Abstract
Although most plant species from algae to flowering plants use blue light for inducing phototropism and chloroplast movement, many ferns, some mosses, and green algae use red as well as blue light for the regulation of these responses, resulting in better sensitivity at low light levels. During their evolution, ferns have created a chimeric photoreceptor (phy3 in Adiantum) between phytochrome (phy) and phototropin (phot) enabling them to use red light effectively. We have identified two genes resembling Adiantum PHY3, NEOCHROME1 and NEOCHROME2 (MsNEO1 and MsNEO2), in the green alga Mougeotia scalaris, a plant famous for its light-regulated chloroplast movement. Like Adiantum PHY3, both MsNEO gene products show phytochrome-typical bilin binding and red/far-red reversibility, the difference spectra matching the known action spectra of light-induced chloroplast movement in Mougeotia. Furthermore, both genes rescue red-light-induced chloroplast movement in Adiantum phy3 mutants, indicating functional equivalence. However, the fern and algal genes seem to have arisen independently in evolution, thus providing an intriguing example of convergent evolution.
Keywords: convergent evolution, Mougeotia
Plants have evolved sophisticated photomovement responses such as chloroplast movement, phototropism, and stomatal opening to optimize photosynthesis. In most plant species these responses are mediated by the blue-light photoreceptor phototropin (phot) (1). However, cryptogams such as ferns, mosses, and green algae, but not angiosperms, also use red light for monitoring the direction of incident light (2). Most of these red-light responses show red/far-red reversibility, indicating phytochrome (phy) involvement. In the case of the fern Adiantum capillus-veneris (Fig. 1A), a chimeric photoreceptor phytochrome 3 (phy3) has arisen, in which the N terminus consists of a phytochrome sensory module attached to an almost complete phot sequence (3). This photoreceptor mediates red-light-induced phototropism and chloroplast movement in polypodiaceous ferns, perhaps conferring an adaptive advantage in low light under dense canopies (4, 5).
Fig. 1.
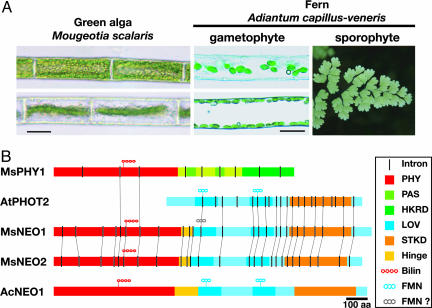
Mougeotia and Adiantum: Chloroplast movements and the associated neochrome. (A) Giant chloroplasts of M. scalaris in their face-on (Left Upper) and side-on (Left Lower) orientations under different light treatment, and those of A. capillus-veneris protonemal cells in which the chloroplast accumulation to the upper (Center Upper) or side (Center Lower) wall are induced by polarized red-light vibrating parallel or perpendicular to the cell axis irradiated from horizontal direction. (Scale bar, 20 μm.) A Right shows Adiantum sporophyte. (B) The domain structure of fern and algal neochromes and canonical plant phytochromes and phototropins. MsNEO1 LOV1 domain has so far failed to bind FMN in vitro, and the other MsNEO LOV domains are unlikely to bind flavins. Introns whose positions are equivalent in each gene are connected with thin lines. PHY, phytochrome photosensory region; HKRD, histidine kinase-related domain; STKD, serine/threonine kinase domain.
Red light also induces chloroplast movement, phototropism, and polarotropism in mosses. Although Physcomitrella patens has four conventional PHY (6) and four PHOT (7) genes, we have been unsuccessful in detecting a chimera resembling Adiantum PHY3. Because such a gene sequence is also absent from the extensive moss EST and nearly complete Physcomitrella genome databases, it is very unlikely that an Adiantum PHY3 homolog is present in mosses. In any case, targeted knockout of PHY and PHOT genes in Physcomitrella has shown that red-light-induced chloroplast movement in Physcomitrella is mediated by canonical phys with phots acting downstream (6, 7).
On this basis it would seem that the AcPHY3 (Ac, Adiantum capillus-veneris) gene arose late in fern evolution. However, we show here that in the filamentous green alga Mougeotia scalaris [a species famous for its phy-mediated chloroplast rotation (8)] (Fig. 1 A) two AcPHY3-like genes are present and, moreover, that they are functionally equivalent to the fern gene in mediating chloroplast movement. Based on this, we propose placing all three chimeric photoreceptors in a new category, neochrome [hence, MsNEO1 (Ms, Mougeotia scalaris), MsNEO2, and Ac-NEO1 (= AcPHY3)]. Comparison of the algal and fern neos suggests that they have arisen independently, providing a most unusual example of convergent evolution.
