Abstract
Disease resistance strategies are powerful approaches to sustainable agriculture because they reduce chemical input into the environment. Recently, Piriformospora indica, a plant-root-colonizing basidiomycete fungus, has been discovered in the Indian Thar desert and was shown to provide strong growth-promoting activity during its symbiosis with a broad spectrum of plants [Verma, S. et al. (1998) Mycologia 90, 896-903]. Here, we report on the potential of P. indica to induce resistance to fungal diseases and tolerance to salt stress in the monocotyledonous plant barley. The beneficial effect on the defense status is detected in distal leaves, demonstrating a systemic induction of resistance by a root-endophytic fungus. The systemically altered “defense readiness” is associated with an elevated antioxidative capacity due to an activation of the glutathione-ascorbate cycle and results in an overall increase in grain yield. Because P. indica can be easily propagated in the absence of a host plant, we conclude that the fungus could be exploited to increase disease resistance and yield in crop plants.
Keywords: root endophyte, powdery mildew, symbiosis, ascorbate, glutathione
Despite a worldwide intensification of agriculture and tremendous progress toward increasing yields in major crops over the last decades, the goal to reduce the problems associated with hunger is far from being reached (1). Major causes for crop losses are abiotic and biotic stresses due to unfavorable climate and plant diseases and pests. Increased plant productivity, therefore, relies on a high chemical input and is achieved at the expense of detrimental effects on the environment (2, 3). Abiotic-stress tolerance can be evoked in crops by the exploitation of worldwide abundant endophytic arbuscular mycorrhiza fungi, which live in reciprocally beneficial relationships with ≈80% of land plants (4). However, mycorrhizal plants, albeit effective against many root diseases (5, 6), often show enhanced susceptibility to biotrophic leaf pathogens (7, 8). On the other hand, ascomycete endophytes have been frequently reported to protect against plant pathogens and pests. Grasses (Poaceae) and fungi of the family Clavicipitaceae have a long history of associations, ranging from mutualism to antagonism (9). These fungi are strictly confined to upper parts of the plant, grow only intercellularly, and exert a rather narrow host range. A critical review of the literature suggests that the beneficial action of these endophytes is based on direct antimicrobial and insecticidal activity due to alkaloid production.
We used the cereal model plant barley (Hordeum vulgare L.) to test whether growth-promoting activity of the recently discovered root-endophytic fungus Piriformospora indica (10) associates with agronomically desirable traits. Discovered in the Indian Thar desert in 1997 (11), P. indica has been recently related to the Sebacinales [ordo nov.] (form genus Rhizoctonia; Hymenomycetes, Basidiomycota) on the basis of an alignment of nuclear DNA sequences from the D1/D2 region of the large ribosomal subunit (12). In contrast to arbuscular mycorrhiza fungi, the fungus can be easily cultivated in axenic cultures, where it asexually forms chlamydospores containing 8-25 nuclei (10). The fungus associates with roots of various plant species, where it promotes plant growth. Hosts include the cereal crops rice, wheat, and barley as well as many Dicotyledoneae, including Arabidopsis (13, 14). Interaction of the endophytic fungus with Arabidopsis roots is accompanied by a considerable requisition of nitrogen from the environment (14). In the interaction with Arabidopsis and tobacco, the fungus stimulates nitrate reduction (15), in contrast to the activity of arbuscular mycorrhiza fungi. We report here on the enormous agronomical potential of the fungus. First, and most importantly, the growth-promoting activity of the fungus resulted in enhanced barley grain yield. Second, P. indica amended tolerance to mild salt stress, and third, P. indica conferred resistance in barley against root and leaf pathogens, including the necrotrophic fungus Fusarium culmorum (root rot) and the biotrophic fungus Blumeria graminis. Thus, interaction of barley with P. indica constitutes a model system for systemic disease resistance in cereals.
Materials and Methods
Plant and Fungal Material, Yield Experiments. Barley was grown in a 2:1 mixture of expanded clay (Seramis, Masterfoods, Verden, Germany) and Oil-Dri (Damolin, Mettmann, Germany) in a growth chamber at 22°C/18°C day/night cycle, 60% relative humidity, and a photoperiod of 16 h (240 μmol·m-2·s-1 photon flux density) and fertilized weekly with 20 ml of a 0.1% Wuxal top N solution (Schering, N/P/K: 12/4/6). Hydroponic cultures contained expanded clay (Seramis, Masterfoods) as substrate. For inoculation with P. indica, 2 g of mycelium were added to 300 g of substrate before sowing. P. indica was propagated in liquid Aspergillus minimal medium (14). For yield evaluations, barley was sown in soil containing P. indica mycelium (4 g in 300 g of substrate) and grown for 4 weeks in the growth chamber. Before transplantation to outdoor conditions, root samples were checked for P. indica infestation. In the beginning of April 2004, when plants reached growth stage (GS) 30 (16), they were transplanted into 6-liter Mitscherlich pots (Stoma, Siegburg, Germany) (six plantlets per pot) and filled with a mixture of a loam (loess) soil and sand (1:2). The preceding crop grown in the soil was potato. Soil nutrient additives were 0.25 g of N, 0.4 g of P, 1.6 g of K, and 0.2 g of Mg; N was applied a second time at a rate of 0.25 g per pot, 2 weeks after planting (GS 32). The fungicide Opus Top (250 g·liter-1 Fenpropimorph and 84 g·liter-1 Epoxiconazole; BASF, Ludwigshafen, Germany) was sprayed at a rate of 1.5 liter·hectare (ha)-1 to control powdery mildew and Rhynchosporium secalis. Both systemic fungicides are transported acropetally via xylem and do not reach roots. Aphids and the cereal beetle Oulema spp. were controlled during anthesis by using the insecticide Karate (100 g·liter-1 lambda-Cyhalothrin, Syngenta Agro, Basel) at a rate of 150 ml·ha-1. The presence of P. indica was monitored microscopically throughout the vegetation period.
