Abstract
Epigenetic inheritance of heterochromatin structure is an important cellular process whose mechanism remains elusive. In this article, we describe the identification of nine enhancers of the silencing defect of a Saccharomyces cerevisiae-PCNA mutant by screening a library of ≈4,700 viable yeast deletion mutants. Of the nine mutants identified, six (hir1, hir3, sas2, sas4, sas5, and sir1) were previously known to reduce silencing synergistically with a mutation in Cac1p, the large subunit of chromatin assembly factor 1 (CAF-1). The predicted gene products that are affected in three other mutants (nam7, msh2, and rtt106) have not been implicated previously in silencing. Characterization of the rtt106Δ allele revealed that it synergistically reduced heterochromatin silencing when combined with a mutation in Cac1p but not with a mutation in Asf1p (a histone H3 and H4 chaperone). Moreover, Rtt106p interacted with histones H3 and H4 both in vitro and in vivo, and it displayed a nucleosome assembly activity in vitro. Furthermore, Rtt106p interacts with CAF-1 physically through Cac1p. These biochemical and genetic data indicate that Rtt106p is a previously uncharacterized histone chaperone connecting S phase to epigenetic inheritance.
Keywords: epigenetic silencing, nucleosome assembly, chromosome assembly factor 1
In eukaryotic cells, DNA is packaged into chromatin. Historically, chromatin has been classified into two cytologically distinguishable forms: euchromatin, which is competent for transcription of most genes, and heterochromatin, which generally silences transcription. Once a gene assumes a silenced state, silencing can be inherited for many mitotic and meiotic divisions (1-3). However, how heterochromatin silencing is inherited during chromosome replication in the S phase of the cell cycle is not well understood.
In the yeast Saccharomyces cerevisiae, silent mating-type loci (HMR and HML), telomeres, and the ribosomal DNA locus form heterochromatin-like structures that result in silencing of genes within or near these regions (4-6). It is hypothesized that inheritance of heterochromatin states is carried out by the replication-coupled chromatin assembly process (7, 8). In this process, parental histones must be transferred to two nascent chromatids, a process that is still enigmatic (9, 10). Moreover, newly synthesized histones must be deposited onto the replicated DNA to form nucleosomes. Deposition of newly synthesized histones onto replicated DNA is facilitated by histone chaperones such as chromatin assembly factor 1 (CAF-1) and anti-silencing factor 1 (Asf1p) (11).
In support of the idea that the DNA replication-coupled chromatin assembly process is involved in epigenetic inheritance, yeast and human CAF-1 are required for heterochromatin formation and silencing in yeast and mammalian cells. Human CAF-1 consists of three subunits, p150, p60, and p48, and it deposits histones H3 and H4 onto DNA after DNA replication or repair to promote nucleosome formation (12-15). In addition to its role in nucleosome formation, human CAF-1 binds to heterochromatin protein 1 (16), a structural component of mammalian heterochromatin. Interference of CAF-1 functions by a dominant negative form of p150 or by small interfering RNA results in slowed progression through S phase and reactivated transcription of a silenced transgene (17-20). These results demonstrate that CAF-1 is a key factor connecting DNA replication to heterochromatin silencing.
The three subunits of yeast CAF-1 are Cac1p, Cac2p, and Cac3p (21). Yeast cells either lacking each subunit of CAF-1 alone or lacking all of them grow at near wild-type rates. However, these mutant cells are partially defective for silencing at telomeres, the HM loci, and the rDNA locus (21-25). These results suggest that additional factors exist in cells to promote nucleosome formation and silencing. Supporting this idea, other histones H3 and H4 chaperones such as Asf1p, Hir1p, and Hif1p have been found to be required for silencing in yeast cells (26-30). Moreover, the cac1Δ asf1Δ or cac1Δ hir1Δ double-mutant cells grow very slowly and exhibit synergistic reduction in silencing. On the other hand, the asf1Δ hir1Δ double mutant reduced silencing to a degree similar to either the asf1Δ or hir1Δ single mutant (26, 27, 31). Thus, there exist at least two histone assembly pathways that are partially redundant in yeast cells.
