Abstract
sorLA (sorting protein-related receptor) is a type-1 membrane protein of unknown function that is expressed in neurons. Its homology to sorting receptors that shuttle between the plasma membrane, endosomes, and the Golgi suggests a related function in neuronal trafficking processes. Because expression of sorLA is reduced in the brain of patients with Alzheimer's disease (AD), we tested involvement of this receptor in intracellular transport and processing of the amyloid precursor protein (APP) to the amyloid β-peptide (Aβ), the principal component of senile plaques. We demonstrate that sorLA interacts with APP in vitro and in living cells and that both proteins colocalize in endosomal and Golgi compartments. Overexpression of sorLA in neurons causes redistribution of APP to the Golgi and decreased processing to Aβ, whereas ablation of sorLA expression in knockout mice results in increased levels of Aβ in the brain similar to the situation in AD patients. Thus, sorLA acts as a sorting receptor that protects APP from processing into Aβ and thereby reduces the burden of amyloidogenic peptide formation. Consequently, reduced receptor expression in the human brain may increase Aβ production and plaque formation and promote spontaneous AD.
Keywords: endocytic receptors, knockout mouse, neurodegeneration, Vps10p-domain receptors
Sorting protein-related receptor (sorLA), also known as LR11, is a 250-kDa type-1 membrane protein of unknown function that is expressed in neurons of the central and peripheral nervous system (1-4). The protein is a member of a family of neuronal receptors that share structural similarity with the vacuolar protein sorting 10 protein (Vps10p), a sorting protein in yeast that transports carboxypeptidase Y from the Golgi to the vacuole (5). Other family members include the proneurotrophin receptor sortilin (6) and the head activator-binding protein in hydra (7). Because sorLA interacts with the family of GGA (Golgi-localizing, γ-adaptin ear homology domain, ARF-interacting) adaptors that shuttle between the Golgi and endosomes/lysosomes, the receptor was proposed to act in intracellular protein trafficking (8). The relevance of such sorLA-mediated protein transport in neurons is unclear at present. However, expression profiling has demonstrated reduction of sorLA expression in the brain of patients suffering from Alzheimer's disease (AD), suggesting a causal role for the receptor in the pathogenesis of this disease (9).
Central to the pathogenesis of AD is the proteolytic processing of a neuronal membrane protein called the amyloid precursor protein (APP). APP follows a complex intracellular trafficking pathway that influences processing to either a soluble fragment sAPPα (nonamyloidogenic) or to sAPPβ and the insoluble amyloid β-peptide (Aβ), the principal component of senile plaques (10). The rate of Aβ production is considered the major risk factor for onset of AD (10). En route through the secretory pathway to the cell surface, most newly synthesized APP molecules are cleaved into sAPPα by α-secretase; however, some precursor molecules are reinternalized from the plasma membrane and delivered to endocytic compartments for β-secretase (and subsequent γ-secretase) processing into sAPPβ and Aβ (10) (see model in Fig. 6). Accordingly, the intracellular transport and localization of APP is a crucial determinant of APP processing and Aβ production. Yet, considerable controversy exists regarding the mechanisms that govern intracellular transport of the precursor protein.
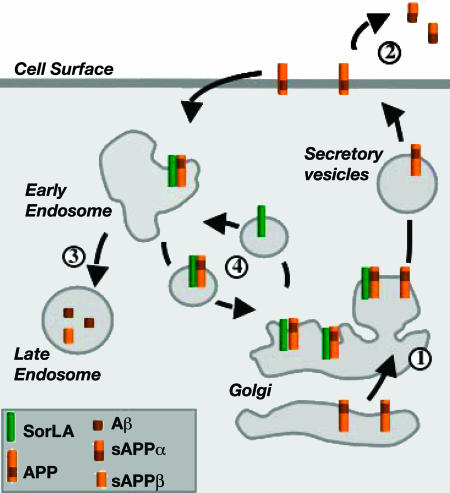
Fig. 6.
Proposed role for sorLA in APP processing. Newly synthesized APP molecules traverse the Golgi (“1”) to the plasma membrane, where some are cleaved to sAPPα (“2”). Nonprocessed precursors endocytose from the cell surface into late endosomal compartments for processing into sAPPβ and Aβ (“3”). SorLA acts as a sorting receptor that traps APP in the Golgi (“1”), reducing the amount of precursors that reach the cell surface for processing. In addition, sorLA may also shuttle APP from early endosomes back to the Golgi, further reducing the extent of Aβ production in late endosomes (“4”). Consistent with this model, overexpression of sorLA in cultured cells further reduces transition of APP to the cell surface and suppresses processing into sAPPα and Aβ, whereas loss of sorLA expression in AD patients and in knockout mice results in accelerated trafficking of APP into processing pathways and in increased production of sAPPα and Aβ.
Here, we have tested the hypothesis that sorLA acts as neuronal sorting receptor that binds APP and regulates its trafficking and proteolytic processing into Aβ, the process causative to AD.
Materials and Methods
Materials. A full description of the materials used in this study is provided in Supporting Methods, which is published as supporting information on the PNAS web site.