Analysis of powdery mildew infections was done in a detached-leaf-segment assay on agar plates containing 0.4% benzimidazole to inhibit leaf senescence. Plants were inoculated with 15 conidia·mm-2 (for macroscopic evaluation) or with 25 conidia·mm-2 (for microscopy) of B. graminis f.sp. hordei, race A6. For gene expression studies, leaves were inoculated with 80 conidia·mm-2. Colonies were counted at 7 days after inoculation. Microscopic inspection of powdery-mildew-infected leaves was done by determining the frequency of the three different interaction types. Cells showing a hypersensitive response were detected by their whole-cell autofluorescence. Successful penetration was ascertained by the detection of haustoria formation or the development of elongated secondary hyphae (17). “Nonpenetrated cells” are those in which fungal penetration attempts were unsuccessful.
For root inoculation with pathogens, oat kernels colonized by F. culmorum strain KF 350 or Cochliobolus sativus were used. Kernels (1 g) were added to 300 g of substrate before sowing. The control pots were amended with 1 g of autoclaved inoculum. For inoculum production, kernels were autoclaved (125°C, 25 min), inoculated with conidia of F. culmorum or C. sativus, and incubated for 1 week at room temperature.
Biochemical Measurements. Ascorbate was determined by using the bipyridyl method (18). Dehydroascorbate reductase activity was assayed spectrophotometrically at 265 nm as reduced glutathione (GSH)-dependent dehydroascorbate oxidation (19). The assay mixture contained 50 mM sodium phosphate buffer (pH 6.5), 0.1 mM Na2EDTA, 20 μM dehydroascorbate, 50 μM GSH, and 20- to 100-μl extracts in a total volume of 2.3 ml. Glutathione concentrations ([GSH] and [oxidized glutathione]) were measured as described in ref. 20 with slight modifications. Briefly, 0.3 g of plant tissue was mixed with 3 ml of 2% sulfosalicylic acid containing 0.15 g of ascorbic acid and 1 mM Na-EDTA per 100 ml. The sample was centrifuged (10,000 × g at 4°C for 10 min), and the supernatant was either used for analysis directly or stored at -20°C. For glutathione determination, 30 μl of supernatant was assayed in a total volume of 1 ml of phosphate buffer containing 0.6 mM 5.5′-dithiobis(2-nitrobenzoate) (DTNB), 0.5 units/ml human glutathione reductase (GR), and 0.3 mM NADPH. The reduction of DTNB to nitrothiobenzoate was determined spectrophotometrically at 412 nm and 25°C and was related to a calibration curve. Recombinant human GR was prepared as described in ref. 21. To measure GR activity, 0.5 g of plant material was mixed with 2 ml of 0.1 M Tris, 1 mM EDTA, and 7.5% wt/vol polyvinylpyrrolidone (pH 7.8). After centrifugation, protein was determined in the supernatant according to Bradford (22) by using BSA as the standard. GR activity was measured spectrophotometrically (for NADPH, ε340 = 6.22 mM-1·cm-1) in 47 mM potassium phosphate, 200 mM KCl, and 1 mM EDTA (pH 6.9) at 25°C (21). A baseline was monitored in the presence of plant extract and 100 μM NADPH to account for NADPH oxidase activity. The reaction was then started with 1 mM glutathione disulfide.
Northern Blot and RT-PCR Analysis. Northern blots were prepared from 10 μg of total RNA extracted from leaves of 3-week-old barley plants as described in ref. 23. Blots were hybridized with 32P-labeled JIP-23 [a 1,100-bp fragment of nucleotide with Entrez database accession no. X98124 (24)]-, BCI-1 [a 1,000-bp fragment of accession no. U56406 (25)]-, and barley PR-5 [a 600-bp fragment of accession no. AJ276225 (26)]-specific probes and, after removal of the probes, hybridized with a 28S rRNA probe to confirm equal transfer of RNA onto the membrane. To exclude the presence of P. indica in leaves from root-infested plants, RT-PCR reactions were performed with leaf material by using primers specific for the P. indica tef gene (27).