In addition to mutations in histone chaperones, distinct mutations in PCNA (encoded by the POL30 gene), a factor important for the DNA replication-coupled chromatin assembly, affect either CAF-1-dependent or Asf1-dependent silencing (32-34). Genetic and biochemical studies demonstrate that the pol30-8 mutant allele prevents CAF-1 from contributing to silencing (32). Genetic evidence suggests that the pol30-79 mutant acts primarily to disable Asf1p-dependent silencing. In an effort to understand how PCNA is involved in silencing, we have made a PCNA mutant, which we call pol30-879, containing the amino acids changes from both pol30-8 and pol30-79. This mutant reduces telomeric silencing more than either pol30-8 or pol30-79 alone. However, silencing at the HM loci is largely intact in the pol30-879 mutant. We have therefore introduced the pol30-879 mutation into a collection of ≈4,700 viable yeast deletion mutants and identified nine enhancers of the silencing defect of pol30-879. We describe the characterization of one of the previously uncharacterized silencing proteins, Rtt106p. The rtt106Δ mutation synergistically reduces silencing with cac1Δ but has little effect on silencing when combined with a deletion of ASF1, suggesting that Rtt106p might function with Asf1p in the same genetic pathway. Like Asf1p, Rtt106p also binds to histones H3 and H4 and CAF-1 and has a nucleosome assembly activity in vitro. These results strongly indicate that Rtt106p is another histone chaperone connecting DNA replication to epigenetic silencing.
Experimental Procedures
Yeast Strains and Plasmids. All of the yeast strains in our experiments except those used for screening are isogenic to W303-1A (leu2-3,112 ura3-1 his3-11, trp1-1, ade2-1 can1-100) (35). Standard yeast media and manipulations were used. See Supporting Experimental Procedures, which is published as supporting information on the PNAS web site, for detailed procedures for screening a library of 4,700 viable yeast deletion mutants and purification of Rtt106p.
Silencing Assays. The telomeric silencing assay and the mating assay used to determine silencing at the HML locus were performed as described in refs. 32 and 36, respectively. Experimental procedures to assay silencing at the HMR locus by using the GFP gene were described in Supporting Experimental Procedures.
Binding Assays to Detect Binding Between Rtt106p and Histones or CAF-1. To test whether Rtt106p binds to histones or Cac1p in vitro, equal amounts of purified GST-Rtt106p or GST-REGα (a human proteasome activator) (37, 38) were incubated with glutathione Sepharose beads for 2 h at 4°C. After the beads were washed three times with 1 ml of binding buffer A300 (25 mM Tris, pH 8.0/10% glycerol/1 mM EDTA/0.01% Nonidet P-40/300 mM NaCl), they were incubated with increasing amounts of core histone octamers purified from HeLa cells or [35S]methionine-labeled Cac1p by using TNT rapid in vitro transcription translation kits (Promega) in 500-μl reactions for 6 h at 4°C. After beads were washed extensively with A300, the bound proteins were eluted by SDS loading buffers, resolved by SDS/PAGE, and visualized by Coomassie blue staining or autoradiography.
To determine the binding between Rtt106p and histones in vivo, Rtt106p was tagged at its C terminus with the tandem affinity purification (TAP) tag. We followed a standard procedure to purify Rtt106p by using the TAP tag (39), and copurified proteins were detected by Western blotting with antibodies against histones H3 or H4. Similar procedures were also performed to study in vivo binding between Rtt106p and CAF-1.
Plasmid DNA Supercoiling Assay. Negatively supercoiled plasmid DNA (pSV011) was incubated with Topoisomerase I from wheat germ at 30°C for 1 h to relax the DNA. Increasing amounts of Rtt106p purified from Escherichia coli, and purified histone octamers were added to the relaxed DNA to allow nucleosome assembly over a 1.5-h incubation at 30°C. The reactions were then stopped by incubating with stop buffer (1% SDS/0.2 mg/ml protease K/20 mM EDTA, pH 8.0) at 30°C for 30 min. After removal of proteins by digestion with proteinase K, DNA was purified by phenol extraction and ethanol precipitation, resolved on a 1% agarose gel, and visualized by ethidium bromide staining.
