Abstract
The p53 tumor suppressor gene is the most frequently mutated gene in human cancers, and germ-line p53 mutations cause a familial predisposition for cancer. Germ-line or sporadic p53 mutations are usually missense and typically affect the central DNA-binding domain of the protein. Because p53 functions as a tetrameric transcription factor, mutant p53 is thought to inhibit the function of wild-type p53 protein. Here, we studied the possible dominant-negative inhibition of wild-type p53 protein by two different, frequently occurring point mutations. The R270H and P275S mutations were targeted into the genome of mouse embryonic stem cells to allow the analysis of the effects of the mutant proteins expressed in normal cells at single-copy levels. In embryonic stem cells, the presence of a heterozygous point-mutated allele resulted in delayed transcriptional activation of several p53 downstream target genes on exposure to γ irradiation. Doxorubicin-induced apoptosis was severely affected in the mutant embryonic stem cells compared with wild-type cells. Heterozygous mutant thymocytes had a severe defect in p53-dependent apoptotic pathways after treatment with γ irradiation or doxorubicin, whereas p53-independent apoptotic pathways were intact. Together these data demonstrate that physiological expression of point-mutated p53 can strongly limit overall cellular p53 function, supporting the dominant-negative action of such mutants. Also, cells heterozygous for such mutations may be compromised in terms of tumor suppression and response to chemotherapeutic agents.
The p53 tumor suppressor protein has been proposed to function in many, diverse cellular processes, such as apoptosis, cell-cycle arrest, DNA repair, recombination, cellular differentiation, and senescence (1). Among other upstream stimuli, DNA damage is a potent activator of p53 function, and p53 is required for DNA damage-induced G1 arrest and apoptosis in many cell types (1), in part, by activating the expression of downstream target genes (1–4). Given these functions, mutation of p53 during tumorigenesis would be expected to lead to inappropriate S-phase entry or survival of damaged cells, possibly promoting genomic instability (1). In addition, in model systems, p53 mutant tumors and cell lines have been relatively resistant to certain chemotherapeutic agents and radiation (5–7). In humans, p53 mutations have been detected in at least 52 different cancer types, and ≈50% of all human tumors carry point mutations in p53 (8, 9). Heterozygous germ-line mutations in p53 predispose individuals to a wide range of tumor types at an early age, a condition known as Li–Fraumeni syndrome (LFS) (10).
The majority of both sporadic and germ-line p53 mutations are missense and occur in the conserved DNA-binding domain in the central portion of the protein. Depending on the tumor type, certain “hotspot” mutations are found, including at codons 273 and 248 (9). These residues make direct contact with the DNA helix and accordingly seem important for the transcriptional activation function of p53 (9). However, virtually all mutations of p53 abolish its ability to bind specific DNA sequences and activate the expression of its target genes (4, 8, 9). Extensive data from studies in vitro and in cell culture suggest that many missense mutations in p53 can inhibit the function of the wild-type protein in a dominant-negative manner, which would indicate that a heterozygous mutation in p53 could result in functional inactivation of cellular p53. To regulate downstream target genes, p53 binds the DNA as a tetrameric protein complex. Mutated protein within this complex is thought to abolish the DNA-binding capacity of the entire complex. Experiments with ectopic expression of wild-type and mutant p53 protein have demonstrated inhibition of DNA-binding activity and transactivation of target genes (11–13). However, conflicting data on this point and general concern about the effects of ectopic expression on the results exist (4 and references therein). Finally, despite possible dominant-negative function of missense p53 mutants, in approximately 50% of human tumors harboring such mutations, the remaining wild-type allele is mutated or lost, suggesting that complete loss of normal p53 can promote tumorigenesis further (8, 9).
Several mouse models have been generated to study p53 (reviewed in ref. 14). Mice homozygous for a deletion in the gene develop tumors (mainly lymphomas) at high incidence and short latency, clearly demonstrating the important role of p53 in tumor suppression. Transgenic overexpression of p53 point mutations (codon 135 Ala to Val or codon 193 His to Pro) resulted in lung adenocarcinomas and osteosarcomas. On a background of germ-line heterozygous p53 deletion, transgenic overexpression of mutant A135V alleles can accelerate tumor development compared with nontransgenic p53+/− mice. However, in p53−/− knockout animals, no effect of the p53 A135V transgene was observed, indicating that the mutant protein affected tumor development by interfering with wild-type p53 (15). Liu et al. (16) described a point mutation of the endogenous p53 gene in mice. Animals heterozygous for a mutation in codon 172 (arginine to histidine substitution) differed from p53+/− mice in tumor spectrum, and tumors metastasized with a high frequency. Moreover, loss of the wild-type p53 allele was rarely observed in these tumors, again supporting a dominant-negative function for the R172H allele. However, one drawback of this model is that the mutant p53 allele also contains an altered splice acceptor site, which leads to significantly reduced expression of the mutant protein.