Materials and Methods
Cloning of M. scalaris Photoreceptor Genes. RT-PCR with primers directed to the phototropin LOV2 and kinase domain-encoding regions was performed by using M. scalaris total RNA. After this, three full-length cDNAs were isolated by 5′ and 3′ RACE (rapid amplification of cDNA ends) and by RT-PCR. The deduced amino acid sequences of two cDNA clones were similar to phototropin (MsPHOTA and MsPHOTB), whereas the third, MsNEO1, showed strong similarity to A. capillus-veneris PHY3 (AcPHY3). The MsNEO2 gene was cloned simultaneously and independently by using a genomic DNA template with degenerate primers directed to phytochrome chromophore-binding and phototropin kinase domains, respectively. Again, the complete coding sequence was obtained with 3′ and 5′ RACE and RT-PCR. This too showed strong similarity to AcPHY3. The cognate genes were cloned via PCR of genomic DNA. Full details of the PCR and cloning procedures are described in Supporting Methods, which is published as supporting information on the PNAS web site.
Expression, Purification, and Determination of Difference Spectrum of Phytochrome Domains of MsNEO1 and MsNEO2 Proteins. For expression of the phytochrome-like region (amino acids 1-418) of MsNEO1 primers 5′-GGAATTCATTAAAGAGGAGAAATTAACTATGTCTTCCCGTCCTGGCCATG-3′ and 5′-GGGAGATCTGTTTGCCGCCAGGGAAATCTCTC-3′ were used to amplify an ≈1.3-kb cDNA product. This was cloned into pCR-BluntII TOPO (Invitrogen), the resulting plasmid was cut with BglII and EcoRI, and the insert was cloned into the corresponding sites of pQE12 (Qiagen) in XL1-blue (Stratagene). An equivalent domain of MsNEO2 (amino acids 1 to 480) was amplified from cDNA by using primers 5′-GGAATTCATTAAAGAGGAGAAATTAACTATGGATCGGACAATCGATATTTCG-3′ and 5′-GGGAGATCTGAGGTGCTTCAGGATCCACTCTGCCAG-3′, resulting in an ≈1.5-kb RT-PCR product. This product was cloned into BglII/EcoRI of pQE12 as described above.
For overexpression, clones were grown in LB containing ampicillin (100 μg/ml) and tetracycline (15 μg/ml) at 37°C until an OD600 of 0.5. The flasks were then cooled at 4°C for 30 min and transferred to 18°C. After addition of isopropyl β-d-thiogalactoside (IPTG) to a final concentration of 50 μM, expression was allowed to proceed for 16 h before collection, washing, and resuspension in cold TESβ buffer [50 mM Tris·Cl, pH 7.8; 5 mM EDTA; 300 mM NaCl; 0.001% (vol/vol) 2-mercaptoethanol] and broken at 4°C with a pressure cell at 120 MPa. Crude extracts were cleared, and the protein was precipitated with ammonium sulfate at 60% saturation. The pellet was resuspended in TISI10 (50 mM Tris·Cl, pH 7.8; 5 mM iminodiacetic acid; 300 mM NaCl; 10 mM Imidazole), and the His6-tagged protein was purified over Ni-NTA (Superflow, Qiagen) by using a ΔKTA system (Amersham Pharmacia). Phycocyanobilin (PCB) adducts were prepared in vitro by using excess chromophore isolated from freeze-dried Spirulina by methanolysis and purified by HPLC. Phytochromobilin (PΦB) adducts were produced in vivo by overexpression of the phytochrome-like regions (see above) together with Synechocystis heme oxygenase-1 (HO1; sll1184) and Arabidopsis thaliana phytochromobilin synthase (HY2) cDNAs cloned as a single operon into pPRO-Lar.A122 (Clontech) in Escherichia coli (9). Cells containing both plasmids were grown at 37°C in LB medium containing kanamycin and ampicillin with vigorous shaking to OD600 ∼ 0.5. Again, the cells were cooled at 4°C for 30 min, and expression was induced by adding 50 μM IPTG and 0.2% (wt/vol) Arabinose (Roth, Karlsruhe, Germany). All subsequent steps were carried out in darkness. Expression proceeded for 18 h at 18°C. Extraction and purification of the His6-tagged protein was carried out as above.