Results
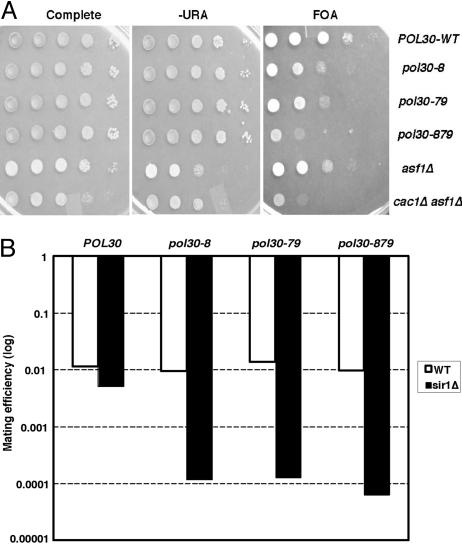
The pol30-879 Mutation Significantly Reduces Telomeric and HM Silencing. PCNA connects DNA replication to epigenetic inheritance of the chromatin state (15, 32). Two classes of PCNA mutants, represented by pol30-8 and pol30-79, respectively, were found to reduce silencing by different mechanisms. Although the pol30-8 mutant prevents CAF-1 from contributing to silencing, pol30-79 disables Asf1p's ability to contribute to silencing (32, 40). We asked how a combination of pol30-8 and pol30-79 mutations would affect heterochromatin-mediated silencing. We made a PCNA mutant, designated pol30-879, containing the following mutations: R61A and D63A (pol30-8) and I126A and L128A (pol30-79). Wild-type POL30, pol30-879, pol30-8, or pol30-79 was integrated at the TRP1 locus as the only functional copy of PCNA in cells, and the effect of each mutant on telomeric silencing was examined by using a standard telomeric silencing assay (41, 42). For this assay, the URA3 gene was integrated at the end of the right arm of chromosome VII. Expression of the URA3 gene at the telomere is variegated: some cells in a population will express the gene, whereas others will not, and each state is semistable and heritable. Because of this mixed population, plating of wild-type cells will yield colony growth on medium lacking uracil or medium containing fluoroorotic acid (FOA, a drug that kills cells expressing URA3). Defects in silencing of the telomeric URA3 reporter gene can be detected as a reduction in growth on FOA. As shown in Fig. 1A, both pol30-8 and pol30-79 reduced telomeric silencing in agreement with ref. 32. Moreover, the pol30-879 mutant reduced telomeric silencing significantly more than either pol30-8 or pol30-79 did. The pol30-879 mutant also reduced silencing of the ADE2 gene at the HMR locus more than either pol30-8 or pol30-79 (data not shown). These results suggest that a combination of the pol30-8 and pol30-79 mutations strongly decreased telomeric silencing and HMR silencing.
Fig. 1.
The pol30-879 mutant reduced heterochromatin silencing. (A) the pol30-879 mutant reduced telomeric silencing. Ten-fold serial dilutions of yeast cells were spotted onto yeast extract/peptone/dextrose medium (complete) to assay cell growth and medium lacking uracil (-URA) or containing fluoroorotic acid (FOA) to assay silencing. (B) The pol30-879 mutant reduced HML silencing in the absence of Sir1p. Wild-type or various PCNA mutant strains with MATa as the mating type were mated with a MATα strain, and the mating efficiency of each strain was determined and plotted on a log scale.
To determine whether the pol30-879 mutant also reduced silencing at the HML locus, we used a quantitative mating assay. A complete loss of silencing at the HMLα locus in a MATa strain renders the strain sterile because of coexpressed α and a genes. As shown in Fig. 1B, the pol30-8, pol30-79, or pol30-879 mutant cells mated as efficiently as wild-type cells, suggesting that none of these mutants impaired HML silencing to a detectable degree by this relatively insensitive assay. Because it has been shown that a PCNA mutant allele (pol30-52) synergistically reduced silencing at the HML locus in the absence of Sir1p (34), we also tested whether the pol30-879 allele affected HML silencing in the absence of Sir1p. As shown in Fig. 1B, pol30-8 or pol30-79 strongly reduced silencing at the HML locus when combined with sir1Δ. Interestingly, the pol30-879 mutant reduced HML silencing to a degree similar to pol30-8 or pol30-79 in the absence of Sir1p even though pol30-879 reduced telomeric silencing more than pol30-8 or pol30-79 alone.
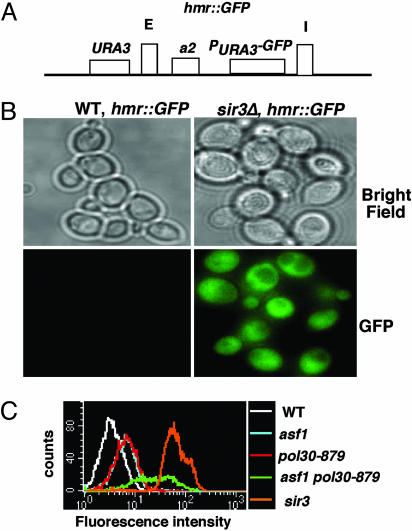
The pol30-879 Mutation Prevents CAF-1 from Contributing to Silencing. We also performed epistasis analysis of pol30-879, cac1Δ, and asf1Δ mutants by using a quantitative HMR silencing assay. In this assay (Fig. 2A), the GFP gene under the control of the URA3 gene promoter (PURA3-GFP) was integrated at the HMR locus and was silenced in wild-type cells (Fig. 2B and ref. 43). Moreover, silencing of the GFP gene depended on SIR3 because deleting the SIR3 gene resulted in GFP expression in all cells examined (Fig. 2B). Because cells expressing GFP could be differentiated and quantified by FACS from those that did not, FACS might be suitable for quantitatively determining silencing at the HMR locus. To test this idea, we first determined the percentage of cells expressing GFP in the sir3Δ mutant or wild type (Fig. 5, which is published as supporting information on the PNAS web site). As shown