Abstract
Rho family GTPase Cdc42 is known to regulate polarity and growth in lower eukaryotes, but its physiologic function in mammals has yet to be determined. Here we have disrupted cdc42gap, a ubiquitously expressed negative regulator of Cdc42, in mice. Cdc42GAP-/- embryonic fibroblasts and various organs displayed significantly elevated Cdc42 activity. The embryonic and neonatal homozygous mice were reduced in size by ≈25-40% and suffered severe growth retardation. Major organs from Cdc42GAP-/- mice were proportionally smaller because of decreased cell number. Basal apoptosis was increased in Cdc42GAP-/- cells and tissues, and this was attributed to altered c-Jun N-terminal kinase apoptotic signals. These results reveal a role of Cdc42GAP in mammalian perinatal growth and implicate the c-Jun N-terminal kinase-mediated apoptosis machinery as a Cdc42 effector pathway in vivo.
Keywords: GTPase-activating protein, Rho GTPases
First identified in Saccharomyces cerevisiae as a gene product involved in bud-site assembly (1), Cdc42 is a member of the Rho GTPase family that has subsequently been implicated in the regulation of multiple cell functions, including actin cytoskeleton reorganization, polarity, intracellular vesicle trafficking, transcriptional activation, and cell cycle progression (2-5). Cdc42 cycles between the GTP-bound, active conformation and the GDP-bound inactive conformation. The dynamic GTP-binding and GDP-hydrolysis cycle of Cdc42 is important for the signal transduction processes and is tightly regulated spatiotemporally in cells by the negative regulators, Rho GTPase-activating proteins (RhoGAPs), and the positive regulators, guanine nucleotide exchange factors (GEFs). RhoGAPs accelerate the intrinsic GTPase activity of Cdc42 to return it to the GDP-bound conformation, whereas Rho GEFs catalyze the GTP-loading to Cdc42 to promote Cdc42-GTP formation (6, 7). Various extracellular stimuli/stresses or intracellular genetic cues may modulate Cdc42 activity by regulating specific RhoGAP and/or GEF activities, among other possible means, to enable it to directly interact with a variety of effectors to elicit biological functions (6, 7).
Genetic studies in lower eukaryotes have indicated an essential role of Cdc42 in organism survival, growth, and development (8). However, conventional gene targeting of cdc42 in mice led to early embryonic lethality (9), hindering the effort to further study its function in mammals. Current knowledge of Cdc42 function in mammalian cells came mostly from studies using overexpression of dominant negative or constitutively active mutants of Cdc42 in clonal cell lines. Although the dominant mutant approach has provided useful insights into the functions of Cdc42 in mammalian cells, it also has significant drawbacks (10, 11). For example, the dominant negative form of Cdc42 can be nonspecific because of its ability to sequester the upstream guanine nucleotide exchange factors that are needed for multiple Rho GTPase functions (12). On the other hand, overexpression of active mutants of Cdc42 also lacks specificity and may tie up individual effectors to disrupt their spatiotemporal actions. It is now appreciated that multiple Cdc42 effectors exist in the cell, and dynamic binding of Cdc42 to each effector is necessary for optimal Cdc42 signaling (13). Therefore, it is highly desirable to develop a genetic approach to clearly define the physiologic function of Cdc42 in mammalian cells and organisms.
In the present studies, we have taken a gene-targeting approach toward a putative Cdc42-specific negative regulator, Cdc42GAP (also termed p50RhoGAP) (14, 15), to generate a Cdc42 gain-of-activity mouse model. Among the large number of RhoGAP family members, Cdc42GAP appears to favor Cdc42 as a substrate over other Rho GTPases in vitro (16, 17), but its physiologic role has yet to be verified in vivo. We found that Cdc42GAP-/- mice have significantly reduced body and organ sizes, similar to that of the p190-B RhoGAP knockouts (18). However, they differ in mechanism, in that the Cdc42GAP knockout causes a specific elevation of Cdc42, not Rho, activity, to affect cell number, rather than cell size, of various organs. Further studies suggest that Cdc42GAP regulates c-Jun N-terminal kinase (JNK) activity to impact cell apoptosis. The results reveal a critical role of Cdc42GAP in mammalian growth in embryonic and neonatal periods and implicate the JNK-mediated apoptosis machinery as a Cdc42 effector pathway in vivo.
Materials and Methods
Generation of Cdc42GAP Knockout Mice and Genotyping. See Supporting Text, which is published as supporting information on the PNAS web site, for the details of the gene targeting strategy. The correctly targeted ES cell clones whose part of exons 3 (including ATG start codon) and 4 were replaced by the Neo gene cassette were microinjected into C57BL6 blastocysts to generate chimeras for germ-line transmission. Genotype analysis was performed by PCR by using primer pairs P3 (locating 1.8 kb upstream of ATG start codon with sequence 5′-CAGTCCTCAGCTTAGTCGGGACTC-3′) and P4 (locating in the 5′-promotor region of the neo gene cassette with the sequence of 5′-TGCGAGGCCAGAGGCCACTTGTGTAGC-3′) for the mutated allele and P1 (1.6 kb upstream of ATG start codon with sequence 5′-GTGATCCTTTGATACAGTTCCTC-3′) and P2 (42 bp downstream of ATG start codon with sequence 5′-CTGGTTCAGAGCTTGGCTGGTG-3′). The deletion of Cdc42GAP was confirmed by Western blotting by using a Cdc42GAP monoclonal antibody (BD Biosciences).
