Abstract
Between 40% and 80% of the variation in human intelligence (IQ) is attributable to genetic factors. Except for many rare mutations resulting in severe cognitive dysfunction, attempts to identify these factors have not been successful. We report a genomewide linkage scan involving 634 sibling pairs designed to identify chromosomal regions that explain variation in IQ. Model-free multipoint linkage analysis revealed evidence of a significant quantitative-trait locus for performance IQ at 2q24.1-31.1 (LOD score 4.42), which overlaps the 2q21-33 region that has repeatedly shown linkage to autism. A second region revealed suggestive linkage for both full-scale and verbal IQs on 6p25.3-22.3 (LOD score 3.20 for full-scale IQ and 2.33 for verbal IQ), overlapping marginally with the 6p22.3-21.31 region implicated in reading disability and dyslexia.
There are substantial individual differences in cognitive abilities, which tend to cluster within individuals. This “positive manifold” formed the basis for the construction of intelligence tests that assess abilities across different domains (e.g., visuospatial abilities, memory, vocabulary, semantics, and symbolic reasoning) that can be summarized into higher-order factors, such as verbal and performance intelligence. Traditional psychometric intelligence tests, such as the Wechsler Adult Intelligence Scale (WAIS) (Wechsler 1997), show high predictive validity, stability across age spans, and substantial heritability (Bouchard and McGue 1981). The identification of genes underlying the genetic variation in human intelligence has thus far been limited to neurological mutations with rather severe cognitive effects (e.g., Pick disease and X-linked mental retardation) (Ramakers 2003; Inlow and Restifo 2004), which are largely Mendelian in nature (Flint 1999). Identifying genes for variation in the range of normal intelligence could provide important clues to the underlying mechanisms of milder but more-prevalent forms of impaired cognitive functioning, which are often associated with autism, schizophrenia, reading disorder, and attention deficit hyperactivity disorder (Goldman-Rakic 1999; Willcutt et al. 2001; Badcock et al. 2005).
A major effort in identifying genetic variants that influence cognitive ability in the normal range was undertaken by Plomin and colleagues (Petrill et al. 1996; Chorney et al. 1998; Fisher et al. 1999a; Plomin et al. 2001, 2004; Butcher et al. 2005). The only association that survived rigorous adjustment for multiple testing was a significant association of a functional polymorphism in the ALDH5A1 gene (MIM 271980) withcognitive ability (Plomin et al. 2004). This association was based on a selected sample (subjects with high IQs vs. subjects with normal IQs), and the effect size was rather small. Thus, in spite of the overwhelming evidence from twin studies of the existence of “genes for human intelligence,” the actual identification of such genes through well-designed candidate-gene or whole-genome allelic association studies has not met with large success. It might be that these pre-HapMap studies did not achieve a high enough SNP density. Generally, association analyses may overlook closely spaced genes that act in concert to affect a trait. In contrast, linkage analysis is more sensitive to such concerted effects. For example, Yalcin et al. (2004) recently suggested such a mechanism for emotionality in mice, in which multiple closely linked genes on chromosome 1 may be needed to explain the solid linkage found in this region (Yalcin et al. 2004).
Here, we report the first genomewide linkage study for intelligence, using two unselected samples consisting of 725 individuals from 329 Australian families, which yield 475 sib pairs, and 225 individuals from 100 Dutch families, which yield 159 sib pairs. In these populations, the contribution of genetic factors to full-scale IQ (FSIQ) was shown to be 0.69 in the Australian sample (Luciano et al. 2004) and 0.86 in the Dutch sample (Posthuma et al. 2001). Heritabilities for verbal IQ (VIQ) and performance IQ (PIQ) were also high (table 1).
Table 1.