in Fig. 2C and Table 1, ≈99.2% of the cells expressed GFP in the sir3 mutant, but only ≈0.3% of cells showed fluorescence in wild-type cells. These results demonstrate that this assay can be used to quantify silencing at the HMR locus. Therefore, we determined the effect of pol30-879, cac1Δ, and asf1Δ alone or in combination on silencing of GFP at the HMR locus (Fig. 2C and Table 1; see also Table 4, which is published as supporting information on the PNAS web site). Surprisingly, the pol30-879 allele by itself had very little effect on HMR silencing. However, 80% of pol30-879 asf1Δ double-mutant cells expressed GFP, i.e., the double-mutant cells reduced silencing at the HMR locus much more than did either single mutant. The percentage of GFP-expressing cells in the pol30-879 cac1Δ double mutant, by contrast, was very close to the single mutant alone (<2%), suggesting that these PCNA and CAF-1 mutations affect silencing through the same mechanism. Together, these results suggest that pol30-879 prevents CAF-1 from contributing to HMR silencing, but it has little effect on Asf1p-dependent silencing. Supporting this idea, the pol30-879 cac1Δ asf1Δ triple mutant reduced HMR silencing to a similar degree as the cac1Δ asf1Δ double mutant (Table 1). It is not clear whether the pol30-879 mutant impacts telomeric silencing in a similar way as it does on HMR silencing because the pol30-879 mutant is very defective in telomeric silencing (Fig. 1), and, consequently, it is impossible to perform epistasis analysis on pol30-879, cac1Δ, and asf1Δ by using the available quantitative telomeric silencing assay described in Table 2. Nonetheless, these results indicate that the pol30-879 mutant displays a phenotype different from that of the cac1Δ asf1Δ mutant, at least at the HMR locus.
Fig. 2.
Epistasis analysis of the pol30-879, cac1Δ, and asf1Δ mutants on silencing at the HMR locus. (A) A schematic representation of the HMR locus with the integrated GFP gene expressed under the control of the URA3 gene promoter (PURA3-GFP). Two silencers, E and I, and the a2 gene at the silent HMR locus are indicated. This altered HMR locus is marked by the URA3 gene integrated outside the silent HMR locus. (B) The expression of GFP depended on the SIR3 gene. (Upper) Bright field images of wild-type or sir3 mutant cells, (Lower) Fluorescence images of wild-type or sir3 mutant cells. (C) FACS analysis of GFP expression in wild-type or various mutants with relevant genotype shown at the right.
Table 1. Expression of GFP in wild-type or various mutants was determined by FACS.
| Genotype | % GFP cells |
|---|---|
| WT | 0.3 |
| cac1 | 0.2 |
| asf1 | 0.9 |
| pol30-879 | 0.8 |
| cac1 pol30-879 | 1.3 |
| asf1 pol30-879 | 79.0 |
| cac1 asf1 pol30-879 | 80.1 |
| cac1 asf1 | 71.1 |
| sir3 | 99.2 |
The percentage of GFP-positive cells from each strain was determined as described in Experimental Procedures. Because of autofluorescence of yeast strains, the percentage of GFP positive cells at low range (<1%) in different mutants is not reliable for comparison (Table 4).
Table 2. rrt106Δ reduced telomeric silencing when combined with the cac1Δ mutant.
| Genotype | Telomeric silencing (FOA%) |
|---|---|
| WT | 0.43 (0.21-0.59) |
| cac1Δ | 0.015 (0.004-0.026) |
| rtt106Δ | 0.28 (0.19-0.4) |
| cac1Δ rtt106Δ | <1.0 × 10−6 |
Fractions of FOA-resistant cells of wild-type and mutant strains were determined. The average values of three independent experiments are shown, and the range of values is shown in parentheses.
Identification of Enhancers of the Silencing Defect of the pol30-879 Mutant. Because the pol30-879 mutant did not affect silencing of the GFP gene at the HMR locus to a significant degree (Fig. 2C and Table 1), we decided to isolate mutants that enhanced the silencing defect of this mutant to identify additional factors functioning with PCNA in silencing. In yeast, loss of heterochromatin silencing has no apparent effect on cell growth. We therefore used the collection of ≈4,700 viable yeast deletion mutants and tested whether any of these mutants enhanced the silencing defect of the pol30-879 allele. We followed a standard approach to obtain double mutant cells containing the pol30-879 mutant and each of the deletion mutants (Supporting Experimental Procedures; see also Fig. 6, which is published as supporting information on the PNAS web site) (44) and analyzed the effect of each double mutant on silencing of the GFP gene at the HMR locus. Nine mutants enhanced the silencing defect of the pol30-879 mutant (Table 5, which is published as supporting information on the PNAS web site).