Biochemical Studies. Complex formation between sorLA and APP was tested by using surface plasmon resonance analysis (11) and molecular mass determination by sedimentation equilibrium technique (12) as previously described and detailed in Supporting Methods.
Immunoprecipitation. Cells were washed with PBS before treatment with membrane-permeable linker dithiobis-succinimidyl-propionate (Pierce) and subsequently lysed in Triton X-100/Nonidet P-40-containing buffer on ice. Immunoprecipitations were performed by using anti-sorLA or anti-APP antiserum and protein G-coupled Sepharose beads.
Cell Studies. SH-SY5Y or CHO cell lines expressing human APP695 were stably transfected with sorLA expression constructs. The amounts of APP and sorLA were determined in cell lysates by Western blotting; amounts of Aβ40 in cell supernatants were determined by ELISA (BioSource International, Camarillo, CA). Cellular localization of proteins was detected by confocal immunofluorescence microscopy using Alexa Fluor 488- or Cy3-conjugated secondary antibodies. Subcellular fractionation of CHO cells by discontinuous iodixanol density gradient was performed according to protocols described in ref. 13. Fluorescence lifetime imaging microscopy (FLIM) was performed on mouse neuroblastoma N2A cells (14) after transient transfection with expression constructs for human sorLA and APP695 as detailed in Supporting Methods.
Surface Biotinylation. SH-SY5Y cells were washed in PBS and treated with membrane-impermeable sulfo-N-hydroxysuccinimidobiotin (Pierce) in PBS for 30 min. The biotin labeling was quenched with Tris·HCl (pH 8.0) before cells were lysed with buffer containing Triton X-100/Nonidet P-40, and the biotinylated cell surface proteins were precipitated with streptavidin-coupled Sepharose (Amersham Pharmacia Biosciences).
SorLA-Deficient Mice. SorLA-deficient mice were generated by gene targeting in embryonic stem cells with mouse lines generated from three independently targeted cell clones. Animals were analyzed on hybrid (129SvEmcTer × C57BL/6N) or (129SvEmcTer × Balb/c) genetic backgrounds with identical results in all strains compared with age- and sex-matched control mice derived from heterozygous breeding. For immunohistology, 4-μm sections of paraffin-embedded tissues were stained for APP (CT695) or Aβ (4G8) by using primary antibodies, followed by peroxidase-conjugated secondary antibodies and detection with diaminobenzidine. Murine Aβ40 and Aβ42 were quantified by ELISA using murine-specific antibodies (15). Subcellular fractionation of murine brain homogenates was carried out by differential centrifugation (OptiPrep application sheet S07, Axis-Shield PoC, Oslo).
Results
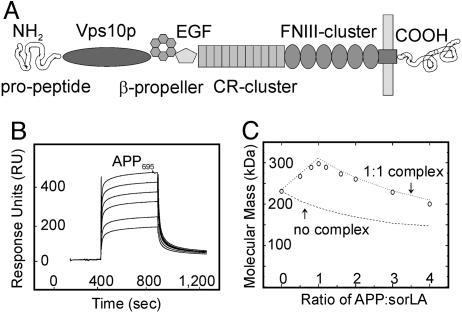
Initially, we tested the ability of sorLA to bind APP in vitro, a finding that may indicate a role for sorLA as an intracellular APP receptor. To do so, we expressed and purified the extracellular domains (ectodomains) of sorLA (Fig. 1A; see also Fig. 7, which is published as supporting information on the PNAS web site) and of the three major APP splice variants (APP695, APP751, and APP770) (Fig. 7) (10). Interaction of the proteins was evaluated by surface plasmon resonance analysis, an assay that records interaction of proteins with a target immobilized on a microchip. When the sorLA ectodomain immobilized on the biosensor chip was incubated with increasing concentrations of APP695, a rapid and reversible increase in response units indicated specific interaction of the two proteins on the chip surface (Fig. 1B). Based on the kinetics, a KD of ≈200 nM was determined by using biaevaluation 3.1 software. Similar binding kinetics were seen for APP751 and APP770 (Fig. 8, which is published as supporting information on the PNAS web site), demonstrating the ability of sorLA to interact with all APP isoforms.
Fig. 1.
APP and sorLA interact in vitro.(A) The structural elements of sorLA are depicted, including (from amino to carboxyl terminus) the propeptide, vacuolar protein sorting 10 protein (Vps10p) domain, β-propeller, epidermal growth factor repeat (EGF), and clusters of complement-type repeats (CR) and fibronectin type III domains (FNIII). (B and C) The sorLA ectodomain used for surface plasmon resonance (SPR) (B) or molecular mass analysis (C) encompassed all elements amino-terminal to the membrane anchor. (B) SPR analysis of binding of a concentration series (0.1, 0.2, 0.5, 1, 2, and 5 μM) of APP695 to sorLA immobilized on the sensor chip (KD = 200 nM). (C) Molecular-weight plot of APP-sorLA complex formation by analytic ultracentrifugation. The sorLA concentration was kept at 0.1 μM, whereas the APP695 concentration varied to give the indicated APP:sorLA input ratios. Circles indicate the actual data points of the observed average molecular masses in the protein mixture at a given APP:sorLA ratio. Lines represent theoretical graphs to be expected for a perfect 1:1 complex (dotted) or no complex formation (dashed).
