3 (GABRB3) gene polymorphisms are not associated with autism in the IMGSA families. Am J Med Genet 88:492–496 [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley JL, Li C, Jiang L, Olson LM, Crockett G, Gainer K, Folstein SE, Haines JL, Sutcliffe JS (2005) Genome-wide and Ordered-Subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Med Genet 6:1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley JL, Olson LM, Dowd M, Amin T, Steele A, Blakely RD, Folstein SE, Haines JL, Sutcliffe JS (2004) Linkage and association analysis at the serotonin transporter (SLC6A4) locus in a rigid-compulsive subset of autism. Am J Med Genet B Neuropsychiatr Genet 127:104–112 [DOI] [PubMed] [Google Scholar]
- McPeek MS (1999) Optimal allele-sharing statistics for genetic mapping using affected relatives. Genet Epidemiol 16:225–249 [DOI] [PubMed] [Google Scholar]
- Miller KJ, Hoffman BJ (1994) Adenosine A3 receptors regulate serotonin transport via nitric oxide and cGMP. J Biol Chem 269:27351–27356 [PubMed] [Google Scholar]
- Nickerson DA, Kolker N, Taylor SL, Rieder MJ (2001) Sequence-based detection of single nucleotide polymorphisms. Methods Mol Biol 175:29–35 [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie AD, Battersby S, Bubb VJ, Fink G, Harmar AJ, Goodwim GM, Smith CA (1996) Polymorphism in serotonin transporter gene associated with susceptibility to major depression. Lancet 347:731–733 [DOI] [PubMed] [Google Scholar]
- Ozaki N, Goldman D, Kaye WH, Plotnicov K, Greenberg BD, Lappalainen J, Rudnick G, Murphy DL (2003) Serotonin transporter missense mutation associated with a complex neuropsychiatric phenotype. Mol Psychiatry 8:933–936 [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE (2004) Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet 5:389–396 [DOI] [PubMed] [Google Scholar]
- Persico AM, Militerni R, Bravaccio C, Schneider C, Melmed R, Conciatori M, Damiani V, Baldi A, Keller F (2000) Lack of association between serotonin transporter gene promoter variants and autistic disorder in two ethnically distinct samples. Am J Med Genet 96:123–127 [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, van Malldergerme L, Penet C, Feingold J, Brice A, Leboyer M (1999) Genome-wide scan for autism susceptibility genes. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim CH, Rutter M (1995) Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet 57:717–726 [PMC free article] [PubMed] [Google Scholar]
- Prasad HC, Zhu C-B, McCauley JL, Samuvel DJ, Ramamoorthy S, Shelton R, Hewlett WA, Sutcliffe JS, Blakely RD. Human serotonin transporter variants display selective insensitivity to protein kinase G and p38 mitogen activated protein kinase. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Galli A, Ramamoorthy S, Risso S, DeFelice LJ, Blakely RD (1997) Protein kinase C activation regulates human serotonin transporters in HEK-293 cells via altered cell surface expression. J Neurosci 17:45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S, Blakely RD (1999) Phosphorylation and sequestration of serotonin transporters differentially modulated by psychostimulants. Science 285:763–766 [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Pingree C, Mason-Brothers A, Jorde L, Jenson WR, McMahon WM, Petersen PB, Mo A, Ritvo A (1989) The UCLA–University of Utah epidemiologic survey of autism: prevalence. Am J Psychiatry 146:194–199 [DOI] [PubMed] [Google Scholar]
- Samuvel DJ, Jayanthi LD, Bhat NR, Ramamoorthy S (2005) A role for p38 mitogen-activated protein kinase in the regulation of the serotonin transporter: evidence for distinct cellular mechanisms involved in transporter surface expression. J Neurosci 25:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schain RJ, Freedman D (1961) Studies on 5-hydroxyindole metabolism in autistic disorder and other mentally retarded children. J Pediatr 58:315–320 [DOI] [PubMed] [Google Scholar]
- Shao Y, Raiford KL, Wolpert CM, Cope HA, Ravan SA, Ashley-Koch AA, Abramson RK, Wright HH, DeLong RG, Gilbert JR, Cuccaro ML, Pericak-Vance MA (2002a) Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Hum Genet 70:1058–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wolpert CM, Raiford KL, Menold MM, Donnelly SL, Ravan SA, Bass MP, McClain C, von Wendt L, Vance JM, Abramson RH, Wright HH, Ashley-Koch A, Gilbert JR, DeLong RG, Cuccaro ML, Pericak-Vance MA, McCoy PA (2002b) Genomic screen and follow-up analysis for autistic disorder. Am J Med Genet 114:99–105 [DOI] [PubMed] [Google Scholar]
- Spielman RS, Ewens WJ (1996) The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet 59:983–989 [PMC free article] [PubMed] [Google Scholar]
- Stone JL, Merriman B, Cantor RM, Yonan AL, Gilliam TC, Geschwind DH, Nelson SF (2004) Evidence for sex-specific risk alleles in autism spectrum disorder. Am J Hum Genet 75:1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadevosyan-Leyfer O, Dowd M, Mankoski R, Winklosky B, Putnam S, McGrath L, Tager-Flusberg H, Folstein SE (2003) A principal components analysis of the Autism Diagnostic Interview–Revised. J Am Acad Child Adolesc Psychiatry 42:864–872 [DOI] [PubMed] [Google Scholar]
- Tordjman S, Gutknecht L, Carlier M, Spitz E, Antoine C, Slama F, Carsalade V, Cohen DJ, Ferrari P, Roubertoux PL, Anderson GM (2001) Role of the serotonin transporter gene in the behavioral expression of autism. Mol Psychiatry 6:434–439 [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Anderson GM, Cook EH Jr (2000) Pharmacogenetics and the serotonin system: initial studies and future directions. Eur J Pharmacol 410:165–181 [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Christian SL, Cook EH Jr (2004) Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet 5:379–405 [DOI] [PubMed] [Google Scholar]
- Weiss LA, Abney M, Cook EH Jr, Ober C (2005) Sex-specific genetic architecture of whole blood serotonin levels. Am J Hum Genet 76:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeargin-Allsopp M, Rice C, Karapurkar T, Doernberg N, Boyle C, Murphy C (2003) Prevalence of autism in a US metropolitan area. JAMA 289:49–55 [DOI] [PubMed] [Google Scholar]
- Yirmiya N, Pilowsky T, Nemanov L, Arbelle S, Feinsilver T, Fried I, Ebstein RP (2001) Evidence for an association with the serotonin transporter promoter region polymorphism and autism. Am J Med Genet 105:381–386 [DOI] [PubMed] [Google Scholar]
- Yonan AL, Alarcón M, Cheng R, Magnusson PKE, Spence SJ, Palmer AA, Grunn A, Hank Juo S-H, Terwilliger JD, Liu J, Cantor RM, Geschwind DH, Gilliam TC (2003) A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet 73:886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C-B, Carneiro AM, Dostmann WR, Hewlett WA, Blakely RD (2005) p38 MAPK activation elevates serotonin transport activity via a trafficking-independent, protein phosphatase 2a-dependent process. J Biol Chem 280:15649–15658 [DOI] [PubMed] [Google Scholar]
- Zhu C-B, Hewlett WA, Feoktistov I, Biaggioni I, Blakely RD (2004) Adenosine receptor, protein kinase G, and p38 mitogen-activated protein kinase-dependent up-regulation of serotonin transporters involves both transporter trafficking and activation. Mol Pharmacol 65:1462–1474 [DOI] [PubMed] [Google Scholar]