Given the importance of p53 mutation in tumor development and therapy, we have constructed genomic p53 point mutations in mouse embryonic stem (ES) cells by homologous recombination. The R270H mutation is the equivalent of human R273H, a common mutation in many tumor types and also present in some LFS patients. The P275S mutation (equivalent to human P278S) is common in skin tumors but not in other tumor types nor in LFS. This system has allowed us to examine the effects of tumor-associated p53 point mutations expressed at single-copy levels in otherwise genetically normal cells. The effects of these mutations were examined on the biochemical and functional properties of wild-type p53 in mutant ES cells and thymocytes derived from them.
Materials and Methods
Cloning of p53 Mutant Targeting Vectors.
The p53.R270H and p53.P275S targeting vectors were constructed by cloning fragments of the murine p53 gene (isolated from strains BALB/c and 129sv) into the vector pBSK+. Both vectors contained an ≈8-kb genomic p53 sequence, extending from the XhoI site in exon 2, to the EcoRI site at the extreme 3′ end. First, a 1.3-kb BamHI/HindIII fragment comprising exons 7–9 was subcloned into pBSK+. Primers were generated harboring the desired mutation (mutated nucleotide in bold):
p53.R270H#1: 5′-GACAGCTTTGAGGTTCATGTTTGTGCCTGCCC-3′
p53.R270H#2: 5′-GGGCAGGCACAAACATGAACCTCAAAGCTGTC-3′
p53.P275S#1: 5′-TGTTTGTGCCTGCTCTGGGAGAGACCGC-3′
p53.P275S#2: 5′-GCGGTCTCTCCCAGAGCAGGCACAAACA-3′
These primers were used to introduce the mutations into the p53 sequence by site-directed mutagenesis with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The presence of the correct mutation was verified by sequencing. A selectable marker cassette (kindly provided by S. Tonegawa, Massachusetts Institute of Technology) was incorporated ≈0.5 kb downstream of the p53 gene. The cassette contains the neomycin-resistance gene and the thymidine kinase (TK) gene, flanked by LoxP sites. A pGK-DTA selection marker (encoding the diphtheria toxin A protein; kindly provided by F. Gertler, Massachusetts Institute of Technology) was cloned at the 3′ end of the vector for negative selection (without drug treatment). The complete inserts of the targeting vectors were sequenced, to exclude the presence of additional (undesired) mutations in the p53 encoding sequences and exon/intron boundaries.
Homologous Recombination Experiments in ES Cells.
The targeting vectors were linearized by digestion with NotI, and electroporated into D3 ES cells by using standard procedures (17). G418-resistant ES cell clones were analyzed for homologous integration of the targeting vector by Southern blot analysis. The Southern blot probe I consists of a 0.5-kb HindIII to KpnI fragment of intron 1 sequence. Clones with homologous integration of the targeting vector were checked for the presence of the mutation by a PCR/digestion-based assay. The following primers were used to amplify the p53 alleles:
p53in7#2: 5′-TTGGGCTTAGGGACGTCTCTTATC-3′ (situated in intron 7)
p53in9#1: 5′-ATGCGACTCTCCAGCCTTGGTA-3′ (situated in intron 9)
The resulting PCR product (486 bp) was digested with either MslI or BstNI. The R270H mutation results in a new MslI site in the PCR product, and the P275S mutation results in loss of a BstNI site compared with the wild-type p53 sequence. In this way, the p53 mutant alleles can be discriminated from the wild-type allele and from each other. Correctly targeted ES cell clones with either the R270H or P275S heterozygous mutation (p53+/R270H or p53+/P275S) were expanded, and electroporated with circular pMC-CreN plasmid (kindly provided by F. Alt, Harvard Medical School, Boston). Clones that had lost the selectable marker cassette were selected for by adding ganciclovir to the culture medium. Loss of the cassette was confirmed by Southern blot analysis by using probe II (BamHI–HindIII fragment). All correctly targeted clones that were used for blastocyst injection procedures were analyzed by reverse transcription–PCR followed by sequencing to check for additional (undesired) mutations. For all assays described here, multiple independent mutant clones were used.
ES Cell Exposure to DNA-Damaging Agents.
For the analysis of transcriptional activation of p53 downstream target genes, ES cells of all genotypes [wild-type (D3 parental cell line), p53+/− (17), p53−/− (kindly provided by R. Jaenisch, Whitehead Institute for Biomedical Research, Cambridge, MA), p53+/R270H and p53+/P275S] were plated on day 1 at a density of 2.5 × 106 per 10-cm dish. At day 3, cells were exposed to a single dose of 500 cGy of γ irradiation (γ-cell irradiator with a Cs source). At 1, 3, and 6 hr after the treatment, cells were isolated by trypsinization and centrifugation, and pellets were frozen for subsequent RNA isolation.