Results and Discussion
P. indica Colonizes Root Cortical Cells and Enhances Yield in Barley. Microscopic inspection of barley plants grown in P. indica-inoculated substrate showed that the fungus enters roots primarily via root hairs (Fig. 1a) and, later, grows intracellularly in the root cortex (Fig. 1b). Hyphae were detected neither in the central part of the roots beyond endodermis (Fig. 1c) nor in stems or leaves. This observation was verified by performing RT-PCR reactions with leaf material using primers specific for the P. indica tef gene (data not shown). During the first 4 weeks of barley development, shoot fresh weight of infested plants was up to 1.65 times higher compared with control plants (see Fig. 2, first two columns). To assess whether this early increase in biomass would also lead to higher grain yields, two barley cultivars, including the elite cultivar Annabell, were tested in Mitscherlich pots in an open-air field station in spring/summer 2004. P. indica-infested Annabell showed an increase in grain yield of 11%, mainly because of a higher number of ears per plant (Table 1). In cultivar Ingrid, the grain yield increase was 5.5% (Table 1). A repetition of the complete set of yield experiments gave similar results, with elevated grain yields observed in the P. indica-infested plants. Notably, P. indica also increased grain yield in soils with a high nitrogen supply (data not shown).
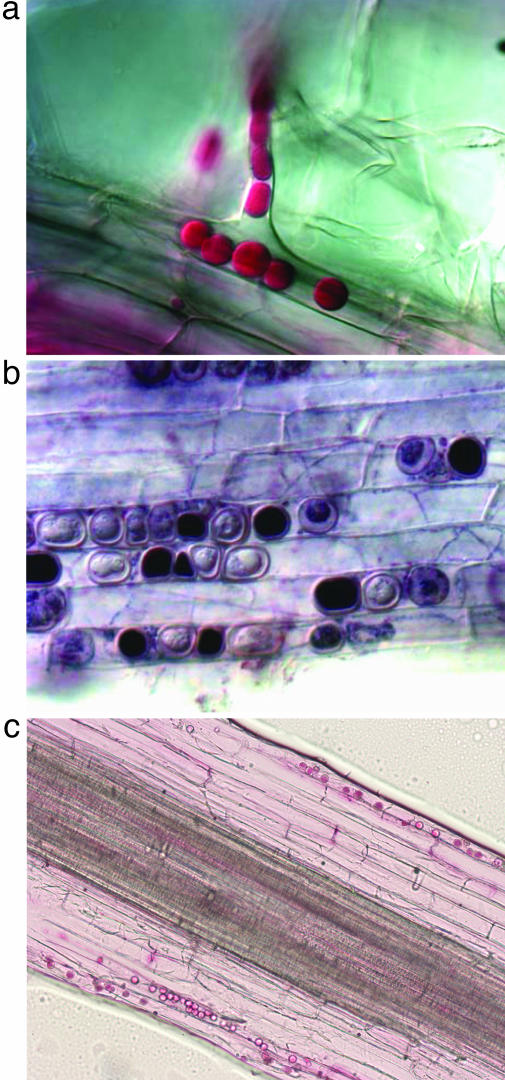
Fig. 1.
Colonization pattern of P. indica in barley roots. (a) Fungal hyphae enter roots via root hairs from 10-day-old plants. The fungus forms pear-shaped chlamydospores within root hairs and proceeds into rhizodermis cells. (b) The fungus grows into the root cortex tissue. (c) Longitudinal section. The fungus was not detected in the central part of the roots beyond the endodermis. Fungal structures were visualized by 0.01% acid fuchsin-lactic acid (28) (red in a and c), or they were stained for mitochondrial respiratory activity by the succinate dehydrogenase assay (29) (black in b).
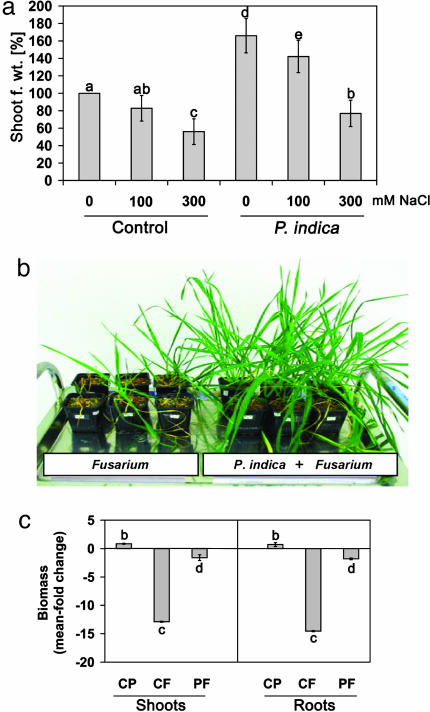
Fig. 2.
Impact of P. indica on salt-stress tolerance and root infections by F. culmorum. (a) Shoot fresh weight of P. indica and control (noninfested) plants was determined in 5-week-old plants that had been grown for the final 2 weeks in the presence of 100 or 300 mM NaCl, in hydroponic culture. Data points are representative of three independent experiments. Error bars, SD. (b) Plant phenotypes demonstrating the protective potential of P. indica toward F. culmorum.(c) Mean fold change of root and shoot weights relative to noninfested (no P. indica or F. culmorum) 4-week-old plants. CP, P. indica-infested; CF, Fusarium-infected; PF, P. indica-infested, Fusarium-infected. Error bars, SD. Columns labeled with the same letter represent not-significantly-different means, according to multiple unpaired Student t tests (P < 0.05), after ANOVA.