To confirm interaction of APP with sorLA in solution, we determined complex formation by molecular mass determination using the sedimentation equilibrium technique (12). In this method, the molecular weight of individual proteins or protein complexes is evaluated by determination of sedimentation coefficients during ultracentrifugation. When the isolated sorLA ectodomain was characterized by ultracentrifugation in the absence of APP, an accurate molecular mass of 230 kDa for the monomeric domain was determined (Fig. 1C; APP:sorLA ratio = 0). When increasing concentrations of the APP695 ectodomain (80 kDa) were titrated into the sorLA solution, the average molecular mass in the protein mixture increased because of complex formation and reached a maximum of 310 kDa (80 + 230 kDa) at an equimolar ratio of both proteins (APP:sorLA ratio = 1). When the molar amounts of APP were further increased (APP:sorLA ratio > 1), the average molecular weight in the protein mixture decreased again, indicating that excess APP molecules did not bind sorLA anymore. This finding is in perfect agreement with formation of a stable 1:1 complex between APP and sorLA.
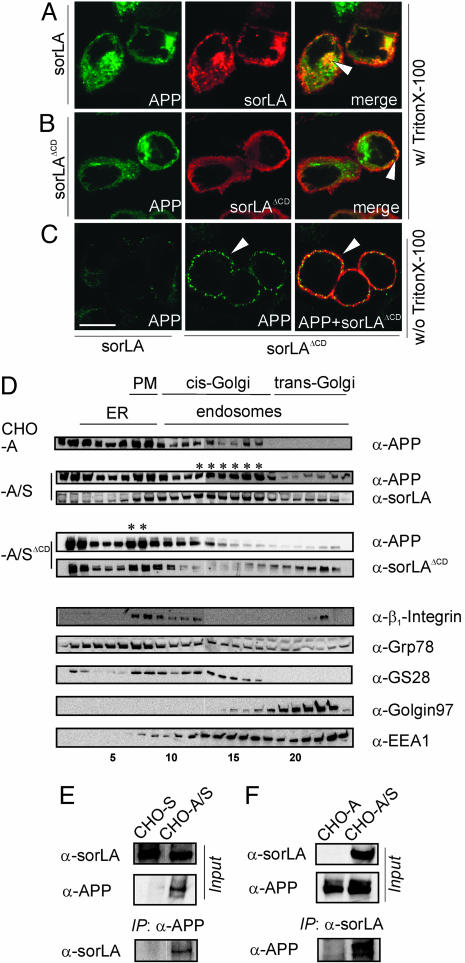
Next, we tested whether the interaction of sorLA and APP also occurred in cells. To do so, we used a CHO cell line that expresses human APP695 (CHO-A), the major neuronal APP variant. CHO-A cells were stably transfected with expression constructs for wild-type sorLA or for a mutant receptor that lacks the cytoplasmic domain (sorLAΔCD). Because of the absence of sorting signals of the cytoplasmic tail [such as GGA (Golgi-localizing, γ-adaptin ear homology domain, ARF-interacting)-binding or endocytosis motifs], sorLAΔCD exhibits an abnormal trafficking pattern and accumulates on the cell surface (Fig. 2B), whereas the wild-type receptor mainly localized to intracellular compartments (Fig. 2A). In cells expressing APP and sorLA (CHO-A/S), both proteins colocalized to vesicular structures and to the perinuclear region (Fig. 2A, arrowhead). This pattern was unique for coexpression with sorLA and was not seen when APP was expressed together with unrelated transmembrane proteins such as the interleukin 2 receptor (Fig. 9B, which is published as supporting information on the PNAS web site). In contrast to the situation in CHO-A/S, in cells transfected with sorLAΔCD (CHO-A/SΔCD), APP colocalized with the mutant receptor to the cell surface (Fig. 2B, arrowhead). The altered subcellular localization of APP in CHO-A/SΔCD compared with CHO-A/S was even more evident by surface staining of cells not permeabilized before antibody application (Fig. 2C). In these cells, APP accumulated on the plasma membrane in a punctuate pattern when coexpressed with sorLAΔCD (Fig. 2C Center and Right), whereas very little surface APP was seen in cells expressing wild-type sorLA (Left). These findings indicated that APP always localized to the same cellular compartments that harbored sorLA (be it intracellular vesicles for sorLA or cell surface for sorLAΔCD).
Fig. 2.