For the analysis of doxorubicin-induced apoptosis, ES cells of all genotypes were plated on day 1 in a six-well tissue culture dish and grown to subconfluence. Cells were treated with 1 or 2 μg/ml doxorubicin (Sigma) for 24 hr. At the indicated time points, both floating and adherent cells were harvested, and pelleted by centrifugation. Cells were washed once with 1× PBS, and stained with annexin V conjugated to FITC and propidium iodide (PI) by following the manufacturer's protocol (PharMingen). Stained cells were analyzed on a Becton Dickinson FACScan machine. The percentage of apoptotic cells was determined by using CELLQUEST software.
RNA Preparation and Northern Blot Analysis.
Total RNA was prepared from ES cells pelleted and frozen in liquid nitrogen by using Ultraspec RNA (Biotecx Laboratories, Houston), according to the manufacturer's instructions. Total RNA was prepared from thymocytes by using Trizol reagent (GIBCO/BRL). Thymocytes were pelleted, immediately resuspended into 0.8 ml of Trizol, and stored at −80°C. Further isolation of RNA was according to the manufacturer's procedure. RNasin (Promega) was added to the RNA samples to prevent degradation.
Northern blotting was performed by using standard procedures. cDNAs corresponding to bax, p21, mdm-2, cyclinG, and gapdh were used as probes (19, 20). Radioactivity was quantified by a PhosphorImager (Molecular Dynamics).
Rag2−/− Blastocyst Complementation Assay.
Rag2−/− blastocysts were isolated from matings between RAG2−/− mice. To obtain chimeras, 6–7 p53+/R270H or p53+/P275S ES cells were injected into each blastocyst.
Contribution of ES cells was determined by coat color. Thymocytes, splenocytes, and cells isolated from the bone marrow were stained with several monoclonal antibodies to analyze reconstitution of the lymphoid lineages by the ES cells. For this procedure, staining with FITC- or phycoerythrin-conjugated monoclonal antibodies directed against CD4, CD8, B220, IgM, and CD3 (PharMingen) were performed by procedures described (21, 22). Flow cytometric analysis was performed by using a FACScan (Becton Dickinson). Animals with thymocyte populations containing less than 70% CD4+/CD8+ double-positive cells were excluded from further analysis.
For the analysis of apoptosis, thymocytes were isolated from wild-type, p53+/−, p53−/−, p53+/R270H/RAG2−/−, and p53+/P275S/RAG2−/− chimeras (age 4–9 weeks) and kept in a PBS/FCS (2%) solution. The nonchimeric mice used were in the same 129sv background as the ES cells. Cells were plated at a density of 1 × 106 per well in 24-well plates in medium [DMEM/Hepes (25 mM, pH 7.2), 5% FCS, penicillin/streptomycin, glutamine]. To induce apoptosis, cells were exposed to (i) a single dose of 500 cGy of γ irradiation with a γ-cell irradiator with a Cs source, (ii) 0.2 μg/ml doxorubicin (Adriamycin), (iii) 1 μM dexamethasone, or (iv) a combination of 10 nM phorbol ester (phorbol 12-myristate 13-acetate) and 500 nM calcium ionophore A23187 (chemicals all obtained from Sigma). Cells were incubated at 37°C, and were, at the indicated time points, analyzed for the amount of apoptosis. For this procedure, cells were washed with cold PBS, and stained with FITC-labeled annexin V antibody (PharMingen) and PI (Sigma). The relative amounts of apoptotic cells were determined at various times by binding of annexin V and subsequent fluorescence-activated cell sorter analysis.
For analysis of expression levels in thymocytes of p53 target genes, thymocytes were isolated and brought into culture as described above at a density of 10 × 106 cells per 10-cm dish. At time 0, cultures were exposed to a single dose of 500 cGy of γ irradiation. Cells were pelleted at 2 or 5 hr after the treatment and resuspended in 0.8 ml of Trizol. RNA was subsequently isolated according to the manufacturer's procedure.
Results and Discussion
Generation of ES Cells and Chimeras with Point Mutations in p53.
To study the possible dominant-negative effect of point mutations in the tumor suppressor gene p53 in a physiological setting, we generated ES cells carrying a mutation in the DNA-binding domain of the p53 protein in a heterozygous state by gene targeting. This approach allows for the analysis of the effects of the mutated alleles when expressed from the p53 promoter at the endogenous locus. With use of gene targeting, we generated ES cells containing a mutation at codon 270 [arginine to histidine (R270H)] or codon 275 [proline to serine (P275S)]. Both mutations have been found frequently in both human and mouse tumors, and the human equivalent of the codon 270 mutation (R273H) is associated with LFS.