Cell/Organ Size and Cell Number Analysis. WT, heterozygous, or homozygous mice and organs were photographed and weighed at different developmental stages. Cell numbers in different organs were also quantified after tissue homogenization. For histological analysis, tissues were fixed in 10% neutralized formalin, dehydrated in 70% ethanol, cleared in xylene, and embedded in paraffin. Slides prepared with 5-μm sections were stained with hematoxylin/eosin. To analyze mouse embryonic fibroblast (MEF) cell size, cells were stained with propidium iodide/RNase and subject to FACS to compare the forward scatter-height histogram after G0/G1 phase gating. MEF and various organ cell sizes were also compared by measuring protein/cell number or protein/DNA ratios, as described (18).
TUNEL and BrdUrd Incorporation Assays. The pregnant mice were injected with 100 μg of BrdUrd per gram of body weight i.p. 1 or 12 h before being killed. The embryonic tissue sections [embryonic day (E)14.5, E15.5, and E18.5] of the respective embryos were prepared as described (19). Tissue sections were stained by TUNEL and counterstained with BrdUrd and Hoechst DNA dye. The slides were analyzed under a Leica fluorescence microscope equipped with deconvolution softwares (Improvision, Lexington, MA).
Western Blotting and Immunofluorescence. MEFs or organs were lysed in RIPA buffer. One hundred micrograms of protein extracts was resolved by electrophoresis and electrotransferred to poly(vinylidene difluoride) membrane. The contents of p21-activated kinase 1 (PAK1), p38, ERK, JNK, c-Jun, or their phosphorylated forms (Cell Signaling Technology, Beverly, MA), c-myc, and BH3 interacting domain death agonist (BID) (Biosource International, Camarillo, CA) were determined by probing with respective antibodies. The cytosolic fraction of the cells was isolated as described (20) and analyzed by an anticytochrome C antibody (Cell Signaling Technology). For actin staining, cells grown on coverglasses were fixed with 3.7% formaldehyde and permeabilized with 0.1% Triton X-100. Cells were blocked with 2% BSA and stained with rhodamine-phalloidin and DAPI.
Effector Domain Pull-Down and JNK Kinase Assays. Relative levels of GTP-bound RhoA, Rac1, or Cdc42 were determined by an effector pull-down assay, as described (11). Briefly, the asynchronously growing WT, heterozygous, or knockout cells or tissues were lysed, and the lysates were probed with the glutathione-agarose-immobilized GST-Rhotekin (for RhoA) or GST-PAK1 (for Cdc42 or Rac1) effector domain. Bound proteins were analyzed by immunoblotting with anti-RhoA, Rac1, or Cdc42 monoclonal antibodies (BD Biosciences). The endogenous JNK activities in tissues/MEFs were assayed after immunoprecipitation of JNK from the tissue/cell lysates by using purified GST-c-Jun as a substrate (19).
Primary MEF Preparation and Cell Proliferation Assays. MEFs were prepared from E13.5 embryos and cultured as described (21). Cell proliferation was determined in triplicate every day by counting the number of cells or by measuring the absorbency at 490 nm by the using CellTiter96AQueous cell proliferation assay kit from Promega.
Cell Cycle and Apoptosis Assays. MEFs were starved for 48 h and restimulated with 10% FBS, and at different time points, cells were subjected to propidium iodide/RNase staining followed by FACS analysis to determine the cell cycle. Cell apoptosis was determined by staining the cells with AnnexinV-cy5-7AAD (BD Biosciences) followed by FACS analysis.
Retroviral Transduction and FACS Analysis. N17Cdc42, Cdc42GAP, and the R305A mutant of Cdc42GAP (Cdc42GAP*) were cloned into retrovirus vector MIEG3; the retrovirus constructs of pRS containing JNK short interfering RNA (siRNA) oligo (pRS-JNK) and the controls were kind gifts from Z. Ronai (Mount Sinai School of Medicine, New York) (22). Production of the recombinant retrovirus in the packaging Phoenix cells and subsequent infection were carried out according to described protocols (21). Cells were harvested 72 h postinfection, and GFP-positive cells (typically 10-30%) were isolated by FACS for further analysis. In the JNK siRNA experiments, cells were first selected with Puromycin (Sigma) (2 μg/ml) for 2 days after infection before immunoblotting and apoptosis assays.