Characteristics of the Australian and Dutch Samples in the Twin-Family Studies of Intelligence
| Characteristic | AustralianSample | DutchSample |
| No. of families | 329 | 100 |
| No. of pairs | 475 | 159 |
| No of. individuals (no. of males) | 725 (360) | 225 (96) |
| Age (years) | 16.4 ± .70 | 39.6 ± 12.46 |
| Mean FSIQ | 112.6 ± 13.05 | 94.2 ± 10.84 |
| Mean VIQ | 111.0 ± 11.39 | 92.4 ± 13.10 |
| Mean PIQ | 112.4 ± 16.50 | 98.3 ± 12.05 |
| Heritability FSIQ | .69 | .86 |
| Heritability VIQ | .72 | .85 |
| Heritability PIQ | .59 | .69 |
The sample of Australian DZ twins and siblings was part of an ongoing study of cognition in adolescents (Wright et al. 2001; Luciano et al. 2004) and was genotyped as part of an earlier study that investigated melanoma risk factors (Zhu et al. 1999). The study was approved by the Human Research Ethics Committee of the Queensland Institute of Medical Research Institute. Informed consent to jointly examine the cognitive and genotype data was obtained from participants or the parents/guardians of participants who were <18 years of age. The Multidimensional Aptitude Battery (MAB) (Jackson 1998) was used to assess IQ and consisted of three verbal subtests (information, vocabulary, and arithmetic) and two performance subtests (spatial ability and object assembly), which were administered by computer and were each timed at 7 min. Scaled scores for VIQ, PIQ, and FSIQ were examined. VIQ, PIQ, and FSIQ were normally distributed. IQ scores assessed with the MAB generally correlate highly with those assessed by the WAIS (Wechsler 1997): r=0.94 for VIQ, r=0.79 for PIQ, and r=0.91 for FSIQ (Jackson 1998).
Genotyping of the Australian sample was performed at two facilities, the Australian Genome Research Facility, by use of the ABI PRISM Linkage Mapping Set v2.5, and the Center for Inherited Disease Research, by use of a marker set based on the Marshfield Genetics version 8 screening set.
Up to 761 autosomal microsatellite markers were typed at approximately equal intervals (average 4.8 cM) across the entire genome, with locations determined from the sex-averaged deCODE map (Kong et al. 2002) and interpolation of unmapped markers (Zhu et al. 2004). Marker heterozygosity ranged from 52.6% to 91.9%. In the Australian sample, both parental genotypes were available for 260 families, one parental genotype for 67 families, and no parental genotypes for 34 families. Parents were typed for 228–784 markers (mean ± SD, 398 ± 101 markers). For twins and siblings, the number of typed markers ranged from 211 to 790, with an average of 601 ± 192 total markers.
The sample of Dutch DZ twins and siblings were part of an ongoing study of cognition in adults (Posthuma et al. 2001; Wright et al. 2001). Subjects were recruited from The Netherlands Twin Register to participate in the cognition study, and they gave written informed consent. The institutional review board of the Vrije Universiteit Medical Centre approved the DNA sampling and cognitive testing. The Dutch adaptation of the WAIS III-R (Wechsler 1997) was used to assess IQ and consisted of four verbal subtests (information, similarities, vocabulary, and arithmetic) and three performance subtests (picture completion, block design, and matrix reasoning). Scaled scores for VIQ, PIQ, and FSIQ were examined. VIQ, PIQ, and FSIQ were normally distributed. In 93 subjects, a genome scan with 369 autosomal markers (with 9.44-cM spacing) was done by the Mammalian Genotyping Service, with microsatellite screening set 10 with few alternative markers. In 132 subjects, a 419-marker genome scan (with 8.34-cM spacing) was performed by the Molecular Epidemiology Section, Leiden University Medical Centre (Heijmans et al., in press). Parents were typed for 344–375 markers (mean 363 ± 6). For offspring, the number of typed markers ranged from 344 to 678, with an average of 389 ± 69 total markers.
Marker locations in the Australian and Dutch data sets were taken from an integrated genetic map with interpolated genetic map positions (see Web Resources section). The positions are in deCODE cM (Kong et al. 2002), estimated via locally weighted linear regression (lo(w)ess) from the build 34.3 (and 35.1) physical map positions and from published deCODE and Marshfield genetic map positions. Mendelian errors were detected using PEDSTATS, and unlikely double recombinants were detected using MERLIN; both were removed using PEDWIPE (Abecasis et al. 2002). Pedigree relationships in the entire sample were checked with the GRR program (Abecasis et al. 2001).
A variance-components (VC) approach was used to evaluate linkage. The VC framework involves partitioning the variance of the trait of interest into components due to covariates, component(s) due to a major QTL, a polygenic component, and unique environmental or random variation. Estimation of the QTL effect requires the use of genetic data in the form of pairwise identity-by-descent (IBD) sharing between siblings. The multipoint probabilities of sharing 0, 1, or 2 alleles IBD were computed separately for each sample by use of a 1-cM grid with the Lander-Green algorithm implemented in MERLIN (Abecasis et al. 2002). These IBD probabilities were used to estimate linkage with the use of Mx software (Neale et al. 1997).
