Abstract
Familial adenomatous polyposis (FAP) is a dominantly inherited colorectal tumor predisposition that results from germ-line mutations in the APC gene (chromosome 5q21). FAP shows substantial phenotypic variability: classical polyposis patients develop more than 100 colorectal adenomas, whereas those with attenuated polyposis (AAPC) have fewer than 100 adenomas. A further group of individuals, so-called “multiple” adenoma patients, have a phenotype like AAPC, with 3–99 polyps throughout the colorectum, but mostly have no demonstrable germ-line APC mutation. Routine mutation detection techniques fail to detect a pathogenic APC germ-line mutation in approximately 30% of patients with classical polyposis and 90% of those with AAPC/multiple adenomas. We have developed a real-time quantitative multiplex PCR assay to detect APC exon 14 deletions. When this technique was applied to a set of 60 classical polyposis and 143 AAPC/multiple adenoma patients with no apparent APC germ-line mutation, deletions were found exclusively in individuals with classical polyposis (7 of 60, 12%). Fine-mapping of the region suggested that the majority (6 of 7) of these deletions encompassed the entire APC locus, confirming that haploinsufficiency can result in a classical polyposis phenotype. Screening for germ-line deletions in APC mutation-negative individuals with classical polyposis seems warranted.
Familial adenomatous polyposis (FAP) (Mendelian Inheritance in Man 175100) is an autosomal dominant predisposition to hundreds or thousands of colorectal adenomas, which accounts for less than 1% of the total colorectal cancer burden (1). The disorder is caused by germ-line mutations in the adenomatous polyposis coli (APC) gene and is classically characterized by more than 100 colorectal adenomas, early onset of colorectal carcinoma, and specific extracolonic features, including congenital hypertrophy of the retinal pigment epithelium, polyposis of the upper gastrointestinal tract, and desmoid tumors (reviewed in ref. 2). In addition to individuals with classical polyposis, there exist attenuated polyposis (AAPC) patients with fewer than 100 adenomas. AAPC patients harbor germ-line mutations in the 5′ and 3′ regions and exon 9 of the APC gene (3, 4). A further group of individuals, so-called “multiple” adenoma patients, have a phenotype like AAPC, with 3–99 polyps throughout the colorectum, but mostly no demonstrable germ-line APC mutation, and often with limited or nonexistent family history of polyposis.
Routine mutation detection techniques [for example, single-strand conformation polymorphism (SSCP) analysis, denaturing gradient gel electrophoresis (DGGE), DNA sequencing, and the protein truncation test (PTT)] identify germ-line APC mutations in approximately 70% and 10% of individuals with classical polyposis and AAPC/multiple adenomas, respectively (4–8). The underlying molecular genetic cause(s) therefore remain(s) to be determined in a considerable proportion of apparently APC mutation-negative patients. The failure to detect germ-line mutations might also, however, be caused by methodological difficulties, as has recently been shown for another colorectal cancer predisposition, hereditary nonpolyposis colorectal cancer, where over 5% of cases in some populations result from exon-spanning genomic deletions in hMSH2 (9). By analogy, such submicroscopic deletions may be missed in a substantial fraction of APC mutation-negative patients with classical polyposis or AAPC/multiple adenomas. So far, only limited data are available, and no comprehensive studies have been performed to establish the frequency of germ-line APC deletions in these patients (10–15).
In this survey, we aimed (i) to develop a real-time quantitative multiplex PCR assay (RQM-PCR) to detect APC germ-line deletions, (ii) to determine their frequency in apparently APC mutation-negative patients with classical disease as well as those with AAPC/multiple adenomas, and (iii) to fine-map all detected APC deletions by using a set of polymorphic markers spanning the entire APC gene.
Patients and Methods
Study Population.