Of these nine mutants, six (sas2, sas4, sas5, hir1, hir3, and sir1) were previously known to affect silencing (45). Moreover, these mutants were known to synergistically reduce silencing in combination with the cac1Δ mutant (23, 27, 46). Therefore, like sir1Δ, the sas and hir mutants were all predicted to be enhancers of the silencing defect of the pol30-879 mutant. The identification of these six genes in our screen confirmed the utility of our screening strategy.
The gene products of three mutants (nam7, msh2, and rtt106) were not previously shown to be involved in silencing. Nam7p is a putative RNA helicase involved in mRNA decay. A region of Nam7p is homologous to S. pombe Hrr1, a component of the RNA-directed RNA polymerase complex involved in RNA interference (RNAi) and heterochromatin silencing (47, 48). However, so far, no sequence homologs of proteins involved in RNAi in higher eukaryotes have been found in S. cerevisiae. Msh2p is best known for its role in DNA repair (49). Future studies are needed to determine how these two genes might affect silencing in yeast.
The rtt106 Deletion Synergistically Reduced Silencing with the cac1Δ. The rtt106 mutant was originally identified in a genetic screen as a mutant that enhanced the retrotransposition of Ty1 elements (50), but the function of Rtt106 was unknown. Mutations in two histone chaperones, CAF-1 and Hir1p, have been shown to affect Ty1 transposition (51). Moreover, the rtt106 mutant also synergistically reduced silencing at the HML locus in combination with the pol30-879 mutant (Table 5). Therefore, we decided to focus on Rtt106p. We deleted the RTT106 gene from our standard genetic background (W303) and tested whether the rtt106Δ mutant affected cell growth and telomeric silencing. The rtt106Δ mutant had no apparent defect in cell growth (data not shown and Fig. 7, which is published as supporting information on the PNAS web site). Moreover, rtt106Δ alone did not affect telomeric silencing to a significant degree compared with wild-type cells (Table 2). We then tested whether the rtt106Δ affected cell growth and silencing with the cac1Δ mutant. The cac1Δ rtt106Δ double mutant grew slower than either single mutant alone when grown from spores (Fig. 7). More importantly, the cac1Δ rtt106Δ double mutant reduced telomeric silencing more than either single mutant alone (Table 2) as revealed by the quantitative telomeric silencing assay. Finally, we tested whether rtt106Δ enhanced the HMR silencing defect in the cac1Δ mutant by using the hmr::GFP silencing assay. As shown in Table 3, the cac1Δ rtt106Δ double mutant significantly reduced HMR silencing compared with the wild type or either single mutant alone. In contrast, combing rtt106Δ with asf1Δ or hir1Δ did not impair silencing beyond the defects seen in each of the single mutants. These genetic data are exactly what would be predicted because pol30-879 synergistically enhanced both the asf1Δ (Fig. 2C and Table 1) and the rtt106Δ (Table 4) silencing defects but not the cac1Δ defect (Table 1). These data indicate that Rtt106p functions primarily in the Asf1p-dependent silencing pathway, which is genetically distinguishable from Cac1p-dependent silencing.
Table 3. rtt106Δ reduced silencing at the HMR locus when combined with the cac1Δ but not other histone chaperone mutations.
| Genotype | % GFP cells |
|---|---|
| WT | 0.2 |
| cac1 | 0.3 |
| asf1 | 0.2 |
| rtt106 | 1.5 |
| hir1 | 0.14 |
| cac1 asf1 | 60.0 |
| cac1 rtt106 | 59.4 |
| cac1 hir1 | 87 |
| hir1 rtt106 | 0.6 |
| asf1 rtt106 | 1.7 |
| asf1 hir1 | 0.2 |
Expression of GFP at the HMR locus in wild-type or various mutant cells with the indicated genotypes at the left was determined as described in Fig. 2. The results presented were from one experiment, and similar results were also obtained from three other independent experiments. We always observed that the cac1Δ asf1Δ mutant reduced silencing to a similar degree as the cac1Δ rtt106Δ mutant did. By contrast, the cac1Δ hir1Δ mutant reduced silencing more than either cac1Δ rtt106 or cac1Δ asf1Δ alone for all four independent experiments performed.