For both data sets, age and sex were included as covariates in the analyses. All parameter estimates were initially allowed to differ between the two data sets. To test for heterogeneity in the variance associated with a putative QTL (σ2q), we ran a second scan in which we restricted σ2q to be the same across both data sets while allowing all other parameters, such as the polygenic effect, to be different. Heterogeneity of the QTL effect across the Dutch and Australian data sets could then be determined by the likelihood-ratio test, where the null model of no heterogeneity included a single QTL effect and the alternative model of heterogeneity included two separate QTL effects, for each genomic position. If no heterogeneity in σ2q is observed, the combined analysis can be used to determine significance of the QTL effect on the basis of the pooled data set. Of all 3,543 genomic positions tested (by use of a 1-cM grid), only 1.2% for PIQ, 3.2% for VIQ, and 0.9% for FSIQ showed a χ2 value >3.84 (asymptotic P=.05) at positions not located under our peak regions.
Significance of genetic variation due to the QTL was evaluated by the likelihood-ratio test, which is a 50:50 mixture of a χ2 distribution with 1 df and a point mass at zero, comparing a model that included the variance associated with a QTL and a model that did not include this component. The LOD score can then be calculated by dividing the test statistic χ2 by 2ln10 (∼4.6). Linkage analyses were also performed using MERLIN, with option –vc for VC linkage analysis, and the results did not show notable differences from the results based on Mx presented here. However, the advantage of Mx is that possible heterogeneity in means, variance, effect of covariates, or polygenic heritability can be included in the statistical model.
To obtain empirical estimates of genomewide significance levels, 1,000 permutations of the data set were performed, with both the family structure and the IBD structure kept intact. These permutations allowed us to account for uneven marker spacing and informativeness and to calculate the probability of observing multiple peaks of a certain height (Churchill and Doerge 1994). The permuted data sets were obtained so that each row in the observed data set represents one family and contains phenotypic data for each individual within that family as well as the precalculated (using MERLIN) IBD probabilities across the whole genome for all pairs within that family. This file is split into a phenotypic file (containing the IQ data as well as age, sex, and country identifier) and a genotypic file (containing the IBD probabilities). Families in the experiment are labeled with unique numbers 1 through n. The phenotypic data are then shuffled by taking a random permutation of the indices 1,…,n and matching the ith phenotypic trait value to the family with an index given by the ith element of permuted indices. This permuted vector of traits is matched with the original (unpermuted) genotypic information for all families. A total of 1,000 permuted data sets were generated under the assumption of no linkage; each permuted data set was then analyzed analogously to the observed data, and the highest peak for each chromosome was recorded. The empirical significance level of an observed LOD score was then estimated by counting the proportion of genome scans containing one or more peaks of that size. The cut-off for suggestive linkage was calculated as the score that was observed, on average, once per genome scan, thus representing the average maximum peak size expected once per genome scan by chance alone (Lander and Kruglyak 1995). The thresholds for suggestive linkage for PIQ, VIQ, and FSIQ were 2.01, 1.70, and 1.86, respectively. The significant linkage threshold was defined as the LOD score occurring in 50 of the 1,000 permutations, corresponding to a probability of 0.05 in a genome scan (Lander and Kruglyak 1995). The thresholds for significant linkage for PIQ, VIQ, and FSIQ were 3.39, 3.05, and 3.22, respectively.