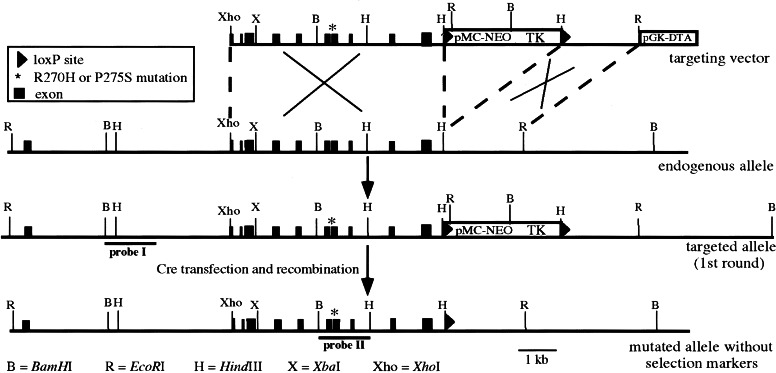
Targeting constructs containing either the R270H or P275S mutation and a neomycin/TK-selectable marker cassette (flanked by LoxP sites) were introduced into the mouse genome by homologous recombination in ES cells (Fig. 1). Homologous recombinant clones were identified by Southern blot analysis of ES cell DNA digested with EcoRI and by using probe I, which is situated outside the targeting vector (Fig. 1 and data not shown). The presence of the mutation was subsequently determined by PCR. Homologous recombination frequencies were 29% and 50% for the R270H and P275S vectors, respectively. Of these homologous clones, 26% (R270H) and 45% (P275S) of the ES cell clones contained the mutation, indicating that in the other clones recombination between the targeting vector and the endogenous p53 allele had occurred between the marker cassette and the mutation in exon 8.
Figure 1.
Generation of p53 point mutated alleles in ES cells. Scheme for targeting one allele of the murine p53 gene in ES cells with a targeting vector containing either the R270H or the P275S mutation (asterisk), and a neo-TK selectable marker cassette flanked by LoxP sites (first selection round). Homologous integration of the vector results in an additional EcoRI site (situated in the promoter of pMC-neo), which is used for Southern blot analysis of neomycin-resistant ES cell clones. In the homologous recombinant clones excision of the neo-TK selectable marker cassette was accomplished by transfection with circular pMC-CreN plasmid (second selection round). The resulting allele differs from the wild-type allele, besides the R270H or P275S mutation, only in the presence of one LoxP site downstream of the coding sequences.
To exclude the possible influence by the selectable marker cassette on the expression of the mutated p53 allele, a second round of ES cell transfections was performed. For each mutation, at least three independent and correctly targeted ES cell clones were retargeted with a Cre-recombinase-expressing plasmid. As a result, the neomycin/TK cassette should be removed from the targeted p53 allele, because it is flanked by LoxP sites (Fig. 1). Excision of the marker cassette was selected for by acquired resistance to ganciclovir, and verified by Southern blot analysis with BamHI-digested ES cell DNA and probe II (Fig. 1 and data not shown). For both mutations several independent clones were obtained lacking the neomycin/TK marker cassette. These clones exclusively harbored the desired point mutation in p53, as detected by direct sequencing of reverse transcription–PCR products spanning the complete p53 coding sequence. Expression levels of the mutant and wild-type p53 allele seemed to be comparable in these cells, as judged by the reverse transcription–PCR sequence signal (not shown). Thus, a single LoxP site downstream of the last exon of p53 present in the mutant allele does not seem to affect expression. For all assays described below, multiple independent mutant ES cell clones were used.
Analysis of ES Cells.
Point-mutated p53 proteins have been reported to have dominant-negative properties against wild-type p53. To test this possibility in this well-controlled cell system, we analyzed the heterozygous p53 mutant ES cells for known p53 functions. As a control, homozygous and heterozygous p53 knockout ES cells were included in the experiments (17).