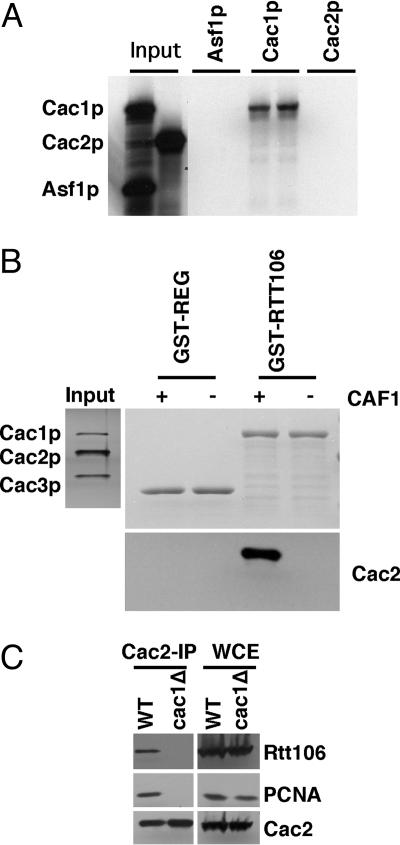
Rtt106p Interacts with CAF-1 Through Cac1p. The above genetic analysis suggests that Rtt106p may interact with CAF-1 or Asf1p physically. To test this idea, we first used a GST-pulldown assay to determine whether GST-Rtt106p bound to in vitro-translated Asf1p or two subunits of CAF-1, Cac1p and Cac2p. As shown in Fig. 3A, only Cac1p, but neither Cac2p nor Asf1p, bound to Rtt106p in vitro. To determine whether Rtt106p interacted with the CAF-1 complex in vitro, we incubated the CAF-1 complex purified from yeast cells with GST-Rtt106p or GST-REGα and detected proteins that bound to GST-Rtt106p or GST-REGα by using antibodies against Cac2p. As shown in Fig. 3B, GST-Rtt106p, but not GST-REGα, pulled down Cac2p in the presence of Cac1p. The data from Fig. 3 A and B suggest that Rtt106p interacts with CAF-1 complex through Cac1p. To test this idea in vivo, we expressed Flag-tagged Rtt106p under the control of its own promoter from a centromere containing plasmid pRS416 in wild-type or cac1Δ mutant cells. We then immunoprecipitated Cac2-TAP from these two strains and determined whether Rtt106p was coprecipitated with Cac2p. As shown in Fig. 3, PCNA, which is known to interact with CAF-1 through Cac1p (32), and Rtt106p were coprecipitated with Cac2p from wild-type, but not from cac1Δ mutant, cells. Thus, Rtt106p interacts with CAF-1 through the large subunit of CAF-1 in vitro and in vivo.
Fig. 3.
Rtt106p interacts with CAF-1 in vitro and in vivo.(A) Rtt106p interacts with Cac1p, but not Cac2p, in vitro. GST-Rtt106p was used to pull down in vitro translated, [35S]methionine-labeled Cac1p, Cac2p, and Asf1p, the bound proteins were resolved by SDS/PAGE, and detected by autoradiography. (B) GST-Rtt106p bound to the CAF-1 complex. GST-Rtt106p was used to pull down the purified CAF-1 complex (Left, silvering staining of the purified CAF-1 complex), and the bound proteins were resolved by SDS/PAGE, detected either by Coomassie staining (Right Upper) or by antibodies against Cac2p (Right Lower). (C) Rtt106p interacts with CAF-1 through Cac1p in vivo. Cac2-TAP was purified from wild-type or the cac1Δ mutant cells, and copurified proteins were detected by antibodies against PCNA, Rtt106p, and Cac2p.
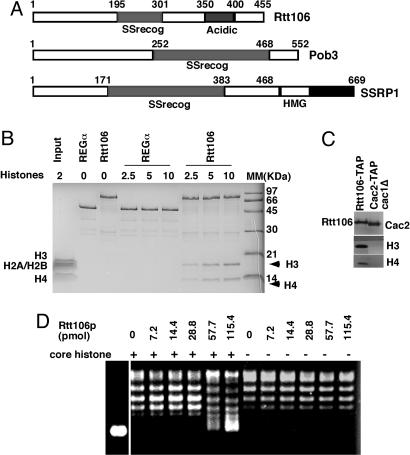
Rtt106p Is a Histone H3 and H4 Binding Protein. The domain structure of Rtt106p is shown in Fig. 4A. A region of the Rtt106p protein is homologous to the single-strand DNA recognition domain in human SSRP1 and yeast Pob3. SSRP1 and Pob3 are subunits of human and yeast FACT, respectively. FACT remodels chromatin to facilitate RNA polymerase II transcription and DNA replication (52-54). Although the FACT complex binds to histones H2A and H2B, SSRP1 interacts with histones H3 and H4 in vitro (52). Therefore, we tested whether Rtt106p could bind to any of the four core histones in vitro. GST-Rtt106p was used to pull down histone octamers purified from HeLa cells. As shown in Fig. 4B, Rtt106p bound to histones H3 and H4 but not to histone H2A and H2B in vitro. Under the same conditions, the negative control protein GST-REGα did not bind any of the histones. These results indicate that Rtt106p binds specifically to histones H3 and H4. To test whether Rtt106p binds to histones H3 and H4 in vivo, we tagged Rtt106p with the TAP tag and purified Rtt106p from yeast cells. Copurified proteins were detected by Western blotting by using antibodies against histones H3 or H4. As shown in Fig. 4C, a fraction of histones H3 and H4 copurified with Rtt106p. The TAP tag was not responsible for this binding because under the same conditions, histones did not copurify with Cac2-TAP in cac1Δ cells (This mutant was used because the association of histones with CAF-1 depends on Cac1p, the large subunit of CAF-1 (H.Z. and Z.Z., unpublished data). Thus, Rtt106p binds histones H3 and H4 in vitro and in vivo.