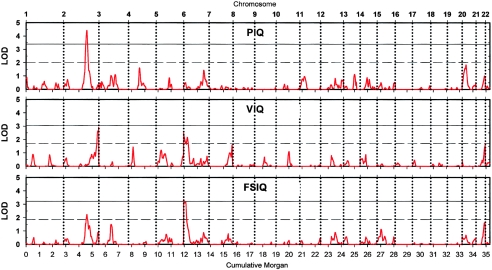
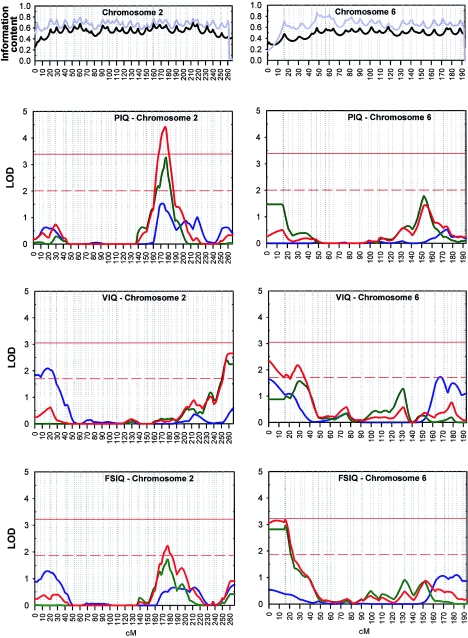
Multipoint analyses revealed significant or suggestive evidence of linkage on two chromosomes (figs. 1 and 2). A single region, on chromosome 2q, revealed significant evidence of linkage to PIQ, at marker D2S2330 (LOD score 4.42). The evidence was reasonably supported by the Australian sample alone (LOD score 3.26) and was marginally supported (Lander and Kruglyak 1995) by the smaller Dutch sample (LOD score 1.53), which peaked at only a 5-cM difference. Heterogeneity tests confirmed that the QTL effects observed in the Australian and Dutch data sets in this region could indeed be combined into a single QTL effect. The borders (defined by a drop of 1 LOD score) of the positive peak at 2q are located at markers D2S142 and D2S2188, which are 16 cM apart (table 2). The shape of the scan for FSIQ score mirrored the findings for PIQ score on chromosome 2q, showing a peak LOD score of 2.23 within the same area.
Figure 1.
Full autosomal genome scans for PIQ, VIQ, and FSIQ based on 634 sibling pairs. Horizontal solid lines denote genomewide empirical thresholds for significant linkage, defined as the LOD score that would be expected to occur by chance with a probability of 0.05 in a whole-genome scan. Horizontal dashed lines denote genomewide empirical thresholds for suggestive linkage, defined as the LOD score that would be expected to occur once by chance in a whole-genome scan (Lander and Kruglyak 1995).
Figure 2.
Genomic regions with suggestive or significant linkage in the Dutch (blue), Australian (green), and combined (red) data sets. Vertical dotted lines represent the positions of the markers. Horizontal solid red lines denote genomewide empirical thresholds in the combined data set for significant linkage, defined as the LOD score that would be expected to occur by chance with a probability of 0.05 in a whole-genome scan. Horizontal dashed red lines denote genomewide empirical thresholds in the combined data set for suggestive linkage, defined as the LOD score that would be expected to occur once by chance in a whole-genome scan (Lander and Kruglyak 1995). Information content, as a measure of entropy in the IBD distribution (Kruglyak et al. 1996), in the Dutch (black) and Australian (gray) data sets is plotted in the top panels.
Table 2.
Genomic Regions with Evidence of Linkage to Intelligence, in a Study of 634 Sib Pairs
|
Peak LOD Score for |
||||||
| GenomicRegion andIQ Measure | CombinedData | DutchSample | AustralianSample | Peak Marker | Location(cM from pter) | 1-LOD Drop Area |
| 2q24.1-31.1: | ||||||
| PIQ | 4.42 | 1.53 | 3.26 | D2S2330 | 173.97 | D2S142–D2S2188 |
| FSIQ | 2.23 | .66 | 1.72 | D2S1776 | 176.74 | D2S142–D2S2364 |
| 6p25.3-22.3: | ||||||
| VIQ | 2.33 | 1.63 | 1.56 | D6S2434 | 32.61 | D6S942–D6S422 |
| FSIQ | 3.20 | .53 | 2.95 | F13A1 | 16.07 | D6S1574–D6S309 |
On chromosome 6, we found suggestive evidence of linkage to FSIQ (LOD score 3.20) and, in the same region, to VIQ (LOD score of 2.33) at marker D6S942. The whole 1-LOD drop area spanned 42 cM (D6S942–D6S422).
Since the correlation between VIQ and PIQ is 0.53, which reflects that 28% of the variance in VIQ is shared with PIQ, we would expect that at least some of the QTLs that are important for VIQ are also important for PIQ. For the 2q24.1-2q31.1 region, linked to PIQ, we did not observe any evidence of linkage to VIQ. In the chromosome 6 region, linked to VIQ, the scan for PIQ showed a modest peak (LOD score 1.47) in the Australian data set in the same region as VIQ, implying that this region may be important for both VIQ and PIQ.