Table 1. Effect of P. indica infestation on yield parameters in barley.
| Cultivar | P. indica | Yield, g per pot− | Straw yield, g per pot− | Harvest index | TGW, g | Ears per pot | Grains per ear |
|---|---|---|---|---|---|---|---|
| Ingrid | − | 50.3 ± 1.56 | 52.4 ± 3.14 | 0.96 ± 0.04 | 48.0 ± 0.90 | 47.8 ± 2.22 | 22.4 ± 0.93 |
| + | 53.1 ± 2.76 | 53.2 ± 1.02 | 0.99 ± 0.06 | 48.1 ± 0.65 | 51.5 ± 1.29* | 22.5 ± 1.86 | |
| Annabell | − | 53.9 ± 3.61 | 46.4 ± 3.46 | 1.17 ± 0.11 | 50.6 ± 1.28 | 47.3 ± 3.50 | 23.1 ± 1.41 |
| + | 59.9 ± 1.73** | 48.0 ± 1.35 | 1.25 ± 0.04 | 50.6 ± 0.96 | 50.0 ± 2.45 | 23.7 ± 0.98 |
To assess whether P. indica infestation affects yields, elite barley cultivar Annabell (Saatzucht Ackermann, Irlbach, Germany) and cultivar Ingrid were grown in soil containing mycelium inoculum of P. indica in Mitscherlich pots at an open-air field station near Marburg, central Hesse, Germany. Values given are means of six pots (with six plants per pot). TGW, thousand-grain weight; * and ** indicate statistically significant differences between infested and noninfested plants (unpaired Student t test; * P < 0.05; **, P < 0.01).
P. indica-Inoculated Plants Are Tolerant to Salt Stress and More Resistant to Root Pathogens. We analyzed the fungus' potential to protect barley from salt stress. When noninfested barley seedlings were exposed for 2 weeks to moderate (100 mM NaCl) and high (300 mM NaCl) salt concentrations in hydroponic culture, they showed increasing leaf chlorosis and reduced growth (Fig. 2a). The detrimental effect of moderate salt stress was completely abolished by P. indica, as shown by the fact that infested plants produced higher biomass than did nonstressed control plants under these conditions. However, under high salt-stress conditions, both noninfested and infested plants exhibited a severe biomass reduction.
We addressed the question of whether P. indica-infested plants would also be more resistant to biotic stress. Barley was grown in soil containing macroconidia of the necrotrophic fungal pathogen F. culmorum, and root and shoot biomass was recorded. We found that P. indica-infested plants are more resistant to root diseases (Fig. 2b). Fusarium root infection caused a >12-fold decrease in root and shoot fresh weight of 4-week-old plants, compared with control plants, which were infested with neither P. indica nor F. culmorum (Fig. 2c). In the presence of P. indica, this devastating effect of F. culmorum infection was strongly diminished. Root and shoot fresh weight was reduced only 2-fold in P. indica-infested plants, compared with the 12-fold decrease in controls with F. culmorum alone. Similar results were obtained when we tested resistance to the root-pathogenic fungus C. sativus (data not shown), which shows a hemibiotrophic nourishment strategy. In axenic culture, P. indica did not exhibit antifungal activity to F. culmorum or C. sativus (data not shown), indicating that the protective potential of the endophytic fungus does not rely on antibiosis. These results show that P. indica exerts beneficial activity against two major cereal pathogens that cause enormous worldwide economic losses (30).