This study examined 203 unrelated individuals clinically diagnosed with colorectal polyposis and in whom no germ-line APC mutation had been identified by using one or more standard mutation detection techniques (SSCP, DGGE, DNA sequencing, PTT). The patients came from diagnostic laboratories in the United Kingdom (Edinburgh, London, Oxford; n = 167), Denmark (Copenhagen; n = 11), Switzerland (Basel; n = 11), Portugal (Lisbon; n = 9), and Italy (Naples; n = 4). Overall, 60 (30%) patients displayed more than 100 colorectal adenomas (classical polyposis), and 143 (70%) patients displayed between 3 and 99 colorectal adenomas (“multiple adenoma”/AAPC phenotype). Additional phenotypic details (gender, age at diagnosis, family history, extracolonic disease, occurrence of colorectal cancer) were available for 165 (81%) individuals. Written informed consent was obtained from all individuals.
Real-Time Quantitative Multiplex (RQM)-PCR.
RQM-PCR determines gene dosage by monitoring PCR amplification in real-time and making use of the 5′ exonuclease activity of Taq DNA polymerase. Besides the standard PCR components, this assay requires an excess of an oligonucleotide probe that is specific to a sequence between the two primers. The probe is labeled with two different fluorophores, a reporter dye [6-carboxyfluorescein (6-FAM) or VIC] at the 5′ end and a quencher dye [6-carboxytetramethylrhodamine (TAMRA)] at the 3′ end. While the probe is intact, the emission of the reporter dye is absorbed by the quencher dye. During the extension phase of each PCR cycle, Taq DNA polymerase cleaves the oligonucleotide probes annealed between the two primers. Probe cleavage separates the reporter from the quencher dye. The resulting increase in reporter emission can be detected by using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) and is directly proportional to the amount of product being generated each PCR cycle. When probes with different reporter dyes are used, different amplicons can be amplified (multiplexed) in the same reaction.
The primers and probes were designed by using primer express software (Applied Biosystems). Exon 14 of APC was chosen as target for the assay, as it is encompassed by the majority of submicroscopic APC germ-line deletions reported to date (10–13). Exon 12 of human serum albumin (Alb), another single-copy gene, was chosen as internal control. Alb is located on chromosome 4q11–q13, a region that is not expected to be deleted or amplified in the germ-line of colorectal adenoma patients because it does not show loss of heterozygosity in colorectal adenomas, implicating a germ-line event. All primer and probe sequences were checked for specificity by using the National Center for Biotechnology Information blast program (www.ncbi.nlm.nih.gov/blast). The APC exon 14 and Alb exon 12 probes were labeled at the 5′ ends with the reporter dyes FAM and VIC, respectively, and the 3′ ends with the quencher dye TAMRA (Applied Biosystems). Sequences of the primers and probes are shown in Table 1.
Table 1.
Primers and probes for the RQM-PCR assay
| Primer/probe name | Sequence (5′ to 3′) | Amplicon size, bp |
|---|---|---|
| APC exon 14 forward | GCCAGACAAACACTTTAGCCATTA | 91 |
| APC exon 14 reverse | TACCTGTGGTCCTCATTTGTAGCTAT | |
| APC exon 14 probe (5′-FAM, 3′-TAMRA) | CTGGACACATTCCGTAATATCCCACCTCC | |
| Alb exon 12 forward | TTGCATGAGAAAACGCCAGTA | 70 |
| Alb exon 12 reverse | GTCGCCTGTTCACCAAGGA | |
| Alb exon 12 probe (5′-VIC, 3′-TAMRA) | TCTGTGCAGCATTTGGTGACTCTGTCAC |
The RQM-PCR assay was optimized by following the instructions in User Bulletin no. 5 (Applied Biosystems). When we used 20 ng of DNA in a 25-μl reaction, the optimal concentrations for APC exon 14 and Alb exon 12 were 50 nM for both forward primers and 300 nM for both reverse primers. The optimal probe concentrations proved to be 200 nM for APC exon 14 and 175 nM for Alb exon 12. DNA concentrations between 10 ng and 50 ng were within the linear dynamic range of the system.