Fig. 4.
The Rtt106p is a histone H3 and H4 chaperone. (A) The structural features of Rtt106p are compared with those of Pob3 and SSRP1, a subunit of yeast and mammalian FACT, respectively. (B) Rtt106p binds to histones H3 and H4 in vitro. Similar amounts of GST-Rtt106p or GST-REGα were used to pull down different amounts of histone octamers (micrograms), and the bound proteins were eluted by using SDS sample buffer, resolved on a SDS/PAGE gel, and visualized by Coomassie staining. The apparent migration differences between histones H3 and H4 that bind to Rtt106 and those in the input sample were likely due to differences in salt concentration of the samples. (C) Rtt106p binds to H3 and H4 in vivo. The experiment was performed as described in Fig. 3C. Rtt106p has nucleosome assembly activity in vitro. Topoisomerase I relaxed plasmid DNA (0.1 pmol) was incubated with Rtt106p alone or with Rtt106p and histones (8 pmol). After digesting away proteins, DNA was resolved on an agarose gel and visualized by ethidium bromide staining.
Rtt106p Promotes Nucleosome Formation in a Reconstitution Assay. Rtt106p contains a 50-residue stretch rich in acidic amino acids (Fig. 4A); this feature is characteristic of many histone chaperones. Therefore, we tested whether Rtt106p promoted nucleosome formation by using a plasmid supercoiling assay. Wrapping DNA around histone cores introduces roughly one negative supercoil per core particle, which can be detected subsequently by electrophoresis of the purified DNA through an agarose gel. In our assays, negatively supercoiled plasmid DNA (pSV011) was first relaxed by topoisomerase I. Histone octamers and increasing amounts of Rtt106p were then incubated with the relaxed DNA to allow nucleosome assembly. After the reaction, DNA was purified, resolved on a 1% agarose gel, and visualized by ethidium bromide staining. As shown in Fig. 4D, supercoiled DNA reappeared with increasing amounts of Rtt106p and histones, and the formation of supercoiled DNA depended on the presence of both histones and Rtt106p. These results indicate that Rtt106p can promote nucleosome formation in a histone chaperone assay in vitro.
Discussion
We have identified nine enhancers of silencing defects of the pol30-879 mutant by screening ≈4,700 viable yeast deletion mutants. This PCNA mutation prevents CAF-1 from contributing to silencing but has little effect on Asf1p-dependent silencing. As predicted from this genetic epistasis, all of the isolated mutants characterized so far synergistically reduce silencing with the cac1Δ mutant. Experiments with the previously uncharacterized protein Rtt106p, one of the factors identified in our screen, reveal that it is a histone H3- and H4-binding protein and possesses nucleosome-assembly activity in vitro.
Boone and colleagues (44) initially devised the synthetic genetic array (SGA) method to study whether combining a deletion in each of the ≈4,700 nonessential yeast genes with a mutation of interest created a double mutant with altered cell growth. Using this method, we have identified 77 deletion alleles that potentially interact genetically with the pol30-879 mutation (Z.Z., unpublished data). In this report, we have taken the method a step further by screening viable double mutants, which were often discarded in SGA screens, for enhancers of the silencing defects of the PCNA mutant by devising the GFP fluorescence-based silencing assay. Our approach relied on the fact that a complete loss of yeast silencing is not detrimental to cell growth. Therefore, mutants enhancing the silencing defect of the pol30-879 mutant are unlikely to overlap completely with those that are synthetically lethal with the PCNA mutant. Indeed, we were able to identify six genes previously known to be involved in silencing by this approach. We anticipate that our approach can be adopted to identify enhancers of the silencing defects of other mutants such as asf1Δ, cdc45-1, and rfc-1. The mechanisms by which these mutants affect silencing remain largely unknown (34). Our results highlight the fact that functional readouts other than growth can be useful in an SGA-screen format and have several potential advantages over simple growth assays. First, synthetic genetic interactions that are not essential for cell growth or that have only weak effects on cell growth might be uncovered by a function-based assay. Second, compared with mutants isolated from SGA screens that measure loss of cell viability of double mutants (synthetic lethality), mutants isolated by a specific functional assay may be more likely to function in the specific cellular process of interest.