For difference spectroscopy, holophytochrome was irradiated with red and far-red (light-emitting diodes at 660 and 730 nm, respectively; 30 nm full width at half maximum), and UV-Vis absorbance spectra (350-800 nm) were measured with a diode array spectrophotometer (Agilent Technologies 8453).
Complementation of Fern neo Mutant by Transient Expression of Two Mougeotia NEO Genes. A synthetic GFP (S65T) driven by the cauliflower mosaic virus 35S promoter (35S-GFP) was used (10). The MsNEO1 and MsNEO2 cDNA fragment covering the interval between the predicted start and stop codons was subcloned into the SalI-NotI sites of 35S-GFP. Complementation tests were performed as described in ref. 4. Briefly, rap2 prothalli (11) were fragmented with a Polytron (PT 1200, Kinematica) and then cultured for 2 to ≈3 weeks. Subjected to osmotic stress by placing prothalli on 0.25 M mannitol media for 1 h, the bombardment was performed by using a Biolistic particle delivery system (PDS-100/He, Bio-Rad). The prothalli were incubated for an additional day in darkness, and GFP-expressing cells were picked up under fluorescence microscopy (Axioplan, Zeiss) and used for microbeam irradiation. For conditions of microbeam irradiation, refer to the figure legends.
Results and Discussion
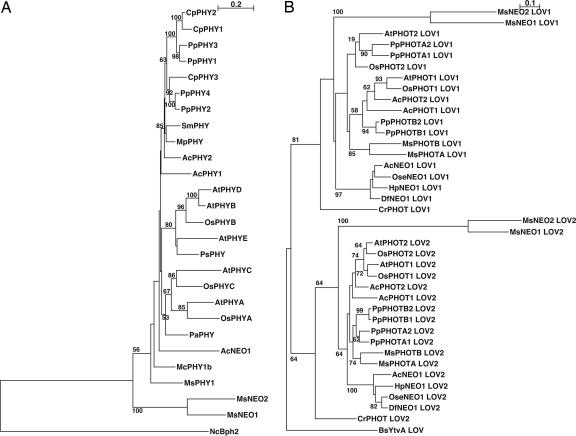
Using a PCR approach, we found two AcPHY3-like genes in the filamentous green alga M. scalaris. Because the products are functionally equivalent (see below), we propose naming this class of PHY-PHOT chimera genes NEOCHROME (NEO, here MsNEO1 and MsNEO2), thereby renaming AcPHY3 as AcNEO1. MsNEO1 and MsNEO2 encode 1,486- and 1,442-aa proteins, respectively, each consisting of a segment similar to the N-terminal sensory module of PHY upstream of an almost complete PHOT-like segment comprising two LOV [light, oxygen, voltage] domains and a serine/threonine kinase domain (Fig. 1B; see also Fig. 5, which is published as supporting information on the PNAS web site). Notably, although the junction point between the PHY domain and the hinge region is exactly the same in all known NEOs, fern representatives are 35 to ≈56 residues longer than those of MsNEOs at this point (Fig. 5). The PHY and each LOV domain of the algal NEOs are ≈60%, and the kinase domain is nearly 80% similar, whereas the algal NEO similarities to those of AcNEO1 and indeed to higher plant PHYs and PHOTs are <40% and 60%, respectively (Table 1). Accordingly, phylogenetic trees based on the PHY N-terminal and PHOT LOV domains indicate that MsNEOs are rather distantly related to other PHYs and PHOTs (Fig. 2).
Table 1. Percent similarity among neochromes, canonical phytochromes, and phototropins.
| MsNEO2
|
AcNEO1
|
Plant PHY or PHOT
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phy | LOV1 | LOV2 | Kinase | Phy | LOV1 | LOV2 | Kinase | Phy | LOV1 | LOV2 | Kinase | |
| MsNEO1 | 57.6 | 53.3 | 63.6 | 78.6 | 39.3 | 39.3 | 40.2 | 59.0 | 37.0∼42.9 | 36.4∼42.1 | 34.6∼43.0 | 50.0∼43.8 |
| MsNEO2 | — | — | — | — | 33.3 | 38.3 | 37.4 | 59.4 | 35.0∼42.3 | 37.4∼43.9 | 33.6∼42.1 | 49.3∼60.3 |
| AcNEO1 | — | — | — | — | — | — | — | — | 41.4∼53.6 | 50.5∼68.2 | 60.7∼73.8 | 48.6∼70.8 |
Fig. 2.