SorLA affects trafficking of APP in CHO cells. (A-C) Detection of APP695 and sorLA in CHO cells by using confocal immunofluorescence microscopy. (A) In permeabilized cells (with Triton X-100), APP695 and wild-type sorLA signals colocalize to intracellular vesicular structures and to the perinuclear region (arrowhead). (B) In stable transfectants coexpressing APP695 and sorLAΔCD, the sorLAΔCD signal is confined to the cell surface, where it colocalizes with APP (arrowhead). (C) In nonpermeabilized cells (without Triton X-100), more APP695 is detected on the surface in cells that express sorLAΔCD (Center, arrowhead), where it colocalizes with the mutant receptor (Right, arrowhead) compared with cells expressing wild-type sorLA (Left). (Scale bar: 10 μm.) (D) Subcellular fractionation of cells expressing APP695 (CHO-A) or APP695 together with wild-type sorLA (CHO-A/S) or sorLAΔCD (CHO-A/SΔCD). Immunodetection of APP and sorLA, as well as marker proteins β1-integrin (plasma membrane, PM), Grp78 (endoplasmic reticulum, ER), GS28 (cis-Golgi), Golgin97 (trans-Golgi network), and EEA1 (early endosomes) in the fractions is depicted. Asterisks mark coaccumulation of APP and sorLA in Golgi or in plasma membrane compartments of CHO-A/S and CHO-A/SΔCD cells, respectively. (E and F) Cells expressing sorLA (CHO-S), APP695 (CHO-A), or both (CHO-A/S) were treated with membrane-permeable cross-linker and proteins immunoprecipitated by using anti-APP (E) or anti-sorLA (F) antibodies. Western blots labeled “Input” show sorLA and APP in total cell extracts before immunoprecipitation. Western blots labeled “IP” demonstrate coimmunoprecipitation of sorLA in anti-APP (E) and APP in anti-sorLA (F) precipitates.
To identify the intracellular compartments that contained APP and sorLA in CHO cells, we applied subcellular fractionation to separate the various cell compartments and to test the presence of the proteins by Western blot (Fig. 2D Upper). As reference, the presence of marker proteins for distinct cell compartments was evaluated in parallel (Fig. 2D Lower). In cells not expressing sorLA (CHO-A), APP mainly localized to fractions containing markers of the endoplasmic reticulum [glucose-related protein 78 (Grp78); fractions 2-8] and the plasma membrane (β1-integrin; fractions 7-9). However, after the cells were stably cotransfected with sorLA (CHO-A/S), APP was also seen in fractions 12-17 (marked with asterisks) that contained sorLA. These fractions were positive for markers of the cis-Golgi [Golgi SNARE 28 (GS28)] and early endosomes [early endosomal antigen 1 (EEA1)], consistent with the vesicular and perinuclear (Golgi) staining for both proteins in intact cells (Fig. 2 A). Thus, expression of sorLA confined a significant portion of APP molecules to Golgi compartments. Again, confinement to the Golgi was not seen when APP was coexpressed with sorLAΔCD. Rather, both proteins accumulated in a distinct peak in the cell surface fraction (Fig. 2D, fractions 7 and 8, asterisks). Because the total amount of APP was not altered in CHO-A compared with CHO-A/S and CHO-A/SΔCD cells (Fig. 9C), the altered subcellular distribution observed for APP by coexpression with sorLA or sorLAΔCD likely represented altered intracellular trafficking rather than increased biosynthesis of the precursor.
To prove that colocalization of APP and sorLA in cells was caused by direct interaction, we applied two methods to evaluate APP and sorLA interaction in intact cells: coimmunoprecipitation (Fig. 2 E and F) and FLIM (Fig. 3). For coimmunoprecipitation, we used CHO cells that express sorLA (CHO-S), APP (CHO-A), or both proteins (CHO-A/S) (“Input” in Fig. 2 E and F). Cells were treated with membrane-permeable cross-linkers that penetrate the undisturbed cellular compartments and link APP- and sorLA-interacting proteins in the context of an intact cell. Thereafter, cell lysates were generated and immunoprecipated with either anti-APP (Fig. 2E) or anti-sorLA (Fig. 2F) antiserum. When anti-APP immunoprecipitates were probed with an anti-sorLA antibody, coprecipitation of sorLA with APP antiserum was detected in CHO-A/S cells that express both proteins but not in CHO-S cells that lack APP (Fig. 2E). Similarly, when anti-sorLA immunoprecipitates were probed with anti-APP antibodies, coprecipitation of APP with sorLA antiserum was detected in CHO-A/S cells but not in CHO-A cells that lack sorLA (Fig. 2F).
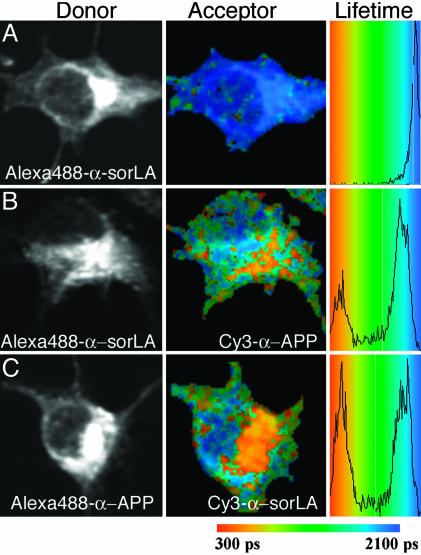
Fig. 3.