A few other regions showed LOD scores that were just below the thresholds for suggestive linkage (i.e., LOD score difference <0.3) (fig. 1): on chromosome 4, VIQ showed a peak LOD score of 1.44 near marker D4S419; on chromosome 7, VIQ showed a peak LOD score of 1.56 near marker D7S3058; on chromosome 20, PIQ showed a peak LOD score of 1.84 near marker D20S851; and on chromosome 21, VIQ and FSIQ showed peak LOD scores of 1.56 and 1.59, respectively, at marker D21S1446. We further observed suggestive linkage for VIQ at 2qter in the Australian data set and at 2pter in the Dutch data set. Replication studies are needed to determine whether these LOD scores were due to chance or are derived from QTLs with small effects.
The 2q24.1-2q31.1 (D2S142–D2S2188) region that shows linkage to PIQ and FSIQ largely overlaps with the 2q21-33 region that has yielded suggestive linkage to autism in at least four independent genomic screens (Philippe et al. 1999; Buxbaum et al. 2001; International Molecular Genetic Study of Autism Consortium [IMGSAC] 2001; Shao et al. 2002). The IMGSAC (2001) reported their highest LOD score (3.74) with language delay in autism-affected families at marker D2S2188, which lies 6 cM from the peak LOD score on 2q, whereas Buxbaum et al. (2001) reported their highest LOD score for phrase speech at marker D2S335, which lies 3 cM away from the peak LOD score found in our study. Recently, Raskind et al. (2005) conducted a genomewide scan for dyslexia and reported evidence of a QTL for speed of phonological decoding efficiency near marker D2S1399 at 2q. This marker lies 13 cM away from our peak marker at 2q with PIQ and lies next to the 1-LOD drop border.
Several positional candidate genes within the 2q24.1-2q31.1 linkage region have been tested for an association to autism, including GAD1 (MIM 605363), HOXD1 (MIM 142987), DLX1 (MIM 600029), DLX2 (MIM 126255), TBR-1 (MIM 604616), RAPGEF4 (MIM 606058), CHN1 (MIM 118423), SLC25A12 (MIM 603667), SCN1A (MIM 182389), SCN2A (MIM 182390), and SCN3A (MIM 182391) (Bacchelli et al. 2003; Weiss et al. 2003; Rabionet et al. 2004; Ramoz et al. 2004). Most promising as a site with possible relevance to IQ is the significant association between the relative risk to autism and two common SNPs in the mitochondrial aspartate/glutamate carrier SLC25A12 gene (Ramoz et al. 2004).
The linkage region on chromosome 6 (6p25.3-6p22.3 [D2S942–D2S422]) overlaps marginally with the region 6p22.3-6p21.3 implicated in reading disability and dyslexia (Cardon et al. 1994; Fisher et al. 1999b; Gayán et al. 1999; Grigorenko et al. 2000; Kaplan et al. 2002; Willcutt et al. 2002; Deffenbacher et al. 2004). Although recently at least five candidate genes were identified that are likely to contribute to linkage with reading disability, those genes lie just outside the region defined by a drop of 1 LOD score on 6p for VIQ and FSIQ scores (Deffenbacher et al. 2004). The ALDH5A1 gene (6p22-6p23), implicated in both cognitive ability (Plomin et al. 2004) and reading disability (Deffenbacher et al. 2004), lies at the border of our 6p region.
Several genes within the 2q and 6p linkage regions have been associated with schizophrenia (NR4A2 [MIM 601828] at 2q24, DTNBP1 [MIM 607145] at 6p22, KIF13A [MIM 605433] at 6p22, and NQO2 [MIM 160998] at 6p25), fragile X syndrome (RANBP9 [MIM 603854] at 6p23), and Bardet-Biedl syndrome (BBS5 [MIM 603650] at 2q31). These disorders are accompanied by rather severe cognitive impairment, but milder variants in these same genes could influence variation in the normal range of cognitive abilities. Recently, evidence was found for linkage at 6p24 to a neurocognitive-deficit subtype of schizophrenia, in which the maximum LOD score occurred at marker D6S309, which is within the region that shows linkage to VIQ and FSIQ (Hallmayer et al., in press). Currently, dysbindin-1 (DTNBP1) at 6p22.3 is the best-supported susceptibility gene for schizophrenia (Talbot et al. 2004). Recent identification of a relationship between dysbindin-1 and hippocampal glutamate neurotransmission, a core concept of leading neurobiological theories of memory and learning, suggests further potential as an IQ gene (Talbot et al. 2004). A second positional candidate gene thought to be involved in memory processes is NRN1 (MIM 607409) at 6p25.1, which plays a role in neuritogenesis in mature brains. Naeve et al. (1997) showed that expression of neuritin, the product of NRN1, is induced by neural activity and by the activity-regulated brain-derived neurotrophic factor and neurotrophin-3. Neuritin is expressed in hippocampal and cortical neurons and is suspected to regulate neuronal plasticity during development and in the adult brain.
