To the Editor:
Parkinson disease [PD (MIM 168600)] is the second most common neurodegenerative disorder, characterized by resting tremor, rigidity, bradykinesia, and gait disturbances. The LRRK2 gene (GenBank accession number AY792511) was recently identified as being responsible for autosomal dominant PD (Paisan-Ruiz et al. 2004; Zimprich et al. 2004). Several groups have reported that a single pathogenic G6055A substitution (G2019S) in the LRRK2 gene was associated with 3%–6% and 1%–2% of familial and sporadic PD, respectively (Di Fonzo et al. 2005; Gilks et al. 2005; Kachergus et al. 2005; Lesage et al. 2005; Nichols et al. 2005). In addition, Kachergus et al. (2005) demonstrated that, in 13 families of European descent, G2019S-mutation carriers shared a small ancestral haplotype, suggestive of a common founder.
In our sample of 198 affected probands from families with PD that is compatible with autosomal dominant inheritance, mostly from France and North Africa, we identified a total of 13 LRRK2 G2019S-mutation carriers, one of whom was homozygous for this mutation. Five were of European descent (two from France and one each from Portugal, Belgium, and The Netherlands), one was from North America, and seven were from North Africa. Surprisingly, one 60-year-old healthy French control individual also carried the same mutation.
To determine whether a common haplotype was also shared by the G2019S carriers in our series, all available family members of the 14 LRRK2-positive families were genotyped for the 17 chromosome 12q microsatellite markers and four SNPs, described elsewhere (Kachergus et al. 2005), that span a 16-Mb region that includes the LRRK2 gene. Seven markers (three microsatellites and four SNPs) were located within the gene. A total of 62 individuals were analyzed: 40 G2019S carriers (23 affected and 17 unaffected) and 22 noncarriers, 2 of whom were affected. The microsatellites were genotyped by multiplexing appropriate labeled primers. The fluorescent PCR products were then pooled for analysis, in two runs, on an ABI 3730 automated sequencer. Results were analyzed with GeneMapper 3.5 software (Applied Biosystems). Two DNA samples from CEPH reference families 1331-01 and 1331-02 were used as external standards, to control for consistency between runs. The four LRRK2 SNPs were genotyped by using the SnapShot multiplex kit (Applied Biosystems), in accordance with the manufacturer’s instructions. Haplotypes were constructed using the Merlin program (Abecasis et al. 2002; Center for Statistical Genetics), and their frequencies were estimated with the expectation-maximization algorithm (Excoffier and Slatkin 1995). The difference in haplotype distribution between mutation carriers and noncarriers was evaluated by χ2 and Fisher’s exact tests. Marker-allele frequencies and marker positions were described elsewhere (Kachergus et al. 2005). The age of the mutation was evaluated by estimating the age of the most recent common ancestor of the G2019S-mutation carriers on the basis of information from their shared haplotypes (Genin et al. 2004).
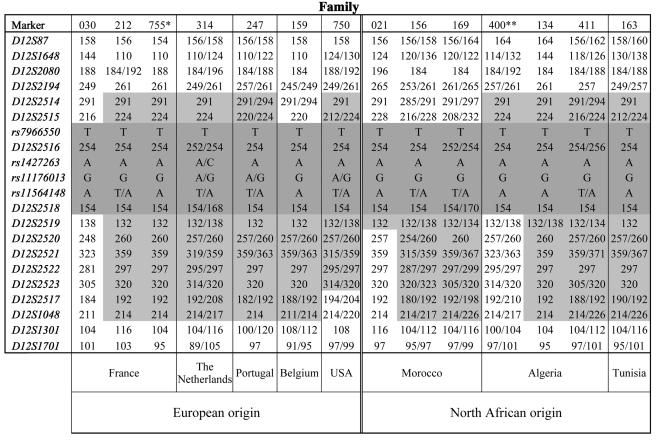
The LRRK2 haplotype that was associated with the G2019S mutation in all families originating from Europe and North Africa (fig. 1) was consistent with that described by Kachergus et al. (2005). However, the shared region was smaller than the 145–154 kb previously published, which reduces the common haplotype to only 60 kb. This minimal haplotype (T-254-A-G-A-154) consisting of six adjacent markers spanning the region between SNP rs7966550 (Entrez SNP) in exon 22 of the LRRK2 gene and microsatellite marker D12S2518 in intron 45 flanking the G2019S mutation in exon 41 was determined in French family 030, in which 29 members in two generations were available for study. Parental phases could also be unambiguously determined in six other families. In the remaining families, the haplotype could not be reconstructed unambiguously from the shared region, but the genotypes were all compatible with the presence of the common haplotype. In the majority of families, the shared region extended farther than the minimal haplotype, particularly 3′ of the LRRK2 gene, and independent of geographical origin. The common haplotype was also shared by five (24.7%) noncarriers of the G2019S mutation. However, the minimal haplotype was significantly more frequent in mutation carriers (7/7) than in noncarriers (5/21) (χ2=12.44; P<.001), even when only the seven families in which the G2019S haplotype could be unambiguously determined were taken into account.