Results and Discussion
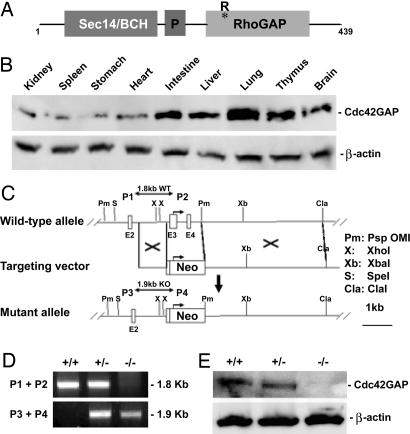
Gene Targeting of Cdc42GAP in Mice. To investigate the physiologic role of Cdc42 in mice, we used gene targeting to disrupt cdc42gap in the mouse embryonic stem cells. Cdc42GAP contains an N-terminal Sec14/BCH domain that is capable of interacting with Cdc42, a central proline-rich SH3-binding motif, and a C-terminal RhoGAP domain that was shown to be catalytically specific toward Cdc42 over other Rho GTPases (16, 17) (Fig. 1A). Among the RhoGAP family members, Cdc42GAP has no closely related homolog in the mouse or human genome. Western blot analysis indicates that Cdc42GAP is ubiquitously expressed in various mouse tissues (Fig. 1B), suggesting a widely used function for Cdc42 regulation. Germ-line transmission of the targeted alleles was achieved after homologous recombination of the targeting construct in the ES cells (Fig. 1C). The resultant homozygous mice were identified by PCR-based genotyping and anti-Cdc42GAP Western blotting of MEF cells derived from the respective embryos (Fig. 1 D and E). Crossbreeding of the Cdc42GAP heterozygous mice gave the birth of pups at the expected Mendelian ratio (Table 1), and the tissue organs of the Cdc42GAP-/- neonatal mice appeared to be normal upon a gross histological examination (Fig. 5, which is published as supporting information on the PNAS web site), suggesting that Cdc42GAP is not essential for embryonic development.
Fig. 1.
cdc42gap gene product structure, tissue distribution, and targeting in mice. (A) Schematic depiction of functional domains of Cdc42GAP. Both the Sec14/BCH and RhoGAP domains could specifically interact with Cdc42, and R305 critical for the GAP activity is indicated. (B) Cdc42GAP is ubiquitously expressed in different mouse organs. Tissues extracts were examined by anti-Cdc42GAP Western blotting. (C) Targeted disruption of the cdc42gap gene. Schematic representations of the 5′ genomic region of the cdc42gap gene (top line), the targeting vector (middle line), and the mutated allele (bottom line). Pm, PspOMI; Xb, XbaI; Cla, ClaI; S, SpeI; X, XhoI; neo, neomycin resistance gene; and E, exon. Arrow indicates ATG starting site. P1-P4 represents the primers used for genotyping. (D) PCR genotyping of WT (+/+), heterozygous (+/-), and homozygous (-/-) mice. (E) An immunoblot of MEFs derived from WT, heterozygous, or homozygous mice was probed with monoclonal antibodies that specifically recognize Cdc42GAP or β-actin.
Table 1.
Offspring obtained from Cdc42GAP+/− × Cdc42GAP+/− crosses
| Stages | Total no. of offspring | −/− | +/− | +/+ |
|---|---|---|---|---|
| E14.5 | 274 | 81 (24.2%)* | 173 (51.8%)* | 80 (24.0%)* |
| E16.5 | 30 | 8 (26.7%)* | 15 (50%)* | 7 (23.3%)* |
| P1 | 970 | 225 (23.2%)* | 495 (51.0%)* | 250 (25.8%)* |
| Live at weaning | 16 (7.1%)† | 490 (99.0%)† | 248 (99.2%)† |
The genotypes of Cdc42GAP+/+, Cdc42GAP+/−, and Cdc42GAP−/− mice are shown as +/+, +/−, and −/−, respectively. The percentage of animals conforming to the Mendelian ratio or the survival rate at weaning is shown in parentheses. P1, postnatal day 1.
Mendelian ratio.
Survival rate at weaning.
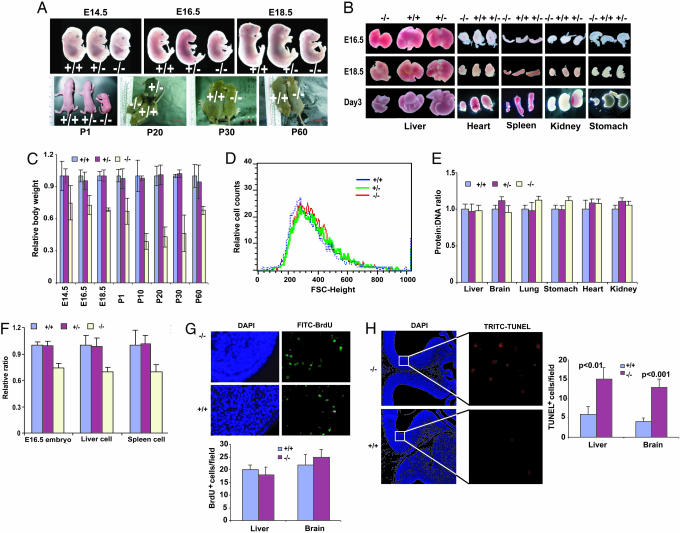
Cdc42GAP Regulates Mouse Embryonic and Neonatal Growth and Organ/Organism Size by Modulating Cell Number. The homozygous Cdc42GAP-/- embryos and neonatal pups were ≈25-40% reduced in body and organ sizes compared with WT (+/+) or heterozygous (+/-) mice (Fig. 2 A and B). Approximately 93% of homozygous pups died before weaning (Table 1), whereas the remaining pups continued to display severe growth retardation (Fig. 2C). The surviving mice could attain adulthood and were fertile despite being ≈30% smaller in body/organ size.