FLIM of APP695 and sorLA interaction in N2A cells. Shown are intensity images of sorLA (A and B) and APP695 (C) immunocytochemistry using donor fluorophore Alexa Fluor 488-conjugated antibodies (Left), pseudocolored FLIM images (Center), and lifetime histograms (Right), indicating shortening of lifetime from blue to orange/red in the absence (A) or presence (B and C) of acceptor fluorophore Cy3. (A) Cells stained for the amino-terminal domain of sorLA with donor fluorophore Alexa Fluor 488 in the absence of acceptor (lifetime of 2,007 ± 12 psec). (B) Cells stained for sorLA with donor fluorophore Alexa Fluor 488 in the presence of acceptor Cy3 on the ectodomain of APP695 (lifetime reduced to 1,365 ± 139 psec). (C) Cells stained for APP with donor Alexa Fluor 488 in the presence of acceptor Cy3 on sorLA (lifetime reduced to 1,481 ± 228 psec).
Direct interaction of APP and sorLA was further confirmed by FLIM (Fig. 3), a proximity assay used to study protein interaction in live cells. FLIM relies on the fact that the fluorescence lifetime of a donor fluorophore tethered to a target protein by means of antibodies (here, anti-sorLA) is shorter in close proximity of an acceptor fluorophore bound by means of antibodies to a binding partner (here, anti-APP). The degree to which lifetime is reduced is directly related to the distance of the two antigens in the cell (see Supporting Methods for details). We performed FLIM on mouse neuronal N2A cells transiently transfected with constructs for APP695 and wild-type sorLA. The cells were stained for the ectodomain of sorLA by donor fluorophore Alexa Fluor 488 and for APP695 with acceptor Cy3. The lifetime of Alexa Fluor 488 at sorLA in the absence of the acceptor Cy3 on APP695 was 2,007 ± 12 psec (Fig. 3A). This lifetime was dramatically shortened to 1,365 ± 139 psec (n = 10; P < 0.0001) when cells were costained for APP with acceptor Cy3 (Fig. 3B). Equivalent results were obtained when acceptor and donor were exchanged (lifetime of 1,481 ± 228 psec versus 1,995 ± 20 psec without acceptor; n = 12; P < 0.0001) (Fig. 3C), confirming close proximity of the sorLA and APP ectodomains.
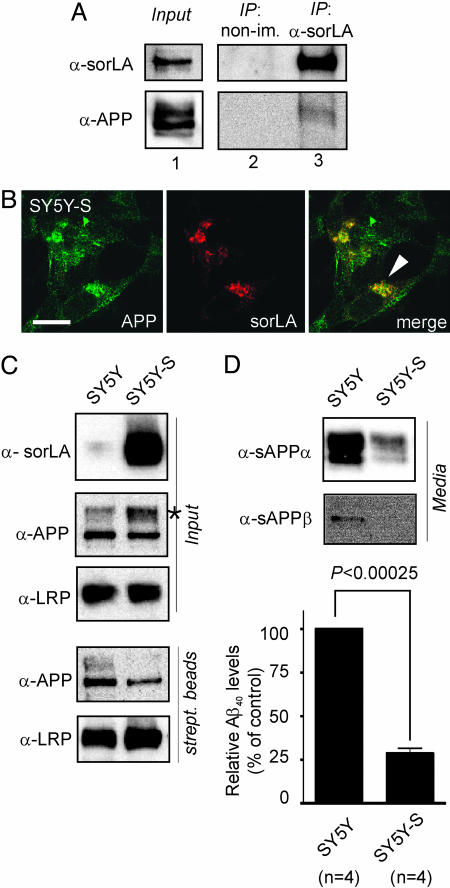
Taken together, findings in CHO and N2A cells demonstrated that sorLA and APP directly interact in living cells and that subcellular localization of sorLA determines trafficking of APP. These data suggested APP as a likely physiological target for sorLA-mediated protein sorting in neurons. Because interaction with sorLA caused accumulation of APP in the Golgi (compare CHO-A and CHO-A/S in Fig. 2D), we investigated whether sorLA-mediated APP transport may affect proteolytic processing of the precursor protein. To do so, we applied the human neuronal cell line SH-SY5Y that is commonly used to study APP processing. Despite low amounts of endogenous sorLA expressed in these cells (“Input” in Fig. 4A), coprecipitation of endogenous APP with anti-sorLA but not with nonimmune IgG was possible (“IP” in Fig. 4A), confirming the findings in CHO cells.
Fig. 4.
SorLA alters processing of APP in SH-SY5Y cells. (A) Nontransfected SH-SY5Y cells were treated with membrane-permeable cross-linker, and proteins were immunoprecipitated by using anti-sorLA antibodies (IP: α-sorLA) or nonimmune serum (IP: non-im.). Lane 1 shows Western blots of endogenous sorLA and APP expression in cell extracts before immunoprecipitation. Also shown are Western blots for sorLA and APP in nonimmune (lane 2) or anti-sorLA (lane 3) immunoprecipitates. (B) Immunodetection of endogenous APP and sorLA in SH-SY5Y cells transfected with a sorLA expression construct (SY5Y-S) indicating colocalization in the perinuclear region (arrowhead). (Scale bar: 10 μm.) (C) Surface biotinylation demonstrating decreased surface localization of APP in cells overexpressing sorLA. Blots labeled “Input” depict sorLA, APP, and LRP expression in cell extracts from parental (SY5Y) and sorLA-transfected (SY5Y-S) neurons. The asterisk indicates accumulation of mature APP in the presence of sorLA. Western blots labeled “strept. beads” show biotinylated APP and LRP in precipitates from streptavidin beads. Overexpression of sorLA reduces cell surface exposure of APP (35.8% of normal) but not of LRP in SY5Y-S compared with SY5Y cells. (D) Levels of sAPPα, sAPPβ, and Aβ40 in medium from SY5Y and SY5Y-S cells as determined by Western blot or ELISA. Aβ40 in SY5Y-S is reduced to 28.6 ± 3.0% (with nontransfected SY5Y levels set at 100%).