Phylogenetic trees of phytochrome sensor domains (A) and phototropin and neochrome LOV domains (B). Numbers indicate local bootstrap percent probabilities (only values >50% are indicated). (Scale bar indicates the evolutionary distance.) Phylogenetic trees were generated by clustalw. Complete species names and accession numbers of protein sequences are provided in Supporting Methods.
The NEO gene structures reveal interesting associations (Fig. 1B; see also Figs. 5 and 6, which are published as supporting information on the PNAS web site). Both MsNEO genes have 26 identically positioned introns, whereas AcNEO1 and some other ferns' NEOs carry no introns (3, 4). With one exception, introns in the PHOT-like region show identical positions to those of Mougeotia or Arabidopsis PHOTs. An intron in both MsNEOs is positioned exactly at the N terminus of LOV1, the point at which the NEO/PHOT alignment breaks down (Fig. 5). These findings imply that an ancestral MsNEO arose as a result of exon shuffling between PHY and PHOT genes. Both MsNEOs contain eight introns within the PHY-like region. Genes of conventional PHYs in fern, gymnosperm, and angiosperm contain no intron within this region, but PHY genes from the green algae Mesotaenium (McPHY1b) (12) and Mougeotia itself (MsPHY1) (13) contain five and three introns, respectively, two of which are identically positioned in the MsNEO genes (Figs. 1B, 5, and 6). The additional intron seen in Physcomitrella PpPHY2 and PpPHY4 (Pp, Physcomitrella patens) genes in this region (6) is positioned differently. The fact that both MsNEO genes have two introns specific to algal PHYs implies that the fusion event (followed by gene duplication) in green algae occurred separately from that in polypodiaceous ferns. Alternative explanations based on a common ancestral NEO gene that was either lost in most plant lineages or transferred laterally are thereby less likely.
The occurrence of a gene encoding a putative chimeric red-/blue-light photoreceptor in Mougeotia is of special interest. Photoorientation of the giant chloroplast (Fig. 1 A) is regulated by light (14, 15). Sophisticated experiments using microbeams and polarized light implied that a gradient of the Pfr form of phy bound in orderly arrays at the cell periphery controls chloroplast orientation (8). Because low levels of blue light result in the same chloroplast “face-on” position as under red light, Haupt (16) proposed a chimeric photoreceptor for blue and red-light absorption. Red/far-red difference spectra (absorption spectra after saturating FR irradiation minus that after saturating R irradiation) of recombinant Mougeotia phy1 with PCB chromophore showed peaks at 646 and 720 nm for Pr and Pfr, respectively (17), whereas action spectra of chloroplast photoorientation and reversion showed maxima at 679 and 717 nm, respectively (18), spectral shifts only partly compatible with PΦB instead of PCB being the native chromophore in Mougeotia. There remains the thorny problem that known phys are cytosolic/nuclear proteins with no known membrane or cytoskeletal associations. It is thus hard to imagine how a conventional phy could show the strong anisotropy implied by Haupt's experiments (8, 16).
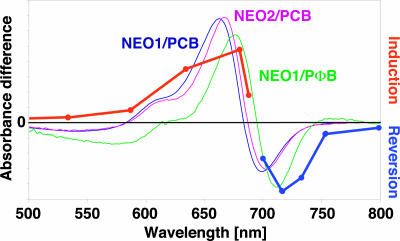
We used recombinant methods to investigate the potential of the MsNEO gene products to act as photoreceptors. The PHY-like domains of both MsNEO1 and MsNEO2 autoassembled with PCB and PΦB were red/far-red photochromic, a feature characteristic of PHY apoproteins. Red-/far-red-light difference spectra showed peaks at 663 and 699 nm (neo1) and 667 and 703 nm (neo2), similar to those reported for PCB-adducts of Adiantum neo (3), whereas PΦB adducts showed peaks at 676 and 714 nm (Fig. 3). Such PCB/PΦB shifts are typical, resulting from the more extensive conjugated π electron system of PΦB (19). Moreover, the spectra of PΦB adducts correspond closely to those of the action spectrum for Mougeotia chloroplast rotation (18) (Fig. 3), consistent with PΦB-neo being the photoreceptor for this response. Because phots are known to be membrane-associated (20), this portion of the NEO molecule might confer a similar property, providing a molecular basis for the system's ability to transduce vectorial light information during chloroplast photoorientation according to Haupt's hypothesis.