Fig. 2.
Cdc42GAP-deficient mice exhibit reduced body and organ sizes that correlate with reduced cell number and increased apoptosis. (A) WT, heterozygous, and homozygous embryos and postnatal mice at different development stages were examined. P1-P6, postnatal day 1-6. (B) Gross appearances of liver, heart, spleen, kidney, and stomach are shown for Cdc42GAP+/+, Cdc42GAP+/-, and Cdc42GAP-/- animals at various embryonic and neonatal stages. (C) Comparison of the growth rate of Cdc42GAP-/- mice with that of WT and heterozygous. Cdc42GAP+/+, Cdc42GAP+/-, and Cdc42GAP-/- mice were weighed from day E14.5 to day P60. (D) The MEFs from Cdc42GAP+/+, Cdc42GAP+/-, and Cdc42GAP-/- mice were analyzed by forward light scattering FACS and the G0/G1-gated mean cell size distributions are shown. (E) The protein/DNA ratios of liver, brain, lung, stomach, heart, and kidney of Cdc42GAP+/+, Cdc42GAP+/-, and Cdc42GAP-/- E17.5 embryos were compared with that of Cdc42GAP+/+.(F) The embryo weights of Cdc42GAP-/- and Cdc42GAP+/- embryos at E16.5 were analyzed as the relative ratio to that of Cdc42GAP+/+ embryos, and the relative liver and spleen cell numbers of the embryos were compared with those of the WT. (G) BrdUrd-labeled E14.5-old WT or Cdc42-/- embryos were fixed and analyzed by anti-BrdUrd staining. A brain section was shown after anti-BrdUrd and DAPI staining (Upper), and various sections of the liver and brain were quantified (Lower). (H) Similar sections of the brain and liver were analyzed by TUNEL assay to reveal apoptotic cells. The section of TUNEL staining was amplified for visualization of positive cells (Left). The quantification represents data obtained from at least three different sections (Right).
Previous gene targeting of p190-B RhoGAP resulted in reduced cell size, leading to smaller organ/organism size by up-regulating Rho activity (18). We examined whether genetic deletion of cdc42gap might have caused a similar effect. FACS analysis of the primary MEFs isolated from heterozygous crosses indicated that the Cdc42GAP-/- cells showed similar cell size as the matching Cdc42GAP+/- or Cdc42GAP+/+ cells (Fig. 2D), and these results were further confirmed by measuring the protein/DNA or protein/cell number ratio of these cells (data not shown). Similarly, the protein/DNA ratios of various organs were indistinguishable among the homozygous, heterozygous, and WT mice (Fig. 2E). These results suggest that the Cdc42GAP knockout mice adopt a cell-size-independent mechanism in achieving the reduced organ/organism size. It appears that the reduction of the body size/weight of the Cdc42GAP knockouts was proportional to the reduction of organ size/weight (data not shown) and to the decrease in cell numbers measured in liver, spleen, and thymus of day E16.5, E18.5, or P3-old embryo/mouse (Fig. 2F and data not shown). We conclude that the reduced organ/organism size in Cdc42GAP-/- mice was due to decreased cell number, not cell size.
Cdc42GAP Knockout Organs Display Increased Apoptotic Activity in Vivo. To examine whether the reduction of cell number of the Cdc42GAP-/- mice is related to an alteration of cell cycle progression or spontaneous apoptosis, we next measured cell cycle and apoptosis activities in various tissues of Cdc42GAP-/- embryos in comparison with those of WT embryos. BrdUrd incorporation of the E14.5 Cdc42GAP-/- fetal liver and brain was quantified, and no detectable differences were observed (Fig. 2G). However, apoptosis of the Cdc42GAP-/- tissues as indicated by TUNEL staining was significantly higher than that of WT (Fig. 2H). Similar observations were made for various embryonic organs at E15.5 and E18.5 (data not shown). These results suggest that the observed cell number reduction of various Cdc42GAP-/- tissues is associated with increased basal apoptosis.