To establish a causal role for sorLA in APP maturation, we compared processing of endogenous APP in normal SH-SY5Y cells (which produce little sorLA) and in cells that overexpress the receptor after transfection with a sorLA expression construct (SY5Y-S). Similar to the situation in CHO cells, APP colocalized with sorLA in a vesicular pattern in cytoplasm and perinuclear regions in SY5Y-S cells (Fig. 4B). Overexpression of sorLA resulted in accumulation of mature APP molecules in SY5Y-S compared with SY5Y cells (asterisk in “Input” in Fig. 4C). APP molecules accumulating in the presence of sorLA in SY5Y-S cells were largely confined to intracellular compartments because the amount of APP present on the cell surface was significantly reduced in these cells. This fact was demonstrated by surface biotinylation of plasma membrane proteins followed by precipitation using streptavidin beads (“strept. beads” in Fig. 4C). By densitometric scanning of Western blots (as the one shown in Fig. 4C), the amount of cell surface-localized APP was reduced to 35.8% (±3.2) in cells overexpressing sorLA (SY5Y-S) compared with parental cells (set at 100%). Surface localization of other neuronal transmembrane proteins, such as the LDL receptor-related protein (LRP), was not affected by sorLA overexpression (Fig. 4C). Remarkably, intracellular accumulation of mature APP in SY5Y-S cells coincided with a significant reduction in the amount of APP processing products compared with parental cells as shown by Western blot for sAPPα and sAPPβ and by ELISA for Aβ (Fig. 4D).
In summary, studies in SH-SY5Y cells established that increasing sorLA levels in neurons sequesters mature APP in intracellular compartments and reduces processing to Aβ. This finding raised the intriguing possibility that in the converse situation, loss of sorLA expression in patients with AD may be responsible for their increased Aβ production. This hypothesis was tested by using biopsies from AD patients (Fig. 5 A and B) and a sorLA-deficient mouse model (Fig. 5 C-H). Patients suffering from AD expressed significantly less sorLA protein in the frontal cortex compared with healthy controls as shown by Western blot analysis (Fig. 5 A and B). Reduction of sorLA levels did not reflect a general loss of neurons because levels of neuronal marker proteins such as LRP, synaptophysin (Fig. 5A), or sortilin (not shown) were unchanged. These findings were consistent in all of the 10 patients (compared with 7 control subjects) characterized in this study.
Fig. 5.
APP metabolism in AD patients and in sorLA-deficient mice. (A and B) SorLA levels are significantly reduced in the frontal cortex gray matter of 10 AD patients compared with 7 healthy subjects as shown by densitometric scanning (B) of Western blots, such as those shown in A. As a control, the levels of neuronal proteins LRP and synaptophysin were documented. n, number of patients. (C) Immunofluorescence detection of endogenous APP and sorLA in primary neurons from wild-type mice indicating partial colocalization in vesicular structures (arrowhead). (D-F) Immunodetection of sorLA (D), APP (E), and Aβ (F) in the frontal cortex of wild-type (+/+) and sorLA-deficient (-/-) mice. Representative data from experiments in three sorLA-/- mice and three control mice are shown. (Scale bar: 100 μm.) (G) Loss of sorLA expression in sorLA-/- mice (α-sorLA) did not affect the total levels of APP (α-APP) but increased processing into soluble APP (α-sAPP) as shown by Western blot analysis of brain homogenates. (H) Quantification of the increase in murine Aβ40 and Aβ42 in cortex extracts from 10-month-old sorLA-/- mice (138.6 ± 12.3% SEM and 135.6 ± 11.1% SEM, respectively) compared with wild-type controls (mean value set at 100%) using ELISA. P values were determined by equal-variance t test. n, number of mice.
To establish a mouse model of impaired sorLA expression), we disrupted the murine gene by using embryonic stem cell technology (as detailed in Fig. 10, which is published as supporting information on the PNAS web site). Mice homozygous for the disrupted sorLA allele lacked receptor expression as shown by immunohistology (Fig. 5D) and Western blot (Fig. 5G) of brain tissue. In wild-type mice, sorLA localized mainly to neurons of the frontal cortex (Fig. 5D), where it partially colocalized with APP in some intracellular vesicles (Fig. 5C, arrowhead). SorLA-containing vesicles stained positive for markers of early endosomes (EEA1) or Golgi (γ-adaptin) (not shown), similar to the situation in CHO (Fig. 2D) and SH-SY5Y cells (Fig. 4B). Subcellular fractionation of brain tissue is notoriously difficult because of the heterogeneity of neuronal cell types. Nevertheless, we were able to confirm colocalization of endogenous APP and sorLA in Golgi-enriched microsomal fractions of crude brain homogenates (Fig. 11, which is published as supporting information on the PNAS web site).