Fig. 3.
Difference spectra of the phytochrome-like domains of MsNEO1 and MsNEO2. The maxima/minima are 663/699 nm for MsNEO1 (PCB), 667/703 nm for MsNEO2 (PCB), and 676/714 nm for MsNEO1 (PΦB). The spectrum of MsNEO2 (PΦB) is not included because of poor expression rate. The action spectra for chloroplast photoorientation in red light and its reversal by far-red light as determined by Haupt (18) are included for comparison.
In phototropins and Adiantum neo both LOV1 and LOV2 domains bind FMN as UV/A-blue-active chromophores (20, 21). A conserved cysteine residue is essential for the subsequent photocycle (21, 22). Interestingly, this residue is represented in LOV1 of MsNEO1 but in none of the other three LOV domains (Fig. 1B; see also Figs. 5 and 7, which are published as supporting information on the PNAS web site). Similarly, 10 other residues thought to be important in FMN-binding or photoactivation (23, 24) are seen only in MsNEO1 LOV1 (Fig. 7). The other LOV domains are thus unlikely to have a role in light absorption. We successfully expressed both calmodulin-binding protein and maltose-binding protein fusions of all four MsNEO LOV domains in E. coli but never observed phot-like UV-Vis absorbance in the purified extracts (data not shown), consistent with the notion that neither MsNEO binds FMN. In view of its primary structure, however, MsNEO1 LOV1 might nevertheless be photoactive in planta. On the other hand, whereas LOV2 is essential for kinase domain activation and physiological responses of phot1 in Arabidopsis (22) and phot2 in Adiantum (25), this is not the case for LOV1. Thus, a possible role of a photoactive MsNEO1 LOV1 remains unclear. The functions of the other three LOV domains are even more obscure. LOV domains are a subclass of the functionally diverse PAS superfamily (26). The helixturn-helix fold characteristic of PAS domains is known or thought to bind cofactors such as FAD or, in the case of the PYP photoreceptor, coumaric acid. Otherwise PAS domains, generally occurring in pairs, are often involved in protein-protein interactions. Higher plant PHYs carry a typical double-PAS domain possibly involved in dimerization. Indeed, PHOT LOV domains dimerize in vitro (27, 28). Thus it is quite possible that the LOV/PAS domains of NEO in Mougeotia have evolved to play a role other than flavin binding. Haupt (16) suggested that a single photoreceptor might mediate both red- and blue-light-induced chloroplast photoorientation in Mougeotia (16). As phytochrome-phototropin chimeras, neos might seem exactly to fulfil the prediction of the hypothesis. This is, however, improbable given that MsNEOs probably cannot perceive blue light in planta. More likely the conventional phototropins, MsPHOTA and MsPHOTB, mediate blue-light-induced chloroplast photoorientation in this alga.
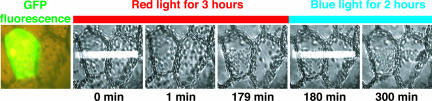
Because neither transgene expression nor mutagenesis techniques are established in Mougeotia, we cannot yet demonstrate directly that neochromes regulate red-light-induced chloroplast movement in that species. Instead, we transiently expressed both MsNEO1 and MsNEO2 cDNAs in the Adi-antum rap2 mutant, in which a lesion in NEO (PHY3) leads to defective red-light-mediated phototropism and chloroplast movement (4, 11). MsNEO1- and/or MsNEO2-mediated rescue of rap2 would indicate functional equivalence. Indeed, when either MsNEO gene was cotransformed with GFP-producing construct, chloroplasts in GFP-expressing cells accumulated in response to red-light irradiation (Fig. 4; see also Movie 1, which is published as supporting information on the PNAS web site) but not those in untransformed cells or in cells transformed with GFP only (4) (data not shown). Blue light irradiation induced the accumulation response in both GFP-positive and -negative cells (that is, in cells transfected with GFP and NEO and in nontransfected cells), showing that the defect in red-light-induced chloroplast movement in the non-transgenic cells does not result from loss of cell viability or damage to the machinery responsible for chloroplast movement. Because transient expression of both MsNEO genes rescue red-light-induced chloroplast movement in rap2 (neo), all three gene products have similar functional properties both in relation to light absorption and signal transduction.