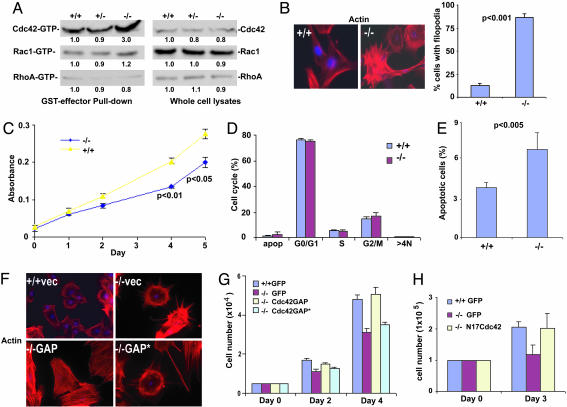
Constitutively Elevated Cdc42 Activity and Slowed Growth Phenotype of Cdc42GAP-/- MEFs. To examine whether the growth and apoptosis defects of the Cdc42GAP-/- mutant are specifically attributed to dysregulation of Cdc42, we have measured the relative Cdc42, Rac1, and RhoA activities in primary MEFs. Cdc42GAP-/- MEFs showed a significantly elevated level of Cdc42-GTP, but not Rac1-GTP or RhoA-GTP, compared with that of the matching heterozygous or WT cells, despite similar Cdc42 protein expression levels (Fig. 3A). In contrast to the WT cells, the elevated Cdc42-GTP level in the homozygous cells did not increase significantly in response to serum or UV challenge (Fig. 6, which is published as supporting information on the PNAS web site), suggesting that Cdc42 activity is constitutively high in these cells and may mimic mitogenic or stress stimulation. Consistent with a specific increase of Cdc42 activity, the Cdc42GAP-/- cells displayed robust spontaneous filopodia and actin microspikes not present in the WT cells (Fig. 3B). Cdc42GAP-/- cells showed similar kinetics as WT in spreading but extended more membrane protrusions in the process (Fig. 7A, which is published as supporting information on the PNAS web site). Furthermore, the homozygous cells appeared slower in adhesion to the fibronectin matrix but reached a similar saturation state 2 h after plating (Fig. 7B). The wound-healing migration rate of the homozygous cells was faster than that of the WT (Fig. 8A, which is published as supporting information on the PNAS web site). Furthermore, the Cdc42GAP-/- cells grew slower than the WT or heterozygous cells (Fig. 3C), and this was associated with an increase of spontaneous apoptosis but not with changes in cell cycle progression (Fig. 3 D and E). Consistent with the cell cycle data obtained by propidium iodide staining, examination of the early passages of the Cdc42GAP-/- cells by nuclear DAPI staining showed a similar number of bi/multinucleated cells as the WT (Fig. 8B). To confirm that the observed changes in actin structure and cell proliferation of Cdc42GAP-/- cells were due to the loss of catalytic GAP activity of Cdc42GAP and to subsequently elevated Cdc42 activity, either WT Cdc42GAP or a GAP domain mutant of Cdc42GAP (R305A) with impaired GTPase-activating activity toward Cdc42 (23) was reconstituted into the Cdc42GAP-/- MEFs for further examination. As shown in Fig. 3 F and G, whereas reconstitution of WT Cdc42GAP could readily reverse actin microspike formation and the slower proliferation phenotypes, the R305A mutant was ineffective. Further, the dominant negative Cdc42 mutant (N17Cdc42) was able to suppress both the actin and the proliferation phenotypes when expressed in an appropriate dose (Fig. 3H and data not shown). These results indicate that the phenotypic changes of actin organization and growth in Cdc42GAP-/- cells are attributed to the deregulation of Cdc42 activity.
Fig. 3.
Cdc42GAP-/- MEFs display constitutively elevated Cdc42 activity, slower growth rate, and increased apoptosis. (A) MEFs from Cdc42GAP+/+, Cdc42GAP+/-, and Cdc42GAP-/- mice were lysed and analyzed by effector domain (GST-PAK1 for Cdc42 and Rac1 activities and GST-Rhotekin for RhoA activity) pulldown assays. The lysate inputs are shown on the right-hand side and the corresponding quantifications are indicated below. Results are representative of three independent measurements. (B) MEFs from Cdc42GAP+/+, Cdc42GAP+/-, and Cdc42GAP-/- mice were seeded on coverslides and stained with Rhodamine-phalloidin and DAPI. The number of cells displaying obvious filopodia extensions was quantified. (C) MEF cells were plated in 96-well plates in triplicates in 10% FBS at 37°C, and at the varying time points, the cell numbers were assayed by using the CellTiter-96AQueous cell proliferation assay kit. (D) MEFs were labeled with propidium iodide/RNase, and the population of the cells at various cell cycle phases was analyzed by FACS. (E) Apoptotic cells were analyzed after Annexin V-7AAD staining. (F) WT or Cdc42GAP-/- cells were transduced with retrovirus expressing GFP, Cdc42GAP, or the R305A Cdc42GAP mutant (Cdc42GAP*). The cells were stained for actin structures. (G) Reconstituted MEFs were subjected to cell proliferation analysis at different time points. (H) WT, Cdc42GAP-/-, or Cdc42GAP-/- cells expressing N17Cdc42 were analyzed for the proliferation properties.