Briefly, genomic DNA was isolated from EDTA-containing blood according to the salting-out method (16). DNA aliquots of 15 ng/μl were prepared for RQM-PCR. The assay was carried out in 96-well optical reaction plates sealed with optical adhesive covers (Applied Biosystems). Each experiment comprised in triplicate 3 normal controls, 1 deletion control, 27 patient samples, and 1 no-template control. Each 25-μl reaction contained 1× TaqMan Universal PCR Master mix (Applied Biosystems), 50 nM APC exon 14 and Alb exon 12 forward primers, 300 nM APC exon 14 and Alb exon 12 reverse primers, 200 nM APC exon 14 probe, 175 nM Alb exon 12 probe, and 2 μl of DNA. To reduce the risk of cross-contamination, samples and controls were added after aliquoting the master mix. The plates were centrifuged at 260 × g for 1 min, and the reactions were performed on an ABI PRISM 7700 Sequence Detection System. The thermal cycling conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and simultaneous annealing and extension at 60°C for 1 min.
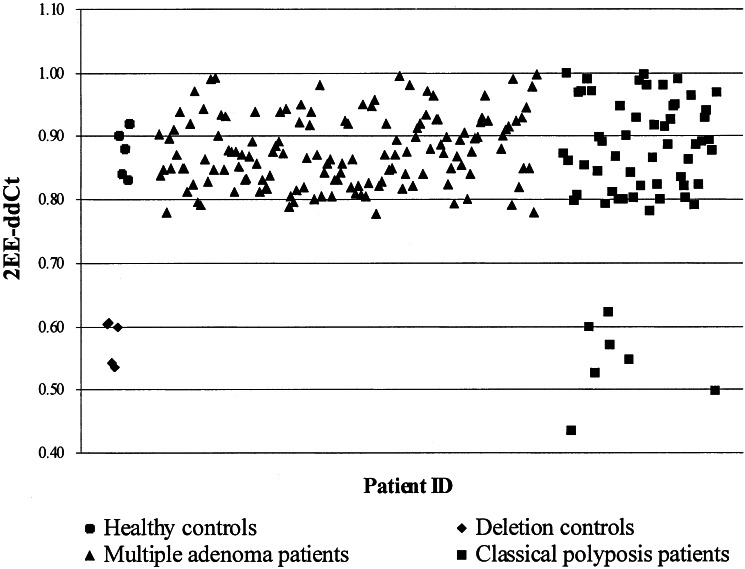
The data obtained were analyzed by using the comparative CT method (as described in User Bulletin no. 2 and ref. 17). After normalization against the internal Alb control, this method allowed determination of APC copy number of a given patient by comparison to a normal calibrator. Although the original CT method relied only on one normal sample as calibrator, we used the mean of three normal samples to obtain a more consistent calibrator value between experiments. The calibrator value (CV) was calculated as follows, with CS denoting the normal calibrator sample: CV = {[ΔCT Alb (CS 1) − ΔCT APC (CS 1)] + [ΔCT Alb (CS 2) − ΔCT APC (CS 2)] + [ΔCT Alb (CS 3) − ΔCT APC (CS 3)]}/3. APC gene copy number was expressed as a 2−(ΔΔCT) value, where ΔΔCT = CV − [ΔCT Alb (patient sample) − ΔCT APC (patient sample)]. ΔCT represented the mean CT value of each sample-triplicate, where CT was defined as the cycle number during the exponential phase at which the amplification plot passed a fixed normalized emission intensity threshold between 0.03 and 0.13 (average 0.08). Samples without APC deletions were expected to give 2−(ΔΔCT) values close to 1, and samples with APC deletions were expected to give 2−(ΔΔCT) values close to 0.5. The reliability of the assay was confirmed by using DNA samples of five unrelated, healthy controls and five unrelated FAP patients with previously characterized APC deletions encompassing exon 14. The healthy controls displayed 2−(ΔΔCT) values ranging from 0.83 to 0.92, whereas APC deletion controls showed 2−(ΔΔCT) values ranging from 0.54 to 0.61 (Fig. 1).