SorLA-deficient mice were viable and fertile with no obvious changes in overall APP levels in the cerebral cortex as observed by immunohistology (Fig. 5E) and Western blot (Fig. 5G). However, an obvious increase in neuron-associated Aβ immunoreactivity (Fig. 5F) and enhanced production of sAPP in brain homogenates (Fig. 5G) was evident at 10 months of age. The increase in Aβ was confirmed by ELISA measurements from cortical brain extracts for endogenous Αβ40 and Aβ42, demonstrating an ≈30% increase in murine Aβ levels in sorLA-deficient versus control mice (Fig. 5H). Consistent with other mouse models of elevated murine Aβ levels (16), increased levels of Aβ did not result in amyloid plaque deposition as tested by thioflavine-S staining (data not shown).
Discussion
Our findings uncovered a neuronal sorting receptor sorLA that interacts with APP and that affects trafficking and proteolytic processing of the precursor protein in the brain. Coexpression with wild-type sorLA confines APP to Golgi compartments and impairs transport to the cell surface and proteolytic processing. Studies in vitro, in living cells, in knockout mouse models, and in patients with AD all support the concept that increased sorLA activity coincides with impaired APP processing and reduced Aβ production, whereas loss of receptor function promotes APP processing and amyloidogenic peptide formation. Based on its homology to sorting receptors that shuttle between the Golgi and endosomes/lysosomes, we envision a model whereby sorLA acts as a sorting receptor for APP that determines transport of the precursor into pathways less favorable for processing (Fig. 6). In particular, sorLA seems to confine APP to the Golgi and to impair its transition to the cell surface, a step that is crucial for conversion into both sAPPα and sAPPβ/Aβ products. Confinement to Golgi compartments may be achieved either by impairing transition of nascent APP molecules en route through the biosynthetic pathway to the cell surface or by rerouting internalized precursors from early endosomes to the Golgi (Fig. 6). The latter process is in line with a proposed role for sorLA in endosome to Golgi trafficking and would prevent transport of APP to late endocytic compartments that harbor β-secretase activity (17).
The central role of the Golgi in APP metabolism is well appreciated; it represents the major site of APP concentration in the cell (18). More importantly, initial processing of APP by α- and β-secretases is intimately associated with a post-Golgi compartment and requires efficient transition of the precursor through this organelle (19, 20). Thus, disrupting Golgi transition of APP blocks processing (21, 22), whereas phorbol ester treatment that enhances membrane shunt from the trans-Golgi network to the plasma membrane increases APP processing (23). The mechanisms that regulate APP trafficking to and from the Golgi are poorly understood, but all of our experimental evidence suggests that sorLA activity represents an important determinant in this process. Conceivably, levels of sorLA activity in individuals may affect the overall kinetics of APP transition and processing in a subtle way but act cumulatively over decades to determine plaque burden and spontaneous AD progression.
Because targeting of APP to distinct subcellular compartments determines processing into amyloidogenic products, much attention has focused on factors that regulate APP trafficking. Previously, LRP (a member of the LDL receptor family) has been implicated in internalization of APP from the cell surface (24); F-spondin, a secreted factor that binds to the extracellular domain of APP, was shown to interfere with processing in vitro (25). Apparently, APP interacts with a number of neuronal proteins; the significance of such interactions for onset and progression of AD in patients remains to be established. The relevance of sorLA for APP processing and AD progression is supported by the observation that the receptor recognizes all APP variants as ligands and that patients with AD exhibit significantly reduced expression of the receptor in the brain (9). The reason for reduced sorLA expression in these individuals remains to be determined, but our findings strongly suggest that low levels of the receptor may be a primary cause of accelerated Aβ production and senile plaque formation. Thus, altered sorLA activity may be an important risk factor for AD, and pharmacological interventions that increase receptor activity may represent a therapeutic approach for treating this devastating disease.
Supplementary Material
Acknowledgments
We thank H. Schulz and S. Schuetz for technical assistance and G. Multhaup (Free University Berlin, Berlin), K. Beyreuther (Zentrum für Molekulare Biologie, Heidelberg), T. K. Bayer (University Clinic, Homburg/Saar, Germany), C. Pietrzik (University of Mainz, Mainz, Germany), and A. J. M. Roebroeck (Katholieke Universiteit Leuven, Leuven, Belgium) for sharing reagents. This work was supported by the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung und Forschung, the Lundbeck Foundation, the Danish Medical Research Council, and the National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: sorLA, sorting protein-related receptor; AD, Alzheimer's disease; APP, amyloid precursor protein; sAPP, soluble APP; Aβ, amyloid β-peptide; FLIM, fluorescence lifetime imaging microscopy; LRP, LDL receptor-related protein.