Given the gradual increase in heritability of IQ from childhood to late adolescence, those genes in our regions that influence brain development may be promising as candidate genes for IQ. For example, TBR-1 (MIM 604616), a neuron-specific T-box transcription factor, plays a critical role in brain development and is specifically expressed in the cortex. It is thought to be a common genetic determinant for the differentiation of early-born glutamatergic neocortical neurons and may provide insights into the functions of these neurons as regulators of cortical development (Hevner et al. 2001, 2002). More specifically, Hevner et al. (2002) showed, using Tbr1 mutant mice, that Tbr1 is critical in the appropriate establishment of precise reciprocal projections between cortical areas and corresponding thalamic nuclei.
In summary, a genome scan for intelligence has revealed two regions with putative evidence of linkage: 2q24.1-2q31.1 and 6p25.3-6p22.3. To identify these regions, we analyzed variation in cognition in healthy subjects. These areas were previously implicated in disorders that are accompanied by cognitive impairment, which supports the idea that trait variation within the normal range might be used to detect genes for (mental) disorders, such as autism, schizophrenia, or reading disability.
Acknowledgments
Financial support was provided by the Netherlands Organization for Scientific Research Spinoza (grant 904-61-090), the National Supercomputing Facilities (grants sg214 and sg217), and the Human Frontiers of Science Program (grant rg0154/1998-B). D.P. was supported in part by the GenomeEUtwin project (European Union contract QLG2-CT-2002-01254). Dutch genotyping was performed by the Center for Medical Genetics at Marshfield (Director, Dr. James Weber) and the Leiden University Medical Centre (Professor Slagboom). Australian phenotype collection was funded by Australian Research Council grants A79600334, A79906588, A79801419, DP0212016, and DP0343921; genotyping was funded by the Australian National Health and Medical Research Council's Program in Medical Genomics (grant NHMRC-219178) and the Center for Inherited Disease Research (CIDR) (Director, Dr. Jerry Roberts) at The Johns Hopkins University, under a grant to Dr. Jeff Trent and N.G.M. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract N01-HG-65403). We thank Anjali Henders, Megan Campbell, and staff from the Molecular Epidemiology Laboratory, for blood processing and DNA extraction of the Australian samples; Peter Visscher, for valuable comments; and Arjen van Bochoven, for support in supercomputer programming.
Web Resources
The URLs for data presented herein are as follows:
- Integrated genetic map with interpolated genetic map positions, http://www2.qimr.edu.au/davidD/master_map.dat
- Mx Home Page, http://www.vcu.edu/mx/
- Mx Scripts, http://www.psy.vu.nl/mxbib/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 [DOI] [PubMed] [Google Scholar]
- ——— (2002) Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Bacchelli E, Blasi F, Biondolillo M, Lamb JA, Bonora E, Barnby G, Parr J, Beyer KS, Klauck SM, Poustka A, Bailey AJ, Monaco AP, Maestrini E, International Molecular Genetic Study of Autism Consortium (IMGSAC) (2003) Screening of nine candidate genes for autism on chromosome 2q reveals rare nonsynonymous variants in the cAMP-GEFII gene. Mol Psychiatry 8:916–924 [DOI] [PubMed] [Google Scholar]
- Badcock JC, Dragovic M, Waters FA, Jablensky A (2005) Dimensions of intelligence in schizophrenia: evidence from patients with preserved, deteriorated and compromised intellect. J Psychiatr Res 39:11–19 [DOI] [PubMed] [Google Scholar]
- Bouchard TJ Jr, McGue M (1981) Familial studies of intelligence: a review. Science 212:1055–1059 [DOI] [PubMed] [Google Scholar]
- Butcher LM, Meaburn E, Dale PS, Sham P, Schalkwyk LC, Craig IW, Plomin R (2005) Association analysis of mild mental impairment using DNA pooling to screen 432 brain-expressed single-nucleotide polymorphisms. Mol Psychiatry 10:384–392 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 [DOI] [PubMed] [Google Scholar]