Figure 1.
RQM-PCR results for 5 healthy controls, 5 known deletion controls, 143 AAPC/multiple adenoma patients, and 60 classical polyposis patients. Patients with two copies of APC exon 14 display 2−(ΔΔCT) values between 0.78 and 1.00, whereas patients with only one copy display values between 0.43 and 0.62 (overall, 2−(ΔΔCT) values were consistent with a normal distribution and all APC deletion patients displayed 2−(ΔΔCT) values >2.9 standard deviations from the mean).
Genotype Analysis.
For all classical polyposis patients, genotypes were determined at nine different polymorphic marker loci. The following markers were intragenic to APC: (i) an A/G polymorphism (National Center for Biotechnology Information single-nucleotide polymorphism cluster ID: rs2019720) located within the promoter region, (ii) an A/T polymorphism (rs1914) located within intron 7, (iii) a T/C polymorphism located within exon 11 (18, 19), (iv) an A/G polymorphism located within exon 15I (20), (v) an A/G polymorphism located within exon 15J (19, 21), and (vi) a T/C polymorphism located within the 3′ untranslated region (19, 22). In addition, all patients with identified APC deletions were genotyped for two polymorphic markers, an A/G (rs748628) and a C/T polymorphism (rs1922665) located about 110 kb and 37 kb 5′ of APC, as well as three microsatellite markers, D5S346, D5S656, and D5S421 located about 32 kb, 396 kb, and 628 kb 3′ of APC, respectively (University of California, Santa Cruz Genome Browser, April 1, 2001 freeze at http://genome.ucsc.edu). Allele frequencies for the rs2019720, rs1914, rs748628, and rs1922665 polymorphisms were estimated using DNA from 28 unrelated, healthy European controls. We used an MJ Research Tetrad PCR machine, and the PCR conditions consisted of 95°C for 5 min, followed by 35 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 1 min, and a final extension step at 72°C for 10 min unless specified otherwise in Table 2. Restriction enzyme digestions, as indicated in Table 2, were performed according to the manufacturer's instructions (New England Biolabs). Sequencing reactions were carried out in forward and reverse orientations by using the ABI BigDye Terminator Ready Reaction Mix and analyzed on an ABI 377 semiautomated sequencer. Pyrosequencing was performed using a PSQ 96 System, PSQ 96 SNP Reagent kit, and PSQ 96 Sample Preparation kit (Pyrosequencing AB) according to manufacturer's instructions.
Table 2.
Primer sequences, annealing temperatures, detection methods applied, and allele frequencies for the APC polymorphisms
| APC region | Primer sequence (5′ to 3′) | Annealing temperature, °C | Detection | Allele 1 (allele frequency) | Allele 2 (allele frequency) | Ref. |
|---|---|---|---|---|---|---|
| rs748628 | CTTTTCTTTTTCTTTTTCt* | 53 | ARMS-PCR | A (0.45) | G (0.55) | † |
| CTTTTCTTTTTCTTTTTCc* | ||||||
| CTTACTACATTCAAGGGGAT | ||||||
| rs1922665 | CTTCCCTGTTCTGCCAATCT | 53 | Pyrosequencing | C (0.46) | T (0.54) | † |
| GCACTGGATGTTCAGAGACG‡ | ||||||
| TCTGTTGGTGGTCTCC§ | ||||||
| Promoter | TGGGGATGAGAGAAAGAGGAGGA | 60 | RsaI digest | A (0.45) | G (0.55) | † |
| CGCAAAAAGCCACTACCACTG | ||||||
| Intron 7 | CAGGTTTGAGCCATCATGC | 60 | Sequencing | A (0.46) | T (0.54) | † |
| ATCCAATCCCTAAGCTTGACTG | ||||||
| Exon 11 | GATGATTGTCTTTTTCCTCTTGC | 55 | RsaI digest | T (0.48) | C (0.52) | 18, 19 |
| CTGAGCTATCTTAAGAAATACATG | ||||||
| Exon 15I | AGTAAATGCTGCAGTTCAGAGG | 56 | BsaJ1 digest | A (0.62) | G (0.38) | 20 |
| CCGTGGCATATCATCCCCC | ||||||
| Exon 15J | CCCAGACTGCTTCAAAATTACC | 55 | Sequencing | A (0.57) | G (0.43) | 19, 21 |
| GAGCCTCATCTGTACTTCTGC | ||||||
| 3′ Untranslated region | GCATTAAGAGTAAAATTCCTCTTAC | 58 | SspI digest | T (0.54) | C (0.48) | 19, 22 |
| ATGACCACCAGGTAGGTGTATT |
Forward primers for A and G allele.