References
- 1.Hermans-Borgmeyer, I., Hampe, W., Schinke, B., Methner, A., Nykjaer, A., Susens, U., Fenger, U., Herbarth, B. & Schaller, H. C. (1998) Mech. Dev. 70, 65-76. [DOI] [PubMed] [Google Scholar]
- 2.Motoi, Y., Aizawa, T., Haga, S., Nakamura, S., Namba, Y. & Ikeda, K. (1999) Brain Res. 833, 209-215. [DOI] [PubMed] [Google Scholar]
- 3.Jacobsen, L., Madsen, P., Moestrup, S. K., Lund, A. H., Tommerup, N., Nykjaer, A., Sottrup-Jensen, L., Gliemann, J. & Petersen, C. M. (1996) J. Biol. Chem. 271, 31379-31383. [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki, H., Bujo, H., Kusunoki, J., Seimiya, K., Kanaki, T., Morisaki, N., Schneider, W. J. & Saito, Y. (1996) J. Biol. Chem. 271, 24761-24768. [DOI] [PubMed] [Google Scholar]
- 5.Marcusson, E. G., Horazdovsky, B. F., Cereghino, J. L., Gharakhanian, E. & Emr, S. D. (1994) Cell 77, 579-586. [DOI] [PubMed] [Google Scholar]
- 6.Nykjaer, A., Lee, R., Teng, K. K., Jansen, P., Madsen, P., Nielsen, M. S., Jacobsen, C., Kliemannel, M., Schwarz, E., Willnow, T. E., et al. (2004) Nature 427, 843-848. [DOI] [PubMed] [Google Scholar]
- 7.Hampe, W., Urny, J., Franke, I., Hoffmeister-Ullerich, S. A., Herrmann, D., Petersen, C. M., Lohmann, J. & Schaller, H. C. (1999) Development (Cambridge, U.K.) 126, 4077-4086. [DOI] [PubMed] [Google Scholar]
- 8.Jacobsen, L., Madsen, P., Nielsen, M. S., Geraerts, W. P., Gliemann, J., Smit, A. B. & Petersen, C. M. (2002) FEBS Lett. 511, 155-158. [DOI] [PubMed] [Google Scholar]
- 9.Scherzer, C. R., Offe, K., Gearing, M., Rees, H. D., Fang, G., Heilman, C. J., Schaller, C., Bujo, H., Levey, A. I. & Lah, J. J. (2004) Arch. Neurol. 61, 1200-1205. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper, B. & Annaert, W. (2000) J. Cell Sci. 113, Part 11, 1857-1870. [DOI] [PubMed] [Google Scholar]
- 11.Andersen, O. M., Christensen, L. L., Christensen, P. A., Sørensen, E. S., Jacobsen, C., Moestrup, S. K., Etzerodt, M. & Thøgersen, H. C. (2000) J. Biol. Chem. 275, 21017-21024. [DOI] [PubMed] [Google Scholar]
- 12.Behlke, J., Ristau, O. & Schonfeld, H. J. (1997) Biochemistry 36, 5149-5156. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y., Tesco, G., Jeong, W. J., Lindsley, L., Eckman, E. A., Eckman, C. B., Tanzi, R. E. & Guenette, S. Y. (2003) J. Biol. Chem. 278, 51100-51107. [DOI] [PubMed] [Google Scholar]
- 14.von Arnim, C. A., Tangredi, M. M., Peltan, I. D., Lee, B. M., Irizarry, M. C., Kinoshita, A. & Hyman, B. T. (2004) J. Cell Sci. 117, 5437-5445. [DOI] [PubMed] [Google Scholar]
- 15.Johnson-Wood, K., Lee, M., Motter, R., Hu, K., Gordon, G., Barbour, R., Khan, K., Gordon, M., Tan, H., Games, D., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 1550-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwata, N., Tsubuki, S., Takaki, Y., Shirotani, K., Lu, B., Gerard, N. P., Gerard, C., Hama, E., Lee, H. J. & Saido, T. C. (2001) Science 292, 1550-1552. [DOI] [PubMed] [Google Scholar]
- 17.Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., Teplow, D. B., Ross, S., Amarante, P., Loeloff, R., et al. (1999) Science 286, 735-741. [DOI] [PubMed] [Google Scholar]
- 18.Caporaso, G. L., Takei, K., Gandy, S. E., Matteoli, M., Mundigl, O., Greengard, P. & De Camilli, P. (1994) J. Neurosci. 14, 3122-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haass, C., Hung, A. Y., Schlossmacher, M. G., Teplow, D. B. & Selkoe, D. J. (1993) J. Biol. Chem. 268, 3021-3024. [PubMed] [Google Scholar]
- 20.Yamazaki, T., Selkoe, D. J. & Koo, E. H. (1995) J. Cell Biol. 129, 431-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peraus, G. C., Masters, C. L. & Beyreuther, K. (1997) J. Neurosci. 17, 7714-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khvotchev, M. & Sudhof, T. C. (2004) J. Biol. Chem. 279, 47101-47108. [DOI] [PubMed] [Google Scholar]
- 23.Xu, H., Greengard, P. & Gandy, S. (1995) J. Biol. Chem. 270, 23243-23245. [DOI] [PubMed] [Google Scholar]
- 24.Pietrzik, C. U., Busse, T., Merriam, D. E., Weggen, S. & Koo, E. H. (2002) EMBO J. 21, 5691-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho, A. & Sudhof, T. C. (2004) Proc. Natl. Acad. Sci. USA 101, 2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.