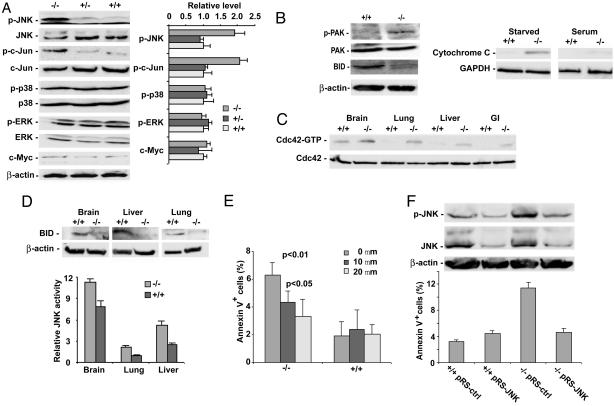
Cdc42GAP Regulates the Basal Apoptosis of Cells Through the JNK Pathway. Cdc42 is known to regulate multiple mitogen-activated protein kinase (MAPK) signaling pathways that might impact on cell growth (24-27), but whether it utilizes particular subtypes of MAPK for growth regulation is unclear (3). We next examined basal ERK1/2, JNK, and p38 MAPK activities in the Cdc42GAP-/- MEFs by using respective phospho-specific antibodies. As shown in Fig. 4A, the extent of JNK phosphorylation and one of its primary substrates, phospho-c-Jun, was significantly increased in Cdc42GAP-/- cells compared with WT or heterozygous cells. An increased JNK activity in Cdc42GAP-/- cells was also seen by an in vitro kinase assay by using c-Jun as a substrate (data not shown). In contrast, neither ERK1/2 nor p38 showed detectable changes in activity among these cells (Fig. 4A). The expression level of c-myc that has been implicated in cell number regulation in mice (28) was not changed either (Fig. 4A). In addition to JNK, the phosphorylation level of the Cdc42 effector, PAK1, was significantly increased in the Cdc42GAP-/- cells (Fig. 4B). Increased JNK activity may lead to spontaneous apoptosis (29-31). Consistent with this possibility, full-length, uncleaved BID was markedly reduced, and cytosolic cytochrome C was significantly elevated in Cdc42GAP-/- cells compared with that of WT cells (20, 30, 31) (Fig. 4B). Further corroborating these results in MEFs, Cdc42 and JNK activities were increased in various tissues of the Cdc42GAP-/- perinatal mice compared with those of WT (Fig. 4 C and D), and the full-length BID levels were correspondingly lower (Fig. 4D). It is therefore possible that Cdc42GAP serves as a regulator of the Cdc42-JNK signaling pathway to affect basal cell survival and apoptosis.
Fig. 4.
Cdc42GAP-/- MEFs or organs exhibit elevated JNK activity that causes increased apoptosis. (A) The whole-cell lysates of WT, heterozygous, and homozygous MEFs were subject to Western blotting by various MAP kinase or phospho-mitogen-activated protein kinase-specific, c-myc, or β-actin antibodies. Relative amounts of phospho-proteins were quantified by densitometry analysis. (B) WT and homozygous cell lysates were probed with anti-phospho-PAK1, BID, or β-actin antibody. The cytosolic fraction of the cells in the presence or absence of serum was analyzed for cytochrome C and GAPDH contents by respective immunoblotting. (C) Various tissues of E16.5 WT or homozygous mice were measured for endogenous Cdc42 activity by the effector domain pulldown assay. (D) Various E16.5 WT or Cdc42GAP-/- tissue lysates were subjected to JNK activity assay by using GST-c-Jun as a substrate and to anti-JNK blotting after normalization of the total protein contents (Lower). Lysates of brain, liver, and lung were also probed with anti-BID or β-actin antibody (Upper). (E) WT or homozygous cells were analyzed for apoptosis by Annexin V-7AAD staining after overnight treatment with a medium containing DMSO or 10 or 20 μM SP600125. (F) WT or homozygous MEFs were infected with retrovirus expressing a control siRNA sequence (pRS-ctrl) or a JNK-specific siRNA (pRS-JNK), and whole-cell lysates were analyzed by anti-phospho-JNK, JNK, or β-actin immunoblotting. The siRNA-treated cells were also examined for apoptotic populations by Annexin V-7AAD staining and FACS analysis.
Because deletion of somatic JNK (JNK1/2) in mice causes early embryonic lethality (30, 32), we used pharmacologic and genetic tools to inhibit JNK signaling in MEFs to address whether the increased JNK activity of the Cdc42GAP knockout cells is critical to the increased apoptosis. Pharmacologic inhibition of JNK activity in the Cdc42GAP-/- cells resulted in a suppression of the apoptotic cell population in a dose-dependent manner (Fig. 4E). Treatment of the Cdc42GAP-/- cells with retrovirus expressing an siRNA construct specific for JNK (JNK1 and JNK2) (22) caused suppression of JNK1/2 expression and decreased phospho-JNK activity (Fig. 4F). The inhibition of JNK expression by siRNA prevented the apoptosis phenotype of the Cdc42GAP-/- cells (Fig. 4F). Interestingly, down-regulation of JNK activity by treatment with the JNK inhibitor or siRNA had no detectable effect on the filopodia formation of Cdc42GAP-/- cells (Fig. 9, which is published as supporting information on the PNAS web site), consistent with previous reports that Rac/Cdc42-regulated actin reorganization might be independent of PAK and JNK activation (25). Together, these results indicate that Cdc42GAP deletion may result in increased activity of the Cdc42-JNK pathway leading to increased cell apoptosis.