See Patients and Methods for details.
5′ Biotinylated reverse primer.
Sequencing primer.
Statistical Analysis.
Statistical comparison of index patients' features (gender, age at diagnosis, polyp number, colorectal cancer occurrence, and extracolonic disease) and molecular genetic data (gene dosage, genotype analysis) was performed by using the χ2 and Fisher's exact tests for categorical variables, or Student's t test for continuous variables, with all of the probabilities reported as two-tailed Ps, considering P < 0.05 to be statistically significant.
Results
Analysis of 60 classical polyposis and 143 AAPC/multiple adenoma patients with no apparent APC germ-line mutation by using the RQM-PCR assay identified 7 (3.4%) individuals harboring APC germ-line deletions encompassing exon 14, with 2−(ΔΔCT) values ranging from 0.43 to 0.62 (Fig. 1). Notably, all APC deletion patients came from the classical polyposis group, accounting for 11.7% (7 of 60) of these cases, compared with none (0 of 143) in the AAPC/multiple adenoma group (P < 0.001, χ2 test). The phenotypic features of the APC deletion patients compared with the remaining APC mutation-negative classical polyposis patients were very similar, with comparable median age at diagnosis (26.5 vs. 30 years) and similar proportions of positive family history (80.0% vs. 85.1%) and extracolonic disease (28.6% vs. 22.6%).
To assess the molecular extent of the APC germ-line deletions identified, six polymorphic markers intragenic to APC and five polymorphic markers flanking the APC gene were typed (Table 3). Six (85.7%) of seven deletion patients were apparently homozygous (suggesting hemizygosity) at markers spanning the coding region of the APC gene, consistent with whole-gene deletions. In three of these cases (602, 236.vi.10, and BAN1773), homozygosity at D5S656 suggested that the deletion might also encompass the MCC (mutated in colorectal cancers) locus. In at least one case (1749-1), the germ-line deletion did not involve the whole gene. Assessment of the polymorphic markers 5′ and 3′ of APC allowed to determine the maximum extent in six (85.7%) of the seven deletion cases and suggested that at least four (57.1%) were restricted to the APC gene (Table 3).
Table 3.
Genotype results for seven classical polyposis patients harbouring an APC exon-14 deletion as determined by RQM-PCR
| ID | 367.vi.4 | 1749-1 | 236.vi.11 | 2350 | BAN1773 | 602 | JS |
|---|---|---|---|---|---|---|---|
| rs748628 | het | hom | het | het | het | hom | het |
| rs1922665 | hom | hom | het | hom | hom | het | het |
| APC promoter | het | het | hom | hom | hom | hom | hom |
| APC intron 7 | hom | het | hom | hom | hom | hom | hom |
| APC exon 11 | hom | het | hom | hom | hom | hom | hom |
| APC exon 14 | del | del | del | del | del | del | del |
| APC exon 15I | hom | hom | hom | hom | hom | hom | hom |
| APC exon 15J | hom | hom | hom | hom | hom | hom | hom |
| APC 3′ untranslated region | hom | hom | hom | hom | hom | hom | hom |
| D5S346 | hom | het | het | hom | hom | hom | hom |
| MCC locus | del? | not del | not del | del? | del? | del? | del? |
| D5S656 | het | het | hom | hom | hom | hom | het |
| D5S421 | het | het | het | hom | het | het | hom |
Bold type indicates areas excluded from the respective APC deletions. het, heterozygosity; hom, homozygosity (suggesting hemizygosity); del, deletion; MCC, mutated in colorectal cancers.