Fig. 4.
Transient expression of MsNEO1 can rescue the Adiantum neo mutant rap2. Before microbeam irradiation, the green fluorescence image was taken. After dark-adaptation overnight, the GFP-fluorescent cell together with the neighboring cell (no GFP fluorescence) was irradiated with a 20-μm microbeam of red light (0.55 μmol·m-2·s-1). Three hours later, a microbeam of blue light (0.38 μmol·m-2·s-1) was applied for an additional 2 h. Times are indicated below.
Although neochromes are now known in several polypodiaceous ferns and in Mougeotia, they are not represented in the genome sequences of any prokaryote, the green alga Chlamydomonas, the red alga Cyanidioschyzon, the diatom Thalassiosira, or the higher plants Arabidopsis, Oryza, or Populus. Furthermore, PCR-based searches or library screening for AcPHY3 homologues in more primitive ferns and the mosses Physcomitrella patens and Ceratodon purpureus were unsuccessful; moreover, such sequences are not present among the >200,000 available moss ESTs or in the currently available ≈109-bp genome sequence data for Physcomitrella (data not shown). Thus, if neochromes are monophyletic they must either have been retained specifically in very few lineages or have been transferred laterally. Detailed examination implies, rather, that they have arisen separately in ferns and algae. Gene rearrangements in different organisms are known to arise via several molecular mechanisms including exon shuffling, duplication, and retrotransposition (29). The present work together with that of Nozue et al. (3) provides an example of chimeric gene products with the same structure and function having independently arisen twice. The complete absence of introns in fern NEO genes suggested that fern NEO genes may be generated by the disruption of preexisting exon-intron structures of PHOT genes by retrotransposons and subsequent fusion with a PHY fragment (30). Because MsNEO genes retain several Mougeotia-specific introns in both PHY- and PHOT-like regions, in addition to an intron positioned at the apparent N terminus of the PHOT-like segment, they might be the product of exon shuffling between PHY and PHOT genes in that species.
Supplementary Material
Acknowledgments
This article is dedicated to Prof. Wolfgang Haupt (University of Erlangen, Erlangen, Germany) on the occasion of his 85th birthday. We thank Wolfgang Haupt, Hironao Kataoka, and Yoshikatsu Sato for Mougeotia cultures; Takatoshi Kagawa for degenerate primers; Yasuo Niwa for GFP vector; Koichiro Tamura for helpful discussion; Masahiro Kasahara for technical advice; and Tina Lang, Andrea Weisert, Yoko Takahashi, and Hidenori Tsuboi for technical assistance. This work was partially supported by German Research Foundation Grant DFG Hu702/2 (to J.H.), Japanese Ministry of Education, Sports, Science, and Technology Grants MEXT 13139203 and 13304061 (to M.W.), Japan Society for the Promotion of Science Grant 16107002 (to M.W.), and a Research Fellowship for Young Scientists (to N.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LOV, light, oxygen, voltage; PΦB, phytochromobilin; PCB, phycocyanobilin.
Data deposition: The Mougeotia photoreceptor cDNA and gene sequences have been deposited in the GenBank database [accession nos. AB206961 and AB206966 (MsNEO1), AB206962/AY878549 and AB206967 (MsNEO2), AB206963 and AB206968 (MsPHOTA), AB206964 and AB206969 (MsPHOTB), and AB206965 and AB206970 (MsPHY1)].
References
- 1.Briggs, W. R. & Christie, J. M. (2002) Trends Plant Sci. 7, 204-210. [DOI] [PubMed] [Google Scholar]
- 2.Suetsugu, N. & Wada, M. (2005) in Handbook of Photosensory Receptors, eds. Briggs, W. R. & Spudich, J. L. (Wiley-VCH, Weinheim, Germany), pp. 349-369.