- Chorney MJ, Chorney K, Seese N, Owen MJ, Daniels J, McGuffin P, Thompson LA, Detterman DK, Benbow C, Lubinski D, Eley T, Plomin R (1998) A quantitative trait locus associated with cognitive ability in children. Psychol Sci 9:159–166 [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD (2004) Refinement of the 6p213 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet 115:128–138 [DOI] [PubMed] [Google Scholar]
- Fisher PJ, Turic D, Williams NM, McGuffin P, Asherson P, Ball D, Craig I, Eley T, Hill L, Chorney K, Chorney MJ, Benbow CP, Lubinski D, Plomin R, Owen MJ (1999a) DNA pooling identifies QTLs on chromosome 4 for general cognitive ability in children. Hum Mol Genet 8:915–922 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999b) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J (1999) The genetic basis of cognition. Brain 122:2015–2031 [DOI] [PubMed] [Google Scholar]
- Gayán J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC (1999) Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS (1999) The physiological approach: functional architecture of working memory and disordered cognition in schizophrenia. Biol Psychiatry 46:650–661 [DOI] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls DL (2000) Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. Am J Hum Genet 66:715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer JF, Kalaydjieve L, Badcock J, Dragovic M, Howell S, Michie PT, Rock D, Vile D, Williams R, Corder EH, Hollingsworth K, Jablensky A. Genetic evidence for a distinct subtype of schizophrenia characterized by pervasive cognitive deficit. Am J Hum Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Beekman M, Putter H, Lakenberg N, van der Wijk HJ, Whitfield JB, Posthuma D, Pedersen NL, Martin NG, Boomsma DI, Slagboom PE. Meta-analysis of four new genome scans for lipid parameters and analysis of positional candidates in positive linkage regions. Eur J Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Hevner RF, Miyashita-Lin E, Rubenstein JL (2002) Cortical and thalamic axon pathfinding defects in Tbr1 Gbx2 and Pax6 mutant mice: evidence that cortical and thalamic axons interact and guide each other. J Comp Neurol 447:8–17 [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL (2001) Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29:353–366 [DOI] [PubMed] [Google Scholar]
- Inlow JK, Restifo LL (2004) Molecular and comparative genetics of mental retardation. Genetics 166:835–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) (2001) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DN (1998) Multidimensional Aptitude Battery II. Sigma Assessment Systems, Port Huron, MI [Google Scholar]
- Kaplan DE, Gayán J, Ahn J, Won TW, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, Smith SD, Gruen J (2002) Evidence for linkage and association with reading disability on 6p21.3-22. Am J Hum Genet 70:1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Luciano M, Wright MJ, Martin NG (2004) Exploring the etiology of the association between birthweight and IQ in an adolescent twin sample. Twin Res 7:62–71 [DOI] [PubMed] [Google Scholar]
- Naeve GS, Ramakrishnan M, Kramer R, Hevroni D, Citri Y, Theill LE (1997) Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc Natl Acad Sci USA 94:2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Boker S, Xie G, Maes HH (1997) Mx software: statistical modeling. Richmond, VA [Google Scholar]
- Petrill SA, Plomin R, McClearn GE, Smith DL, Vignetti S, Chorney MJ, Chorney K, Thompson LA, Detterman DK, Benbow C, Lubinski D, Daniels J, Owen MJ, McGuffin P (1996) DNA markers associated with general and specific cognitive abilities. Intelligence 23:191–203 [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, and the Paris Autism Research International Sibpair Study (1999) Genome-wide scan for autism susceptibility genes. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Plomin R, Hill L, Craig IW, McGuffin P, Purcell S, Sham P, Lubinski D, Thompson LA, Fisher PJ, Turic D, Owen MJ (2001) A genome-wide scan of 1842 DNA markers for allelic associations with general cognitive ability: a five-stage design using DNA pooling and extreme selected groups. Behav Genet 31:497–509 [DOI] [PubMed] [Google Scholar]