Expression Levels of p53-Responsive Genes upon DNA Damage.
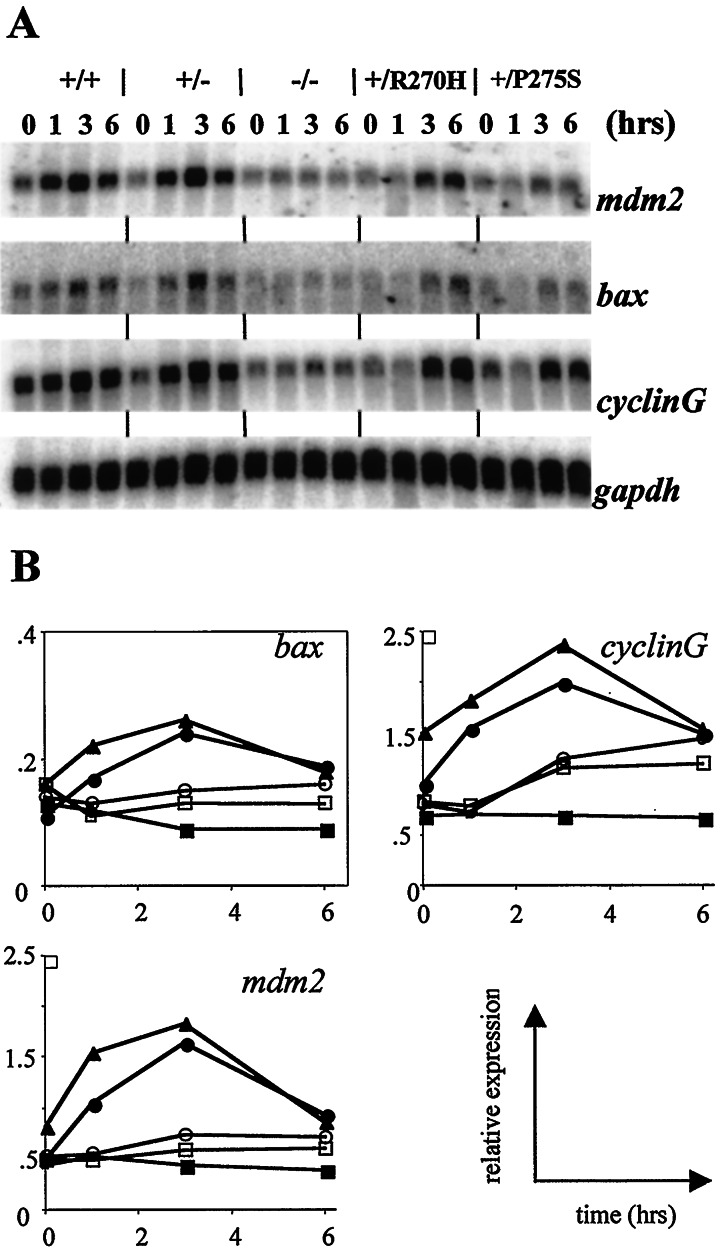
To test what the effect of specific mutations will be on the transcriptional activation of known p53 target genes, we treated ES cells heterozygous for either the R270H or the P275S mutation with a single dose of γ irradiation (500 cGy), an agent known to induce p53 in many cell types. RNA was isolated at several time points after the treatment, and expression levels of p53 targets bax, mdm2, and cyclinG were determined. As is shown in Fig. 2, expression levels of bax, cyclinG, and mdm2 rapidly increase on exposure to γ irradiation in wild-type ES cells. Induction in wild-type cells seems to reach a maximum at 3 hr after treatment, and decrease again afterward. p53+/− ES cells were indistinguishable for wild-type cells in this assay. Cells lacking p53 function (p53−/−) do not show induction of the three target genes, demonstrating the p53 dependence of the effect. ES cells harboring either the R270H or P275S point mutation showed a clearly diminished activation of p53 targets response after exposure to γ irradiation (Fig. 2). Although induction of bax, cyclinG, and mdm2 RNA was observed, the response was delayed and peak levels were reduced compared with controls. Thus, we can conclude that single-copy expression of point-mutant p53 is capable of dominant inhibition of p53 function, at least at the level of transcriptional activity.
Figure 2.
Expression levels of p53 target genes in ES cells upon treatment with γ irradiation. (A) Wild-type D3 (+/+); p53+/− (+/−); p53−/− (−/−); p53+/R270H (+/R270H); and p53+/P275S (+/P275S) ES cells were grown for 2 days, and exposed to a single dose of γ irradiation (500 cGy). At 1, 3, and 6 hr after the treatment, RNA was isolated. Northern blots were probed with bax, cyclinG, mdm2, and gapdh cDNA probes. The 0-hr time points represent untreated cells, isolated at the same time as the 3-hr time point after γ irradiation. (B) Quantitation of Northern blot signals normalized for expression levels of gapdh (used as a loading control). Note that in the bax graph, the scale of the y axis is different from the other axes. ▴, Wild-type D3; ●, p53+/−; ■, p53−/−; ○, p53+/R270H; □, p53+/P275S.
Apoptosis.
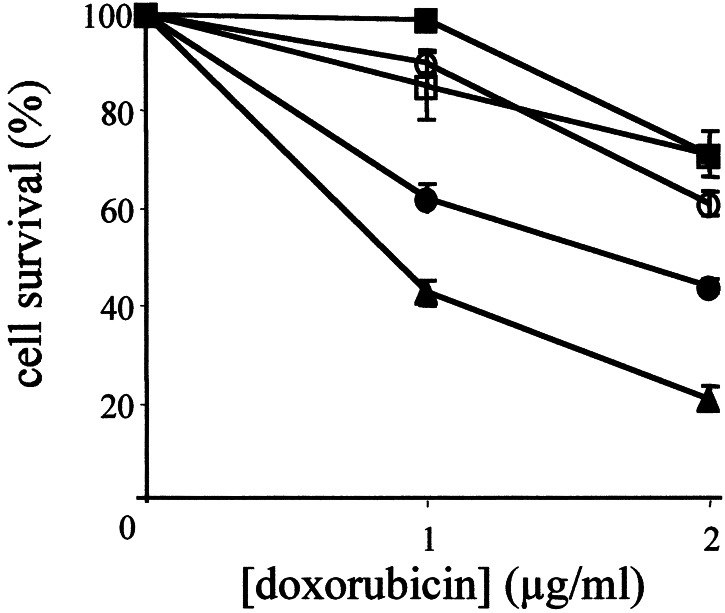
To examine the apoptotic response of p53 mutant ES cells, we treated them with doxorubicin, a chemotherapeutic compound that induces double-strand breaks. Cells were treated with two different doses of doxorubicin (1 or 2 μg/ml), and examined for the percentage of apoptotic cells 24 hr later by using annexin V staining. As shown in Fig. 3, both doses of doxorubicin caused a high percentage of wild-type cells to undergo apoptosis (57% at 1 μg/ml and 79% at 2 μg/ml). In contrast, cells completely lacking p53 were completely resistant to 1 μg/ml, and only 29% of these cells were dead after exposure to 2 μg/ml doxorubicin. The p53+/− cells had a partial response, demonstrating an effect of gene dosage in this pathway. The apoptotic response of the p53+/R270H and p53+/P275S ES cells was highly similar to that of p53−/− cells. One day after treatment with 1 μg/ml doxorubicin, only 10–15% of the mutant cells had undergone apoptosis, and at a dose of 2 μg/ml 39% (p53+/R270H) and 29% (p53+/P275S) ES cells had died. Thus, in this assay for the cellular effects of p53 function, the presence of a single mutated allele potently inhibited wild-type p53.