- 3.Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H., Yeh, K.-C., Lagarias, J. C. & Wada, M. (1998) Proc. Natl. Acad. Sci. USA 95, 15826-15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai, H., Kanegae, T., Christensen, S., Kiyosue, T., Sato, Y., Imaizumi, T., Kadota, A. & Wada, M. (2003) Nature 421, 287-290. [DOI] [PubMed] [Google Scholar]
- 5.Schneider, H., Schuettpelz, E., Pryer, K. M., Cranfill, R., Magallón, S. & Lupia, R. (2004) Nature 428, 553-557. [DOI] [PubMed] [Google Scholar]
- 6.Mittmann, F., Brücker, G., Zeidler, M., Repp, A., Abts, T., Hartmann, E. & Hughes, J. (2004) Proc. Natl. Acad. Sci. USA 101, 13939-13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasahara, M., Kagawa, T., Sato, Y., Kiyosue, T. & Wada, M. (2004) Plant Physiol. 135, 1388-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haupt, W. (1999) Prog. Bot. 60, 3-36. [Google Scholar]
- 9.Landgraf, F. T., Forreiter, C., Hurtado Pico, A., Lamparter, T. & Hughes, J. (2001) FEBS Lett. 508, 459-462. [DOI] [PubMed] [Google Scholar]
- 10.Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M. & Kobayashi, H. (1999) Plant J. 32, 455-463. [DOI] [PubMed] [Google Scholar]
- 11.Kadota, A. & Wada, M. (1999) Plant Cell Physiol. 40, 238-247. [Google Scholar]
- 12.Lagarias, D. M., Wu, S.-H. & Lagarias, J. C. (1995) Plant Mol. Biol. 29, 1127-1142. [DOI] [PubMed] [Google Scholar]
- 13.Winands, A. & Wagner, G. (1996) Plant Mol. Biol. 32, 589-597. [DOI] [PubMed] [Google Scholar]
- 14.Senn, G. (1908) Die Gestaltsund Lageveränderungen der Pflanzen-Chromatophoren (Engelmann, Leipzig, Germany).
- 15.Mosebach, G. (1958) Planta 52, 3-46. [Google Scholar]
- 16.Haupt, W. (1971) Z. Pflanzenphysiol. 65, 248-265. [Google Scholar]
- 17.Jorissen, H. J. M. M., Braslavsky, S. E., Wagner, G. & Gärtner, W. (2002) Photochem. Photobiol. 76, 457-461. [DOI] [PubMed] [Google Scholar]
- 18.Haupt, W. (1959) Planta 53, 484-501. [Google Scholar]
- 19.Falk, H. (1989) The Chemistry of Linear Oligopyrroles and Bile Pigments (Springer, Berlin).
- 20.Briggs, W. R., Christie, J. M. & Salomon, M. (2001) Antiox. Redox Signaling 3, 775-788. [DOI] [PubMed] [Google Scholar]
- 21.Crosson, S., Rajagopal, S. & Moffat, K. (2003) Biochemistry 42, 2-10. [DOI] [PubMed] [Google Scholar]
- 22.Christie, J. M., Swartz, T. E., Bogomolni, R. A. & Briggs, W. R. (2002) Plant J. 32, 205-219. [DOI] [PubMed] [Google Scholar]
- 23.Salomon, M., Christie, J. M., Knieb, E., Lempert, U. & Briggs, W. R. (2000) Biochemistry 39, 9401-9410. [DOI] [PubMed] [Google Scholar]
- 24.Nozaki, D., Iwata, T., Ishikawa, T., Todo, T., Tokutomi, S. & Kandori, H. (2004) Biochemistry. 43, 8373-8379. [DOI] [PubMed] [Google Scholar]
- 25.Kagawa, T., Kasahara, M., Abe, T., Yoshida, S. & Wada, M. (2004) Plant Cell Physiol. 45, 416-426. [DOI] [PubMed] [Google Scholar]
- 26.Taylor, B. L. & Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomon, M., Lempert, U. & Rüdiger, W. (2004) FEBS Lett. 572, 8-10. [DOI] [PubMed] [Google Scholar]
- 28.Nakasako, M., Iwata, T., Matsuoka, D. & Tokutomi, S. (2004) Biochemistry 43, 14881-14890. [DOI] [PubMed] [Google Scholar]
- 29.Long, M., Betrán, E., Thornton, K. & Wang, W. (2003) Nat. Rev. Genet. 4, 865-875. [DOI] [PubMed] [Google Scholar]
- 30.Buzdin, A. A. (2004) Cell. Mol. Life Sci. 61, 2046-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.