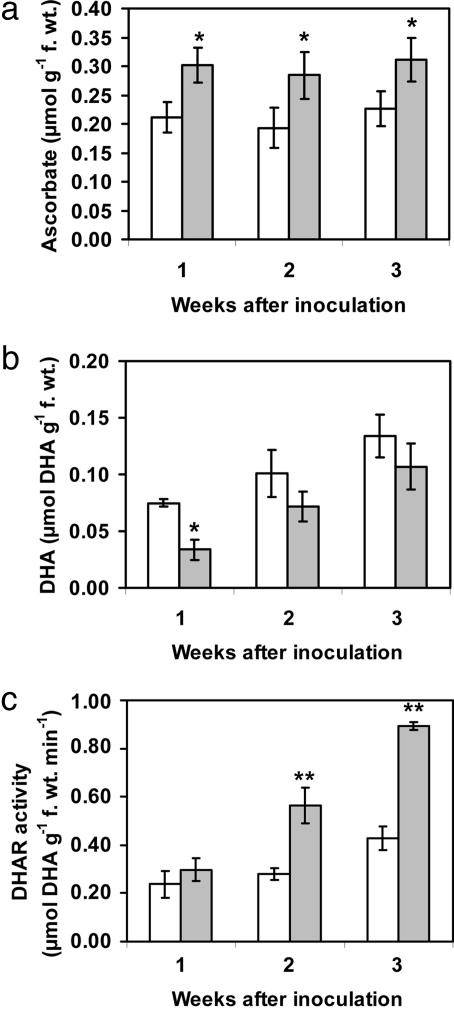
P. indica-Infested Roots Show Higher Antioxidant Capacity. The beneficial protective activity exerted by P. indica against root pathogens with necrotrophic nourishment strategies prompted us to ask whether the antioxidant status of the infested roots was altered by the endophyte. Because ascorbic acid is a major antioxidant buffer and free-radical scavenger (31), we recorded root concentrations of this compound during the first 3 weeks of P. indica infestation. Ascorbate levels were consistently higher 1, 2, and 3 weeks after root infestation (Fig. 3a), whereas levels of dehydroascorbate were reduced (Fig. 3b). At the same time, the activity of ascorbate recycling dehydroascorbate reductase increased during root infestation, reaching 2.2-fold higher levels 3 weeks after infestation with P. indica (Fig. 3c). Concomitantly, we found slightly (but not significantly) enhanced total glutathione concentrations (Table 2). Corresponding GR activity was only slightly enhanced early after infestation (1 week; Table 3). It can be reasoned that higher antioxidant levels protect roots from cell death provoked by root pathogens F. culmorum and C. sativus. Because production of reactive oxygen species and host-cell killing is a prerequisite for successful fungal development and pathogenesis of necrotrophic nourishment (32), we speculate that elevated ascorbate levels could be the reason for the observed control of necrotrophic pathogens in barley roots.
Fig. 3.
Ascorbate, dehydroascorbate (DHA) content, and DHA reductase (DHAR) activity in P. indica-infested roots. Ascorbate content (a), DHA content (b), and DHAR activity (c) were measured in roots of 1-, 2-, and 3-week-old P. indica-infested (shaded columns) and control (free of P. indica, open columns) barley plants. Values are means of three or four samples. Similar results have been obtained with three independent sets of experiments. Error bars, SD. Within each frame, * and ** indicate statistically significant differences between roots of infested and noninfested plants (unpaired Student t test; *, P < 0.05; **, P < 0.01).
Table 2. Glutathione content in P. indica-infested barley roots and leaves.
| Time after sowing, weeks
|
[Glutathione], nmol·g−1 f.wt. in roots
|
[Glutathione], nmol·g−1 f.wt. in leaves
|
||||
|---|---|---|---|---|---|---|
| Control | P. indica | % of control | Control | P. indica | % of control | |
| 1 | 200 ± 42 | 205 ± 16 | 103 | 405 ± 67 | 403 ± 54 | 99.5 |
| 2 | 230 ± 32 | 273 ± 35 | 119 | 370 ± 78 | 467 ± 48 | 126.3 |
| 3 | 223 ± 46 | 309 ± 49 | 139 | 510 ± 54 | 798 ± 113 | 156.5* |
Total glutathione (GSH and oxidized glutathione) was measured according to procedures described in ref. 20. Values are means of three samples. Similar results were obtained with three independent sets of experiments. *, P < 0.05; for intercohortal analyses the unpaired Student t test was applied. f.wt., fresh weight.
Table 3. GR activity in P. indica-infested barley roots and leaves.
| Time after sowing, weeks
|
GR activity in roots, milliunits/mg protein
|
GR activity in leaves, milliunits/mg protein
|
||||
|---|---|---|---|---|---|---|
| Control | P. indica | % of control | Control | P. indica | % of control | |
| 1 | 24.7 ± 11.0 | 32.6 ± 15.9 | 132 | 11.5 ± 9.2 | 22.8 ± 9.0 | 198* |
| 2 | 44.3 ± 9.8 | 44.6 ± 11.5 | 101 | 7.9 ± 4.2 | 22.4 ± 6.0 | 284** |
| 3 | 47.5 ± 7.5 | 37.5 ± 12.2 | 79 | 9.5 ± 5.0 | 16.2 ± 6.0 | 171 |
Total GR activity was measured according to procedures described in ref. 20. Values are means of three samples. Similar results were obtained with two independent sets of experiments. *, P < 0.05; **, P < 0.005; for intercohortal analyses, the unpaired Student t test was applied.