We then assessed whether or not our analysis was likely to have failed to detect any large intragenic deletions 5′ or 3′ of exon 14 in 50 of the patients with classical FAP. Thirteen (26.0%) of these patients were homozygous for all intragenic markers. Two (4.0%) additional patients were homozygous for the three intragenic markers 5′ of exon 14. There was no evidence for an over-representation of homozygotes at any of the polymorphisms studied (data not shown). Although we cannot exclude the possibility that our analysis failed to identify a small number of patients with APC deletions, our data suggest that we did not miss large numbers of deletions and, therefore, such changes cannot account for all of the APC mutation-negative patients with classical FAP.
Discussion
Applying the RQM-PCR assay for the detection of APC exon-14 germ-line deletions, we identified 7 deletion patients in a set of 60 classical polyposis and 143 AAPC/multiple adenoma patients with no apparent APC germ-line mutation. Although previous studies of germ-line APC deletions have not consistently reported classical disease in these patients, all of our deletion patients displayed classical polyposis, exhibiting more than 100 adenomas. In contrast to the multiple adenoma subgroup (3–99 polyps), where no deletion could be detected, the frequency of APC deletions in patients with classical polyposis amounted to 11.7% of mutation-negative cases. Fine-mapping of the detected APC deletions (including available affected and unaffected relatives) by using six intragenic, polymorphic markers suggested a whole-gene deletion in six of seven patients (85.7%; Table 3).
Our results make an interesting comparison with previous data from AAPC patients. It has been suggested that in some cases, AAPC can result from instability of the mutant APC protein (23). van der Luijt et al. (24), for example, reported that truncating germ-line APC mutations in the 3′ half of exon 15 (codons 1862 and 1987) resulted in unstable protein, as determined by Western blotting, and in an AAPC phenotype. Because an unstable protein and a whole-gene deletion might be expected to be functionally equivalent, this difference in disease severity suggests the presence of residual mutant APC protein not detectable by Western blotting in these AAPC patients.
Of our seven APC-deletion patients, three had had formal polyp counts at colectomy (25). These counts (800, 1,425, and 1,899) were extremely similar to those typically reported for FAP patients with truncating APC mutations between codons 168 and 1250 or codons 1400 and 1580 approximately (26). Perhaps APC mutations between codons 168 and 1250 or codons 1400 and 1580 are functionally equivalent to null changes. Germ-line mutations between codons 1250 and 1400, the so-called mutation cluster region (MCR), are associated with more severe colonic polyposis (26, 27), possibly indicating that the mutant protein is more effective at causing tumorigenesis. There are alternative explanations for the phenotypic similarity between patients with APC deletions and those with truncating mutations between codons 168 and 1250 or codons 1400 and 1580. For example, the “second hit” at APC may be rate-limiting for tumorigenesis, and hence the main determinant of disease severity, given the evidence to show that most FAP polyps must acquire a truncating mutation in the MCR for optimal tumorigenesis (28). Germ-line deletion patients and those with truncating mutations outside the MCR must acquire such mutations through a second hit in the MCR, and thus would be expected to have similar disease severity.
The RQM-PCR assay that has been developed for this study proved to be a fast and reproducible method to detect gene dosage at APC exon 14. Given its potential for high-throughput analysis, its reliability as well as the small amounts of DNA needed (30 ng per reaction), this assay may be of use in a routine diagnostic setting. In view of the appreciable frequency (11.7%) of APC deletions in APC mutation-negative classical polyposis patients, this technique may be particularly valuable for this group of patients. Disadvantages of the technique, however, may include the high costs for the detection device and the consumables as well as the laborious assay setup. Although our assay will detect whole-gene APC deletions, it cannot exclude partial deletions 5′ and 3′ of exon 14.