Figure 1.
Genotypes for chromosome 12q12 markers in the disease haplotype in subjects from 14 LRRK2-positive families. The sequences of the microsatellites and SNPs were from the GDB Human Genome Database and Entrez SNP, respectively. An asterisk (*) indicates an unaffected individual from the control population who carried the G2019S mutation. A double asterisk (**) indicates a homozygous G2019S carrier. For families in which phase could not be unambiguously determined, both alleles are shown. The haplotype shared by all G2019S carriers is highlighted in gray.
This finding, in addition to previous data, extends the potential founder effect observed in Norwegians, Irish, Polish, and Americans of European descent to other European countries (France, Belgium, Portugal, and The Netherlands) and to three countries in North Africa (Algeria, Morocco, and Tunisia). The widespread distribution of the founder effect in Europe and its extension to North Africa prompted us to evaluate the time at which the mutation event might have occurred. Using patients for whom extended haplotypes could be unambiguously determined in our study (from families 030, 755, 021, and 400) and the study of Kachergus and his collaborators (2005) (from families P-063 and 1120), we estimated that the mutation occurred 29 generations earlier (95% CI 15–55). If a generation is defined as 25 years, the mutation occurred 725 (95% CI 375–1375) years ago, in the 13th century.
In conclusion, we have extended the founder effect for the LRRK2 G2019S mutation to other European countries and to North Africa, and we have reduced the common ancestral haplotype to a 60-kb region. The most recent common ancestor probably lived ∼725 years ago.
Acknowledgments
We are grateful to the patients and their families. We thank Drs. Pierre Pollak, François Tison, Myriem Tazir, and Joao Guimaraes for their contribution to the recruitment and to the evaluation of patients. This project was supported by the European Grant APOPIS (European Union contract LSHM-CT-2003-503330) and INSERM/Association Français contre les Myopathies Cohortes et Collections 2001. Members of the French Parkinson’s Disease Genetics Study Group are Y. Agid, A.-M. Bonnet, M. Borg, A. Brice, E. Broussolle, P. Damier, A. Destée, A. Dürr, F. Durif, E. Lohmann, M. Martinez, C. Penet, P. Pollak, O. Rascol, F. Tison, C. Tranchant, M. Vérin, F. Viallet, M. Vidailhet, and J.-M. Warter (deceased).
Web Resources
The accession number and URLs for data presented herein are as follows:
- Center for Statistical Genetics, http://www.sph.umich.edu/csg/abecasis/Merlin/
- Entrez SNP, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=snp
- GDB Human Genome Database, http://www.gdb.org/
- Genbank, http://www.ncbi.nlm.nih.gov/Genbank/ (for Homo sapiens LRRK2 [accession number AY792511])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PD) [PubMed]
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Di Fonzo A, Rohe CF, Ferreira J, Chien HF, Vacca L, Stocchi F, Guedes L, Fabrizio E, Manfredi M, Vanacore N, Goldwurm S, Breedveld G, Sampaio C, Meco G, Barbosa E, Oostra BA, Bonifati V, Italian Parkinson Genetics Network (2005) A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson’s disease. Lancet 365:412–415 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 [DOI] [PubMed] [Google Scholar]
- Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L (2004) Estimating the age of rare disease mutations: the example of triple-A syndrome. J Med Genet 41:445–449 10.1136/jmg.2003.017962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW (2005) A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet 365:415–416 [DOI] [PubMed] [Google Scholar]
- Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, Gibson JM, Ross OA, Lynch T, Wiley J, Payami H, Nutt J, Maraganore DM, Czyzewski K, Styczynska M, Wszolek ZK, Farrer MJ, Toft M (2005) Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet 76:672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Ibanez P, Lohmann E, Agid Y, Dürr A, Brice A (2005) The G2019SLRRK2 mutation in autosomal dominant European and North African Parkinson’s disease is frequent and its penetrance is age-dependent. Neurology 64:182615911835 [Google Scholar]
- Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, Michaels VE, Reed T, Rudolph A, Shults CW, Singleton A, Foroud T, and the Parkinson Study Group—PROGENI Investigators (2005) A single LRRK mutation accounting for greater than 5% of familial Parkinson disease. Lancet 365:410–412 [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, de Munain AL, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44:595–600 10.1016/j.neuron.2004.10.023 [DOI] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44:601–607 10.1016/j.neuron.2004.11.005 [DOI] [PubMed] [Google Scholar]