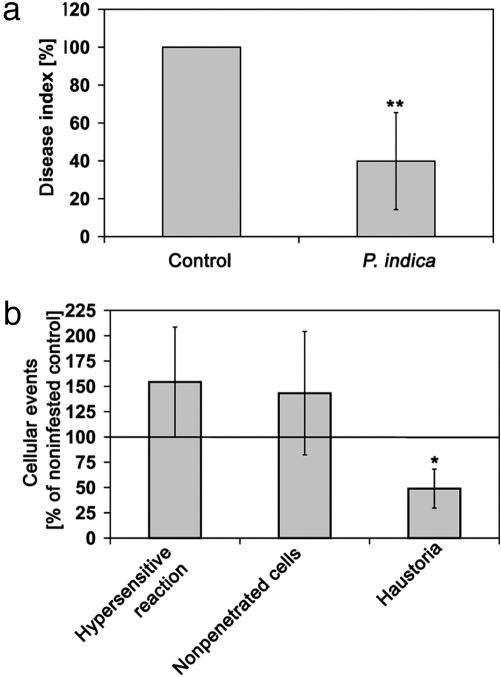
P. indica Induces Systemic Disease Resistance. We wanted to know whether root infestation by P. indica protects barley leaves from fungal infections. Because the fungus does not infest leaves, protection against leaf pathogens would require a systemic response emanating from the root that has not been described for a basidiomycete in cereals. We recorded the outcome of P. indica infestation on leaf infections by the biotrophic barley powdery mildew fungus, B. graminis f.sp. hordei, which belongs to the order Erysiphales, comprising pathogenic fungi of a wide range of crop plants. We found a reduction in powdery mildew infection in P. indica- infested plants (Fig. 4a). The relative decrease of disease, assessed by powdery mildew colony numbers, was 48% in the second-youngest leaves (data not shown) and 58% in the youngest leaves of 3-week-old P. indica-infested plants. Importantly, by microscopic analysis, we uncovered higher frequencies of a hypersensitive reaction, including a host-cell-death response, as well as a cell-wall-associated defense, confirming that the pathogen is arrested by an active plant response (Fig. 4b). Because of these plant responses, the biotrophic fungus cannot establish nutrition organs (haustoria). Together with the biochemical data (see Tables 2 and 3), these data substantiate that the systemic plant response, leading to a reduction of powdery mildew infection, is the result of induced resistance rather than being caused by antibiotics secreted by P. indica.
Fig. 4.
Systemic disease resistance conferred by P. indica. (a) Severity of powdery mildew infection (disease index) was calculated as colonies produced by B. graminis on the youngest leaf of 3-week-old barley plants (cultivar Ingrid), with roots either not infested (Control) or infested (P. indica). (b) Cellular responses to powdery mildew attack were evaluated by counting cells showing an active defense response, a hypersensitive response of the whole cell (Hypersensitive reaction), a local defense stopping a penetration attempt (Nonpenetrated cell), or a successful penetration (Haustoria), visible as the successful formation of a fungal haustorium in the cell. Error bars, SD. * and ** indicate statistically significant differences between leaves of infested and noninfested plants (unpaired Student t test; *, P < 0.05; **, P < 0.01).
We determined systemically elevated antioxidants in leaves from P. indica-infested plants. Initially, we measured ascorbate but did not detect consistent major changes due to P. indica infestation. However, when we analyzed the glutathione pool (GSH and oxidized glutathione), constituting a major cellular thiol-disulfide redox buffer, an enhanced foliar antioxidant capacity was found (Table 2). Strikingly, GR activity (Table 3) was also enhanced in leaves during the first 3 weeks of P. indica infestation, corroborating systemic induction of antioxidant capacity by P. indica. Notably, enhanced GSH concentrations are also associated with resistance to powdery mildew infections in barley, as mediated by major resistance genes (33). Hence, P. indica might cause systemic resistance by ameliorating the antioxidative capacity of barley plants. Consistently, it has been shown recently that fungal and algal partners in a lichen mutually benefit from the interaction by an enhanced glutathione-based antioxidative capacity (34), enabling a higher protection against stress.
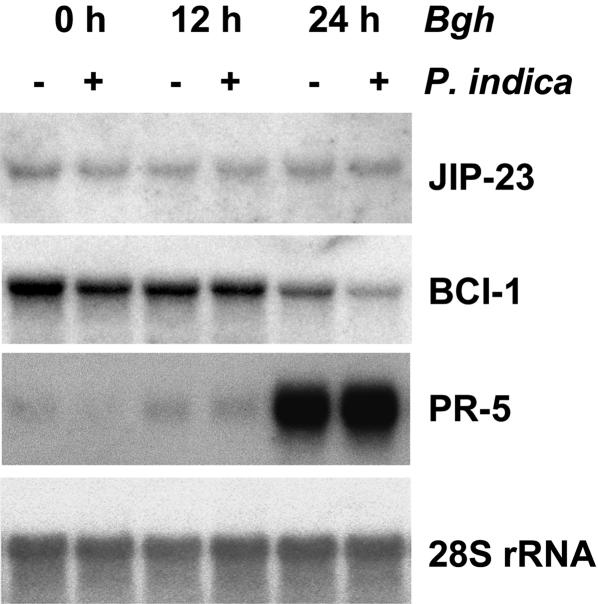
To determine whether any of the known cereal defense pathways was activated by P. indica, we tested expression of selected marker genes in barley leaves. Initial experiments using RT-PCR and cDNA microarrays indicated that P. indica does not induce expression of large sets of pathogenesis-related or jasmonic-acid (JA)-induced genes (data not shown). Northern blot experiments confirmed that gene markers indicative for salicylic acid (SA) (35) and JA (36) accumulation were not expressed. As displayed in Fig. 5, P. indica-induced systemic disease resistance is not associated with elevated foliar steady-state mRNA levels of the barley chemically induced gene 1 (BCI-1), encoding a 13-lipoxygenase (25). BCI-1 and its cereal orthologues are strongly responsive to SA, to resistance-inducing chemicals mimicking SA, such as acibenzolar-S-methyl, and to JA (25, 37, 38). Consistently, the jasmonate-induced protein gene 23 (JIP-23), expression of which is sensitive to exogenous jasmonate and elevated levels of endogenous jasmonate (24), did not respond to P. indica infestation. Moreover, the pathogenesis-related gene 5 (PR-5), indicative for pathogen infections (26, 39), was strongly expressed 24 h after powdery mildew inoculation, although essentially to the same extent in both control and P. indica-infested plants. These results indicate that P. indica might induce systemic disease resistance by an as yet unknown signaling pathway.