Figure 3.
Doxorubicin-induced apoptosis in ES cells. Wild-type D3 (▴), p53+/− (●), p53−/− (■), p53+/R270H (○), and p53+/P275S (□) ES cells were grown to subconfluency, and exposed to either 1 or 2 μg/ml doxorubicin in the culture medium. After 24 hr, the cells were stained with PI and annexin V antibody, and the numbers of viable cells were determined by fluorescence-activated cell sorter analysis. Data are averages of four independent experiments.
Analysis of Thymocytes by the Rag2−/− Blastocyst Complementation Assay.
The transcriptional and cell death data from ES cells were consistent with a dominant-negative function for these point-mutated alleles of p53. We next sought to examine the effect in another cell system. In thymocytes, the apoptotic response on exposure to various agents has been particularly well characterized, and both p53-dependent and independent pathways have been identified (23, 24). To obtain thymocytes heterozygous for the p53 point mutations, we injected heterozygous p53 mutant ES cells (several independent ES cell clones of each mutation) into Rag2−/− blastocysts. Because Rag2−/− mice do not form mature B and T cells (22, 25), lymphocytes are derived exclusively from the ES cells in the resulting chimeras.
Reconstitution of the Lymphoid System.
First, we analyzed the reconstitution of the thymus in the chimeras. The total number of thymocytes isolated from the thymus was on average the same between the chimeras and, more importantly, did not differ substantially from age-matched, nonchimeric wild-type, p53+/− and p53−/− animals (not shown). Flow cytometric analysis of the thymocytes isolated from mice of the different genotypes revealed normal percentages of CD4+CD8+ double-positive (DP) cells (86%, 82%, and 88% for wild-type, p53+/R270H, and p53+/P275S animals, respectively; data not shown). In addition, the profile of CD4−CD8− double-negative (DN), CD4+ and CD8+ single-positive cells was equal to that of wild-type, p53+/− and p53−/− mice. For the cell death experiments, chimeras with low thymocyte numbers or low (<70%) DP numbers were not used for the apoptotic analyses. Finally, spleens and bone marrow of the chimeras consisted of normally differentiated T and B cells, as analyzed by staining the cells with CD4 and CD8; B220 and CD3 and IgM antibodies (not shown). In conclusion, ES cells with a heterozygous point mutation in p53 reconstitute the T and B cell compartments normally in a Rag2−/− blastocyst complementation assay.
p53-Dependent Apoptosis in Thymocytes.