In conclusion, we have developed a RQM-PCR assay to detect APC exon 14 deletions. When this technique was applied to a set of apparently APC mutation-negative polyposis patients, germ-line deletions were exclusively found in individuals with classical polyposis. Fine-mapping of the region suggested that the majority of these deletions encompass the entire APC locus. Given the frequency of 11.7%, screening of APC mutation-negative individuals with classical polyposis for germ-line deletions seems warranted.
Acknowledgments
We thank the patients for their participation in this study, and their respective doctors and pathologists for contributing clinical information; in particular, A. Alam (Imperial Cancer Research Fund, London), I. Frayling (Addenbrooke's Hospital, Cambridge, U.K.), W. Friedl (University of Bonn), and G. Taylor (St. James's University Hospital, Leeds, U.K.) for kindly providing control samples. We also thank the Imperial Cancer Research Fund Equipment Park and Genotyping Facility for their excellent technical support. This work was supported by the Imperial Cancer Research Fund and by grants from the Boehringer Ingelheim Foundation (to O.M.S.) and the Swiss Foundation for Medical-Biological Grants (to K.H.).
Abbreviations
- APC
adenomatous polyposis coli
- FAP
familial adenomatous polyposis
- AAPC
attenuated adenomatous polyposis coli
- RQM-PCR
real-time quantitative multiplex PCR
- Alb
albumin
References
- 1.Lynch H T, de la Chapelle A. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 2.Sieber O M, Tomlinson I P M, Lamlum H. Mol Med Today. 2000;6:462–469. doi: 10.1016/s1357-4310(00)01828-1. [DOI] [PubMed] [Google Scholar]
- 3.Spirio L, Olschwang S, Groden J, Robertson M, Samowitz W, Joslyn G, Gelbert L, Thliveris A, Carlson M, Otterud B, et al. Cell. 1993;75:951–957. doi: 10.1016/0092-8674(93)90538-2. [DOI] [PubMed] [Google Scholar]
- 4.Lamlum H, Al Tassan N, Jaeger E, Frayling I, Sieber O, Reza F B, Eckert M, Rowan A, Barclay E, Atkin W, et al. Hum Mol Genet. 2000;9:2215–2221. doi: 10.1093/oxfordjournals.hmg.a018912. [DOI] [PubMed] [Google Scholar]
- 5.Miyoshi Y, Ando H, Nagase H, Nishisho I, Horii A, Miki Y, Mori T, Utsunomiya J, Baba S, Petersen G, et al. Proc Natl Acad Sci USA. 1992;89:4452–4456. doi: 10.1073/pnas.89.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong J G, Davies D R, Guy S P, Frayling I M, Evans D G. Hum Mutat. 1997;10:376–380. doi: 10.1002/(SICI)1098-1004(1997)10:5<376::AID-HUMU7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 7.Heinimann K, Thompson A, Locher A, Furlanetto T, Bader E, Wolf A, Meier R, Walter K, Bauerfeind P, Marra G, et al. Cancer Res. 2001;61:7616–7622. [PubMed] [Google Scholar]
- 8.Cottrell S, Bicknell D, Kaklamanis L, Bodmer W F. Lancet. 1992;340:626–630. doi: 10.1016/0140-6736(92)92169-g. [DOI] [PubMed] [Google Scholar]
- 9.Wijnen J, van der Klift H, Vasen H, Khan P M, Menko F, Tops C, Meijers Heijboer H, Lindhout D, Moller P, Fodde R. Nat Genet. 1998;20:326–328. doi: 10.1038/3795. [DOI] [PubMed] [Google Scholar]