Fig. 5.
Expression of potential marker genes for known resistance pathways. Shown are Northern blots of 10 μg of total RNA from leaves of 3-week-old barley plants with roots infested (+) or not infested (-) with P. indica. Plants were harvested either directly or at 12 or 24 h after inoculation with powdery mildew (B. graminis f.sp. hordei, race A6). Blots were hybridized with radio-labeled JIP-23, BCI-1, and PR-5 probes. After removal of the probe, they were subsequently hybridized with a 28S rRNA probe to confirm equal transfer of RNA onto the membrane.
Together, we show here that P. indica-infested barley is more resistant to abiotic and biotic stress and that the reprogrammed metabolic state, which includes an enhanced antioxidant capacity and an activation of the glutathione-ascorbate cycle, does not negatively affect grain yield. Our findings challenge the notion that plant resistance to pathogens and pests induced by microorganisms necessarily involves internal, physiological, and ecological costs resulting in yield losses (40, 41). Because P. indica, unlike arbuscular mycorrhiza fungi, can easily be propagated on a large scale in axenic culture in the absence of a host plant (13), we suggest consideration of this endophyte as a tool for sustainable agriculture. Exploitation of P. indica may not only complement crop-growing strategies, but may also serve as a model system to study molecular traits affecting disease resistance and grain yields in cereals.
Acknowledgments
We thank Ajit Varma for discussions and support in the initial stage of the project, Markus Kolmer and Wolfgang Friedt for support in the yield experiments at the Rauischholzhausen experimental station, Sachin Deshmukh for performing RT-PCR to detect P. indica in barley tissues, Carin Jansen for help in preparing microscopic images, and B. Harrach for support in antioxidant measurements. This work was supported by Deutsche Forschungsgemeinschaft Grants FOR 343 and SFB 299.
Abbreviations: GR, glutathione reductase; GSH, reduced glutathione.
References
- 1.Food and Agriculture Organization of the United Nations (2004) The State of Food and Agriculture 2003-04, www.fao.org/documents/show_cdr.asp?url_file=/docrep/006/Y5160E/Y5160E00.HTM.
- 2.Chapin, F. S., III, Zavaleta, E. S., Eviner, V. T., Naylor, R. L., Vitousek, P. M., Reynolds, H. L., Hooper, D. U., Lavorel, S., Sala, O. E., Hobbie, S. E., et al. (2000) Nature 405, 234-242. [DOI] [PubMed] [Google Scholar]
- 3.Parmesan, C. & Yohe, G. (2003) Nature 421, 37-42. [DOI] [PubMed] [Google Scholar]
- 4.Newman, E. I. & Reddell, P. (1987) New Phytol. 106, 745-751. [DOI] [PubMed] [Google Scholar]
- 5.Azcón-Aguilar, C. & Barea, J. M. (1996) Mycorrhiza 6, 457-464. [Google Scholar]
- 6.Borowicz, V. A. (2001) Ecology 82, 3057-3068. [Google Scholar]
- 7.Gernns, H., von Alten, H. & Poehling, H.-M. (2001) Mycorrhiza 11, 237-243. [Google Scholar]
- 8.Shaul, O., Galili, S., Volpin, H., Ginzberg, I., Elad, Y., Chet, I. & Kapulnik, Y. (1999) Mol. Plant-Microbe Interact. 12, 1000-1007. [DOI] [PubMed] [Google Scholar]
- 9.Schardl, C. L., Leuchtmann, A. & Spiering, M. J. (2004) Annu. Rev. Plant Biol. 55, 315-340. [DOI] [PubMed] [Google Scholar]
- 10.Verma, S., Varma, A., Rexer, K.-H., Hassel, A., Kost, G., Sarabhoy, A., Bisen, P., Bütenhorn, B. & Franken, P. (1998) Mycologia 90, 896-903. [Google Scholar]
- 11.Varma, A., Singh, A., Sudha, S., Sharma, J., Roy, A., Kumari, M., Rana, D., Thakran, S., Deka, D., Sahay, N. S., et al. (2001) in Mycota IX, ed. Hock, B. (Springer, Heidelberg), pp. 125-150.