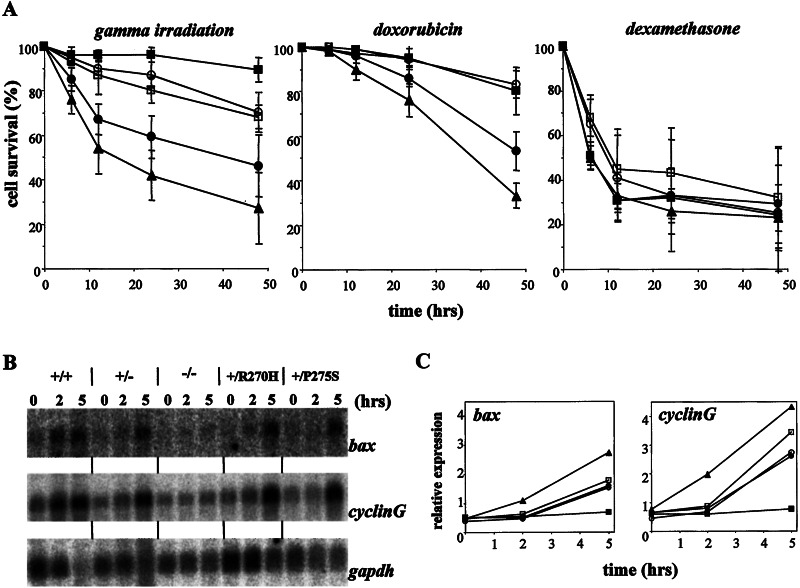
Immature wild-type DP thymocytes undergo apoptosis upon exposure to certain DNA-damaging agents. For γ irradiation and doxorubicin, efficient apoptosis requires a functional p53 pathway (23, 24). To determine whether a heterozygous point mutation in p53 can affect this process, we exposed wild-type, p53+/−, p53−/−, p53+/R270H, and p53+/P275S thymocytes to both types of DNA-damaging agents and measured percentages of apoptotic cells over time with annexin V and PI staining. As shown in Fig. 4A, as expected p53−/− thymocytes were highly resistant to DNA-damage-induced apoptosis. Wild-type thymocytes died rapidly, and p53+/− thymocytes were somewhat resistant to these agents. Significantly, after γ irradiation, thymocytes heterozygous for either the R270H or P275S mutation were more resistant to cell death than either wild-type or p53+/− cells, but did display a slightly higher degree of apoptosis than p53−/− cells. A similar pattern of responses was observed with thymocytes of all five genotypes after several doses of γ irradiation (not shown). After exposure to doxorubicin the apoptotic response of the heterozygous mutants was even more similar to that of p53−/− thymocytes, and again, p53+/− thymocytes showed an intermediate response (Fig. 4A). These results show clearly that thymocytes heterozygous for a point mutation in p53 have a defect in their apoptotic response after the induction of DNA damage, providing perhaps the best evidence to date for the dominant-negative effects of certain tumor-associated p53 mutations.
Figure 4.
The effect of heterozygous point mutations in p53 on apoptosis in thymocytes. (A) p53-dependent and -independent apoptosis. Thymocytes of mice of all different genotypes were isolated (▴, wild type; ●, p53+/−; ■, p53−/−; ○, p53+/R270H; □, p53+/P275S) and exposed in vitro to γ irradiation (500 cGy), doxorubicin (0.2 μg/ml), or dexamethasone (1 μM). At the time points indicated, thymocytes were stained with annexin V and PI. The relative percentage of viable cells (negative for both PI and annexin V) for each sample is shown. All values are normalized to the number of cells remaining viable in untreated cultures derived from the same animal stained simultaneously. Data are representatives of ≥2 independent experiments (i.e., mice). (B) Northern blot of p53 target genes in thymocytes after γ irradiation (500 cGy). Two or 5 hr after the treatment, RNA was isolated, and Northern blots were probed with bax, cyclinG, and gapdh cDNA probes. (C) Quantitation of Northern blot signals normalized for expression levels of gapdh (used as a loading control). ▴, Wild type; ●, p53+/−; ■, p53−/−; ○, p53+/R27OH; and □, p53+/P275S.
Expression of p53-Responsive Genes in Thymocytes upon DNA Damage.
We next examined the induction of p53 target genes in treated thymocytes. As shown in Fig. 4B, thymocytes show a p53-dependent induction of bax and cyclinG RNA upon γ irradiation, with levels rising at 2- and 5-hr time points. In p53+/− thymocytes, the induction of these genes was delayed and reduced. The induction of bax and cyclinG RNA in thymocytes heterozygous for the R270H or P275S mutation was comparable to p53+/− cells, despite the clear difference in DNA-damage-induced apoptosis between cells of these genotypes (Fig. 4 B and C). Thus, the kinetics or extent of induction of at least these target genes cannot fully account for the dominant-negative affects of these p53 point mutations in inhibition of p53-dependent apoptosis. However, given the uncertainty about the specific target genes required for p53-dependent apoptosis, it remains possible that critical target genes are underexpressed in the mutant cells, accounting for the phenotype. Indeed, the analysis of the transcriptional profile of these cells might help define p53 targets required for apoptosis. In addition, other functions of p53, including transcriptional repression (1), might be affected by the presence of the point mutant protein.
p53-Independent Apoptosis.
To establish whether the inhibition of DNA damage-induced apoptosis by the p53 point mutations was specific for the p53 pathway or indicative of a general apoptotic defect, we treated the thymocytes of different genotypes with stimuli that induce p53-independent apoptosis (23, 24). Isolated thymocytes were exposed in vitro to dexamethasone (1 μM; Fig. 4A) or a combination of phorbol 12-myristate 13-acetate/ionomycin (10 nM/500 nM, data not shown). Wild-type, p53+/−, and p53−/− thymocytes all died rapidly after these treatments, confirming the lack of requirement for p53 function. Thymocytes harboring either the R270H or the P275S mutation were comparable to the thymocytes of the other three genotypes, indicating that the p53-independent apoptotic pathways tested are intact in these cells.