- Plomin R, Turic DM, Hill L, Turic DE, Stephens M, Williams J, Owen MJ, O'Donovan MC (2004) A functional polymorphism in the succinate-semialdehyde dehydrogenase (aldehyde dehydrogenase 5 family member A1) gene is associated with cognitive ability. Mol Psychiatry 9:582–586 [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJC, Boomsma DI (2001) Perceptual speed and IQ are associated through common genetic factors. Behav Genet 31:593–602 [DOI] [PubMed] [Google Scholar]
- Rabionet R, Jaworski JM, Ashley-Koch AE, Martin ER, Sutcliffe JS, Haines JL, Delong GR, Abramson RK, Wright HH, Cuccaro ML, Gilbert JR, Pericak-Vance MA (2004) Analysis of the autism chromosome 2 linkage region: GAD1 and other candidate genes. Neurosci Lett 372:209–214 [DOI] [PubMed] [Google Scholar]
- Ramakers GJA (2003) Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci 25:191–199 [DOI] [PubMed] [Google Scholar]
- Ramoz N, Reichert JG, Smith CJ, Silverman JM, Bespalova IN, Davis KL, Buxbaum JD (2004) Linkage and association of the mitochondrial aspartate/glutamate carrier SLC25A12 gene with autism. Am J Psychiatry 161:662–669 [DOI] [PubMed] [Google Scholar]
- Raskind WH, Igo RP, Chapman NH, Berninger VW, Thomson JB, Matsushita M, Brkanac Z, Holzman T, Brown M, Wijsman EM (2005) A genome scan in multigenerational families with dyslexia: identification of a novel locus on chromosome 2q that contributes to phonological decoding efficiency. Mol Psychiatry (http://www.nature.com/mp/journal/vaop/ncurrent/index.html) (electronically published March 8, 2005; accessed 27 June 2005) [DOI] [PubMed] [Google Scholar]
- Shao Y, Raiford KL, Wolpert CM, Cope HA, Ravan SA, Ashley-Koch AA, Abramson RK, Wright HH, DeLong RG, Gilbert JR, Cuccaro ML, Pericak-Vance MA (2002) Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Hum Genet 70:1058–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, Hahn CG, Siegel SJ, Trojanowski JQ, Gur RE, Blake DJ, Arnold SE (2004) Dysbindin-1 is reduced in intrinsic glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest 113:1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D (1997) WAIS-III Wechsler Adult Intelligence Scale. Psychological Corporation, San Antonio [Google Scholar]
- Weiss LA, Escayg A, Kearney JA, Trudeau M, MacDonald BT, Mori M, Reichert J, Buxbaum JD, Meisler MH (2003) Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry 8:186–194 [DOI] [PubMed] [Google Scholar]
- Willcutt EG, Pennington BF, Boada R, Ogline JS, Tunick RA, Chhabildas NA, Olson RK (2001) A comparison of the cognitive deficits in reading disability and attention-deficit/hyperactivity disorder. J Abnorm Psychol 110:157–172 [DOI] [PubMed] [Google Scholar]
- Willcutt EG, Pennington BF, Smith SD, Cardon LR, Gayan J, Knopik VS, Olson RK, DeFries JC (2002) Quantitative trait locus for reading disability on chromosome 6p is pleiotropic for attention-deficit/hyperactivity disorder. Am J Med Genet 114:260–268 [DOI] [PubMed] [Google Scholar]
- Wright MJ, De Geus EJC, Ando J, Luciano M, Posthuma D, Ono Y, Hansell N, Van Baal GCM, Hiraishi K, Hasegawa T, Smith G, Geffen G, Geffen L, Kanba S, Miyake A, Martin NG, Boomsma DI (2001) Genetics of cognition: outline of collaborative twin study. Twin Res 4:48–56 [DOI] [PubMed] [Google Scholar]
- Yalcin B, Willis-Owen SA, Fullerton J, Meesaq A, Deacon RM, Rawlins JN, Copley RR, Morris AP, Flint J, Mott R (2004) Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet 36:1197–1202 [DOI] [PubMed] [Google Scholar]
- Zhu G, Duffy DL, Eldridge A, Grace M, Mayne C, O'Gorman L, Aitken JF, Neale MC, Hayward NK, Green AC, Martin NG (1999) A major quantitative-trait locus for mole density is linked to the familial melanoma gene CDKN2A: a maximum-likelihood combined linkage and association analysis in twins and their sibs. Am J Hum Genet 65:483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Evans DM, Duffy DL, Montgomery GW, Medland SE, Gillespie NA, Ewen KR, Jewell M, Liew YW, Hayward NK, Sturm RA, Trent JM, Martin NG (2004) A genome scan for eye color in 502 twin families: most variation is due to a QTL on chromosome 15q. Twin Res 7:197–210 [DOI] [PubMed] [Google Scholar]