- 12.Weiss, M., Selosse, M.-A., Rexer, K.-H., Urban, A. & Oberwinkler, F. (2004) Mycol. Res. 108, 1003-1010. [DOI] [PubMed] [Google Scholar]
- 13.Varma, A., Verma, S., Sudha, Sahay, N., Bütehorn, B. & Franken, P. (1999) Appl. Environ. Microbiol. 65, 2741-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peskan-Berghöfer, T., Shahollari, B., Giong, P. H., Hehl, S., Markert, C., Blanke, V., Kost, G., Varma, A. & Oelmüller, R. (2004) Physiol. Plant 122, 465-477. [Google Scholar]
- 15.Sherameti, I., Shahollari, B., Venus, Y., Altschmied, L., Varma, A. & Oelmüller, R. (2005) J. Biol. Chem., in press, 10.1074/jbc.M500447200. [DOI] [PubMed]
- 16.Zadoks, J. C., Chang, T. T. & Konzak, C. F. (1974) Weed Res. 14, 415-421. [Google Scholar]
- 17.Hückelhoven, R. & Kogel, K.-H. (1998) Mol. Plant-Microbe Interact. 11, 292-300. [Google Scholar]
- 18.Knörzer, O. C., Durner, J. & Böger, P. (1996) Physiol. Plant 97, 388-396. [Google Scholar]
- 19.Klapheck, S., Zimmer, I. & Cosse, H. (1990) Plant Cell Physiol. 31, 1005-1013. [Google Scholar]
- 20.Becker, K., Gui, M., Traxler, A., Kirsten, C. & Schirmer, R. H. (1994) Histochemistry 102, 389-395. [DOI] [PubMed] [Google Scholar]
- 21.Nordhoff, A., Bücheler, U. S., Werner, D. & Schirmer, R. H. (1993) Biochemistry 32, 4060-4066. [DOI] [PubMed] [Google Scholar]
- 22.Bradford, M. M. (1976) Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 23.Logemann, J., Schell, J. & Willmitzer, L. (1987) Anal. Biochem. 163, 16-20. [DOI] [PubMed] [Google Scholar]
- 24.Hause, B., Demus, U., Teichmann, C., Parthier, B. & Wasternack, C. (1996) Plant Cell Physiol. 37, 641-649. [DOI] [PubMed] [Google Scholar]
- 25.Besser, K., Jarosch, B., Langen, G. & Kogel, K.-H. (2000) Mol. Plant Pathol. 1, 277-286. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, M., Jungling, S. & Knogge, W. (1993) Mol. Plant-Microbe Interact. 6, 745-754. [DOI] [PubMed] [Google Scholar]
- 27.Bütehorn, B., Rhody, D. & Franken, P. (2000) Plant Biol. 2, 687-692. [Google Scholar]
- 28.Kormanik, P. P. & McGraw, A.-C. (1982) in Methods and Principles of Mycorrhizal Research, ed. Schenck, N. C. (Am. Phytopathol. Soc., St. Paul, MN), pp. 37-45.
- 29.Weber, R. W. S., Wakley, G. E. & Pitt, D. (1998) Mycologist 12, 174-179. [Google Scholar]
- 30.Kumar, J., Schäfer, P., Hückelhoven, R., Langen, G., Baltruschat, H., Stein, E., Nagarajan, S. & Kogel, K.-H. (2002) Mol. Plant Pathol. 3, 185-195. [DOI] [PubMed] [Google Scholar]
- 31.Pignocchi, C. & Foyer, C. H. (2003) Curr. Opin. Plant Biol. 6, 379-389. [DOI] [PubMed] [Google Scholar]
- 32.Govrin, E. M. & Levine, A. (2000) Curr. Biol. 10, 751-757. [DOI] [PubMed] [Google Scholar]
- 33.Vanacker, H., Carver, T. L. & Foyer, C. H. (2000) Plant Physiol. 123, 1289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kranner, I., Cram, W. J., Zorn, M., Wornik, S., Yoshimura, I., Stabentheiner, E. & Pfeifhofer, W. (2005) Proc. Natl. Acad. Sci. USA 102, 3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryals, J., Lawton, K. A., Delaney, T. P., Friedrich, L., Kessmann, H., Neuenschwander, U., Uknes, S., Vernooij, B. & Weymann, K. (1995) Proc. Natl. Acad. Sci. USA 92, 4202-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhagen, B. W. M., Glazebrock, J., Zhu, T., Chang, H.-S., van Loon, L. C. & Pieterse, C. M. J. (2004) Mol. Plant-Microbe Interact. 17, 895-908. [DOI] [PubMed] [Google Scholar]
- 37.Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K.-H., Oosterdorp, M., Staub, T., Ward, E., Kessmann, H., et al. (1996) Plant Cell 8, 629-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaffrath, U., Freydl, E. & Dudler, R. (1997) Mol. Plant-Microbe Interact. 10, 779-783. [Google Scholar]
- 39.Vallélian-Bindschedler, L., Métraux J.-P. & Schweizer, P. (1998) Mol. Plant-Microbe Interact. 11, 702-705. [Google Scholar]
- 40.Brown, J. K. M. (2003) Trends Genet. 19, 667. [DOI] [PubMed] [Google Scholar]
- 41.Heil, M. (2002) Curr. Opin. Plant Biol. 5, 345-350. [DOI] [PubMed] [Google Scholar]