Conclusion
We have generated Cdc42GAP null mice that show a gain of Cdc42 activity in a variety of tissues and cells. Characterizations of Cdc42GAP-/- MEFs demonstrate that elevated Cdc42 activity, not related Rho GTPases such as Rac1 or RhoA, is specifically responsible for the observed growth defect and spontaneous filopodia formation. Elevated endogenous Cdc42 activity causes an increase of PAK1 and JNK activities among its effector pathways and induces JNK-mediated spontaneous apoptosis in various embryonic and neonatal tissues and cells. This effect, to some extent, mimics the stress (e.g., UV)-induced responses in JNK activation and the accompanying activation of the JNK-regulated apoptotic machinery, including BID and cytochrome C (30, 31). The increased basal apoptotic activity, not cell cycle alteration, in various tissues and cells may in part be responsible for the reduced cell number of Cdc42GAP null animals. These observations are consistent with the possibility that cells in various organs of Cdc42GAP-/- mice can reach a different homeostasis from WT mice by balancing proliferation and apoptosis rates, leading to smaller organ/organism size.
Given the potentially large number of RhoGAPs that might be involved in regulating Cdc42 activity, it is intriguing that removal of the ubiquitously expressed Cdc42GAP can result in a global effect on Cdc42 activity and consequent growth retardation in mice. One possibility is that because many RhoGAPs are expressed in specific tissues and play roles in a cell type- and/or pathway-specific manner, the ubiquitously expressed Cdc42GAP may serve as a global regulator of Cdc42 in JNK-mediated processes. This is not unlike that of the reported function of p190-B RhoGAP that appears to globally regulate cell sizes of various tissues and organs by controlling Rho activity and the CREB transcription pathway (18). Another possibility is that the quantitative level or intracellular distribution pattern of Cdc42 activity in each cell type that is subject to regulation by multiple RhoGAPs (including Cdc42GAP) may dictate the outcome of cellular responses in morphogenesis. This awaits comparison of the Cdc42GAP-/- phenotypes with that of other Cdc42-specific RhoGAP knockout mice.
Much has been speculated about a role of Cdc42 in regulating mammalian cell growth and a number of effector pathways downstream of Cdc42 have been suggested to mediate growth regulation based on data derived from using dominant mutants in clonal cell lines. Our results demonstrate that Cdc42 indeed has a role in animal perinatal growth. However, elevated Cdc42 activity in primary cells displayed a slowed, rather than accelerated, proliferation rate by modulating the JNK-mediated apoptotic machinery, which was not expected from previous studies in NIH 3T3 fibroblasts or a number of other cell types that found a growth-stimulatory effect of various activating mutants of Cdc42 mediated by intracellular vesicle trafficking, cell cycle promotion, or growth factor receptor turnover (24, 25). Our studies further highlight the necessity of examining the function and mechanism of the signaling of Cdc42 in primary cell settings as well as using genetic approaches.
Supplementary Material
Acknowledgments
We thank Dr. Ze'ev Ronai (Mount Sinai School of Medicine, New York) for providing the JNK siRNA constructs. We also appreciate expert technical assistance by James F. Johnson and Clara Blair. This work was supported by National Institutes of Health Grants CA 105117 and GM 60523 (to Y.Z.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GAP, GTPase-activating protein; RhoGAPs, Rho GTPase-activating proteins; MEF, mouse embryonic fibroblast; En, embryonic day n; siRNA, short interfering RNA; Pn, primer n; BID, BH3 interacting domain death agonist; PAK, p21-activated kinase; JNK, c-Jun N-terminal kinase.