- 10.Mandl M, Caspari R, Jauch A, Boker T, Raschke H, Sengteller M, Propping P, Friedl W. Hum Genet. 1996;97:204–208. doi: 10.1007/BF02265266. [DOI] [PubMed] [Google Scholar]
- 11.De Rosa M, Scarano M I, Panariello L, Carlomagno N, Rossi G B, Tempesta A, Borgheresi P, Renda A, Izzo P. Eur J Hum Genet. 1999;7:695–703. doi: 10.1038/sj.ejhg.5200344. [DOI] [PubMed] [Google Scholar]
- 12.Su L K, Steinbach G, Sawyer J C, Hindi M, Ward P A, Lynch P M. Hum Genet. 2000;106:101–107. doi: 10.1007/s004399900195. [DOI] [PubMed] [Google Scholar]
- 13.Flintoff K J, Sheridan E, Turner G, Chu C E, Taylor G R. J Med Genet. 2001;38:129–132. doi: 10.1136/jmg.38.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodmer W F, Bailey C J, Bodmer J, Bussey H J, Ellis A, Gorman P, Lucibello F C, Murday V A, Rider S H, Scambler P, et al. Nature (London) 1987;328:614–616. doi: 10.1038/328614a0. [DOI] [PubMed] [Google Scholar]
- 15.Hodgson S V, Coonar A S, Hanson P J, Cottrell S, Scriven P N, Jones T, Hawley P R, Wilkinson M L. J Med Genet. 1993;30:369–375. doi: 10.1136/jmg.30.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller S A, Dykes D D, Polesky H F. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aarskog N K, Vedeler C A. Hum Genet. 2000;107:494–498. doi: 10.1007/s004390000399. [DOI] [PubMed] [Google Scholar]
- 18.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 19.Almeida R, Morton N, Fidalgo P, Leitao N, Mira C, Rueff J, Monteiro C. Clin Genet. 1996;50:483–485. doi: 10.1111/j.1399-0004.1996.tb02717.x. [DOI] [PubMed] [Google Scholar]
- 20.Davies S M, Snover D C. Hum Genet. 1994;93:329–330. doi: 10.1007/BF00212032. [DOI] [PubMed] [Google Scholar]
- 21.Kraus C, Ballhausen W G. Hum Genet. 1992;88:705–706. doi: 10.1007/BF02265305. [DOI] [PubMed] [Google Scholar]
- 22.Heighway J, Hoban P R, Wyllie A H. Nucleic Acids Res. 1991;19:6966. doi: 10.1093/nar/19.24.6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Luijt R B, Vasen H F, Tops C M, Breukel C, Fodde R, Meera Khan P. Hum Genet. 1995;96:705–710. doi: 10.1007/BF00210303. [DOI] [PubMed] [Google Scholar]
- 24.van der Luijt R B, Meera Khan P, Vasen H F, Breukel C, Tops C M, Scott R J, Fodde R. Hum Genet. 1996;98:727–734. doi: 10.1007/s004390050293. [DOI] [PubMed] [Google Scholar]
- 25.Crabtree M D, Tomlinson I P M, Talbot I C, Phillips R K. Gut. 2001;49:540–543. doi: 10.1136/gut.49.4.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crabtree, M. D., Tomlinson, I. P. M., Hodgson, S. V., Neale, K., Phillips, R. K. S. & Houlston, R. S. (2002) Gut, in press. [DOI] [PMC free article] [PubMed]
- 27.Nugent K P, Phillips R K, Hodgson S V, Cottrell S, Smith-Ravin J, Pack K, Bodmer W F. Gut. 1994;35:1622–1623. doi: 10.1136/gut.35.11.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamlum H, Ilyas M, Rowan A, Clark S, Johnson V, Bell J, Frayling I, Efstathiou J, Pack K, Payne S, et al. Nat Med. 1999;5:1071–1075. doi: 10.1038/12511. [DOI] [PubMed] [Google Scholar]