Conclusions
The analysis of mouse ES cells and thymocytes heterozygous for tumor-associated p53 point mutations demonstrates that single-copy expression of altered p53 protein can substantially inhibit wild-type p53 function, which is consistent with a dominant-negative function of the mutant alleles. These data support the model in which tumor cells progressively lose p53 function through the acquisition of such p53 mutations, often followed by loss of the wild-type p53 allele. These studies also reveal assay-specific differences in the effects of these mutations, which may help account for some of the previous conflicting data on this point. As discussed above, a disparity occurred between the induction of p53 target genes in mutant thymocytes treated with DNA-damaging agents and the inhibition of apoptosis in these cells. In addition, as judged by standard in vitro p53 DNA-binding assays using extracts from unstimulated wild-type and mutant ES cells, the presence of point mutant p53 did not significantly affect overall p53 DNA-binding activity (data not shown). Given that these assays are performed in the presence of p53 antibody to stabilize the protein–DNA complex, it is possible that the effects of the mutations were masked. It will be necessary to assay p53 DNA binding in vivo under different conditions (and possibly at different promoters) to assess accurately the effects of these p53 mutations.
The ability of these point-mutant p53 alleles to affect p53-dependent apoptosis potently has important implications for understanding the course of tumorigenesis in sporadic cancers with p53 mutations and in patients with LFS. Specifically, these data show clearly that emerging tumor cells that acquire this sort of p53 point mutation will have a selective advantage under conditions that induce p53-dependent apoptosis, including after endogenous DNA damage events or DNA damage induced by chemotherapy or radiation. Indeed, the data underscore the functional difference between the acquisition of a p53 missense versus loss-of-function mutation in the gene in the course of tumor development. These results also raise the interesting possibility that LFS patients carrying certain types of p53 mutations may have less severe side effects of treatment with some forms of chemotherapy or radiation, given that many of these effects are secondary to p53-dependent apoptosis in normal cell types. It will be interesting to examine the effects of these and other p53 point mutations on tumorigenesis in the mouse, and possible gain-of-function properties of certain p53 mutations.
Acknowledgments
We thank Laura Attardi, Timo Breit, and Harry van Steeg for helpful discussions and critical reading of the manuscript. We thank Denise Crowley and Roderick Bronson for histological and pathological analysis of mice, Jan de Wit for performing the γ-induced cell death assays in ES cells, and Tara Schmidt for breeding and maintenance of the RAG2−/− mouse colony. We are grateful to Gigi Lozano for the mdm-2 probe, to Jackie Lees for the cyclinG probe, to Frank Gertler for the pGK-DTA plasmid, to Susumu Tonegawa for the loxP-neo-TK-loxP selectable marker cassette, and to Fred Alt for the pMC-CreN plasmid. This work was supported in part by funding from the Dutch Cancer Society (to A.d.V.). T.J. is an Associate Investigator at the Howard Hughes Medical Institute.
Abbreviations
- ES cell
embryonic stem cell
- LFS
Li–Fraumeni syndrome
- PI
propidium iodide
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Cox L S, Lane D P. BioEssays. 1995;17:501–508. doi: 10.1002/bies.950170606. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb T M, Oren M. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 4.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 5.Lowe S W, Bodis S, McClatchey A, Remington L, Ruley H E, Fisher D E, Housman D E, Jacks T. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 6.Aas T, Borresen A L, Geisler S, Smith-Sorensen B, Johnsen H, Varhaug J E, Akslen L A, Lonning P E. Nat Med. 1996;2:811–814. doi: 10.1038/nm0796-811. [DOI] [PubMed] [Google Scholar]
- 7.Delia D, Goi K, Mizutani S, Yamada T, Aiello A, Fontanella E, Lamorte G, Iwata S, Ishioka C, Krajewski S, et al. Oncogene. 1997;14:2137–2147. doi: 10.1038/sj.onc.1201050. [DOI] [PubMed] [Google Scholar]
- 8.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 9.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 10.Malkin D. Biochim Biophys Acta. 1994;1198:197–213. doi: 10.1016/0304-419x(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 11.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 12.Milner J, Medcalf E A, Cook A C. Mol Cell Biol. 1991;11:12–19. doi: 10.1128/mcb.11.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milner J, Medcalf E A. Cell. 1991;65:765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- 14.Attardi L D A, Jacks T. Cell Mol Life Sci. 1999;55:48–63. doi: 10.1007/s000180050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower L A. Nat Genet. 1995;9:305–311. doi: 10.1038/ng0395-305. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, McDonnell T J, Montes de Oca Luna R, Kapoor M, Mims B, El-Naggar A K, Lozano G. Proc Natl Acad Sci USA. 2000;97:4174–4179. doi: 10.1073/pnas.97.8.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 18.Macleod K, Hu Y, Jacks T. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 19.Macleod K, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 20.Hurford R K, Cobrinik D, Lee M, Dyson N. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 21.Nacht M, Strasser A, Chan Y R, Harris A W, Schlissel M, Bronson R T, Jacks T. Genes Dev. 1996;10:2055–2066. doi: 10.1101/gad.10.16.2055. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Lansford R, Stewart V, Young F, Alt F W. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 24.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wyllie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 25.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]