References
- 1.Adams, A. E., Johnson, D. I., Longnecker, R. M., Sloat, B. F. & Pringle, J. R. (1990) J. Cell Biol. 111, 131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Aelst, L. & D'Souza-Schorey, C. (1997) Genes Dev. 11, 2295-2322. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, D. I. (1999) Microbiol. Mol. Biol. Rev. 63, 54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erickson, J. W. & Cerione, R. A. (2001) Curr. Opin. Cell Biol. 13, 153-157. [DOI] [PubMed] [Google Scholar]
- 5.Etienne-Manneville, S. & Hall, A. (2002) Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- 6.Zheng, Y. (2001) Trends Biochem. Sci. 26, 724-732. [DOI] [PubMed] [Google Scholar]
- 7.Moon, S. Y. & Zheng, Y. (2003) Trends Cell Biol. 13, 13-22. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, D. I. & Pringle, J. R. (1990) J. Cell Biol. 111, 143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, F., Ma, L., Parrini, M. C., Mao, X., Lopez, M., Wu, C., Marks, P. W., Davidson, L., Kwiatkowski, D. J., Kirchhausen, T., et al. (2000) Curr. Biol. 10, 758-765. [DOI] [PubMed] [Google Scholar]
- 10.Feig, L. A. (1999) Nat. Cell Biol. 1, E25-E27. [DOI] [PubMed] [Google Scholar]
- 11.Wang, L., Yang, L., Luo, Y. & Zheng, Y. (2003) J. Biol. Chem. 278, 44617-44625. [DOI] [PubMed] [Google Scholar]
- 12.Debreceni, B., Gao, Y., Guo, F., Zhu, K., Jia, B. & Zheng, Y. (2004) J. Biol. Chem. 279, 3777-3786. [DOI] [PubMed] [Google Scholar]
- 13.Bishop, A. L. & Hall, A. (2000) Biochem. J. 348, 241-255. [PMC free article] [PubMed] [Google Scholar]
- 14.Barfod, E. T., Zheng, Y., Kuang, W. J., Hart, M. J., Evans, T., Cerione, R. A. & Ashkenazi, A. (1993) J. Biol. Chem. 268, 26059-26062. [PubMed] [Google Scholar]
- 15.Lancaster, C. A., Taylor-Harris, P. M., Self, A. J., Brill, S., van Erp, H. E. & Hall, A. (1994) J. Biol. Chem. 269, 1137-1142. [PubMed] [Google Scholar]
- 16.Zhang, B., Wang, Z. X. & Zheng, Y. (1997) J. Biol. Chem. 272, 21999-22007. [DOI] [PubMed] [Google Scholar]
- 17.Low, B. C., Seow, K. T. & Guy, G. R. (2000) J. Biol. Chem. 275, 37742-37751. [DOI] [PubMed] [Google Scholar]
- 18.Sordella, R., Classon, M., Hu, K. Q., Matheson, S. F., Brouns, M. R., Fine, B., Zhang, L., Takami, H., Yamada, Y. & Settleman, J. (2002) Dev. Cell 2, 553-565. [DOI] [PubMed] [Google Scholar]
- 19.Kuan, C. Y., Whitmarsh, A. J., Yang, D. D., Liao, G., Schloemer, A. J., Dong, C., Bao, J., Banasiak, K. J., Haddad, G. G., Flavell, R. A., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 15184-15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, X., Kim, C. N., Yang, J., Jemmerson, R. & Wang, X. (1996) Cell 86, 147-157. [DOI] [PubMed] [Google Scholar]
- 21.Guo, F. & Zheng, Y. (2004) Mol. Cell. Biol. 24, 1426-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhoumik, A., Jones, N. & Ronai, Z. (2004) Proc. Natl. Acad. Sci. USA 101, 4222-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassar, N., Hoffman, G. R., Manor, D., Clardy, J. C. & Cerione, R. A. (1998) Nat. Struct. Biol. 5, 1047-1052. [DOI] [PubMed] [Google Scholar]
- 24.Olson, M. F., Ashworth, A. & Hall, A. (1995) Science 269, 1270-1272. [DOI] [PubMed] [Google Scholar]
- 25.Lamarche, N., Tapon, N., Stowers, L., Burbelo, P. D., Aspenstrom, P., Bridges, T., Chant, J. & Hall, A. (1996) Cell 87, 519-529. [DOI] [PubMed] [Google Scholar]
- 26.Wu, W. J., Erickson, J. W., Lin, R. & Cerione, R. A. (2000) Nature 405, 800-804. [DOI] [PubMed] [Google Scholar]
- 27.Wu, W. J., Tu, S., Cerione, R. A., Erickson, J. W. & Lin, R. (2003) Cell 114, 715-725. [DOI] [PubMed] [Google Scholar]
- 28.Trumpp, A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G. R. & Bishop, J. M. (2001) Nature 414, 768-773. [DOI] [PubMed] [Google Scholar]
- 29.Na, S., Li, B., Grewal, I. S., Enslen, H., Davis, R. J., Hanke, J. H. & Flavell, R. A. (1999) Oncogene. 18, 7966-7974. [DOI] [PubMed] [Google Scholar]
- 30.Tournier, C., Hess, P., Yang, D. D., Xu, J., Turner, T. K., Nimnual, A., Bar-Sagi, D., Jones, S. N., Flavell, R. A. & Davis, R. J. (2000) Science 288, 870-874. [DOI] [PubMed] [Google Scholar]
- 31.Deng, Y., Ren, X., Yang, L., Lin, Y. & Wu, X. (2003) Cell 115, 61-70. [DOI] [PubMed] [Google Scholar]
- 32.Kuan, C. Y., Yang, D. D., Samanta Roy, D. R., Davis, R. J., Rakic, P. & Flavell, R. A. (1999) Neuron 22, 667-676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.