The utility of this approach is underscored by the identification of Rtt106p as a protein involved in silencing. We have provided several lines of evidence supporting the hypothesis that Rtt106p is a histone H3 and H4 chaperone. First, Rtt106p interacts with histones H3 and H4 in vitro and in vivo. Second, Rtt106p possesses nucleosome-assembly activity in vitro. Third, although the lack of Rtt106p has little effect on cell growth or silencing, the rtt106Δ cac1Δ double mutant cells show a synergistic slow growth phenotype and a synergistic loss of silencing. Because CAF-1 is a histone H3 and H4 chaperone functioning in S phase, these results suggest that Rtt106p may also function as a histone chaperone in S phase. Supporting this idea, the expression of RTT106, like that of CAF-1 or Asf1p, two histone chaperones known to function in S phase, peaks in S phase (30, 55).
Rtt106p shares several properties with Asf1p. First, both Asf1p and Rtt106p are histone H3 and H4 binding proteins. Second, Rtt106p functions in the same genetic pathway as Asf1p in silencing. The role of Asf1p in telomeric silencing depends on its interaction with Hir1p (56). Because Rtt106p does not bind Asf1p, at least in vitro (Fig. 3), and because Rtt106p functions in the same genetic pathway as Hir1p in silencing, it is possible that Rtt106p, like Asf1p, interacts with Hir1p to mediate silencing. Third, both Asf1p and Rtt106p interact with CAF-1. Interestingly, Asf1p interacts with CAF-1 through the second subunit of CAF-1 (Cac2p) (33), whereas Rtt106p interacts with CAF-1 through the large subunit of CAF-1 (Cac1p). Asf1p from yeast to human cells, although unable to promote nucleosome formation preferentially onto replicated DNA by itself, enhances the ability of CAF-1 to promote nucleosome formation onto replicated DNA (31, 40, 57). In the future, it would be interesting to determine how Rtt106p functions in the DNA replication coupled nucleosome assembly assay in the presence or absence of CAF-1 and/or Asf1p.
Despite their shared properties, Asf1p and Rtt106p appear to have distinct functions in cells. Cells lacking Asf1p exhibit a slow growth phenotype (30), yet deleting the RTT106 gene has no apparent effect on cell growth (Fig. 7). The underlying mechanisms by which Rtt106p and Asf1p function distinctly in cells are under investigation. Asf1p is known to interact with a variety of proteins. For instance, Asf1p interacts with the checkpoint kinase Rad53p, and this interaction regulates the association of Asf1p with histones H3 and H4 in yeast cells (58, 59). Moreover, Asf1p interacts with RFC, a clamp loader (60), and is required to maintain replication fork stability (61). Furthermore, Asf1p interacts with the SAS histone acetyltransferase (62). Thus, it would be interesting to determine whether Rtt106p and Asf1p interact with different proteins to perform their distinct functions.
In human cells, there are two Asf1 sequence homologs, Asf1a and Asf1b. Both Asf1a and Asf1b stimulate CAF-1 to deposit histones H3 and H4 for nucleosome formation in vitro (57). Moreover, both Asf1a and Asf1b are in the CAF-1 as well as HIRA-containing complexes (10). HIRA, the yeast homolog of Hir1p, functions in the replication-independent nucleosome assembly processes (10, 63). Despite their shared properties, Asf1a and Asf1b appear to have distinct functions in cells. For instance, Asf1a, but not Asf1b, is required for formation of senescence-associated heterochromatin foci (64). No yeast sequence homolog of Asf1p has been reported. It is possible that Rtt106p is the functional homolog of one of the mammalian Asf1 proteins. Supporting this idea, we cannot find sequence homologs of Rtt106p from higher eukaryotes even though the sequence homolog of Rtt106p can be identified in S. pombe (data not shown). Alternatively, Rtt106p may be a unique histone chaperone in yeast cells. Future studies on the mechanisms of Rtt106p in silencing and nucleosome assembly should help to determine how Rtt106p functions with CAF-1 and Asf1p in silencing and nucleosome assembly.
Supplementary Material
Acknowledgments
We thank Dr. Boone (University of Toronto, Toronto) for plasmids and protocols of SGA screen; Sheng Xuetong (University of Texas M. D. Anderson Cancer Center, Houston) for the FLAG-containing plasmid; Alain Verreault (Cancer Research UK, Hertfordshire, U.K.) for antibodies against H3 and H4; Kristi Simmons and Drs. Rui-Ming Xu and Martin Rechsteiner for critical reading of the manuscript; and Drs. Mark McNivan and Franklyn G. Prendergast for using fluorescence microscopes and ArrayScan in their laboratories. M.H. is supported by National Institutes of Health Grant GM46904.
Abbreviations: CAF-1, chromatin assembly factor 1; SGA, synthetic genetic array; TAP, tandem affinity purification.
References
- 1.Pillus, L. & Rine, J. (1989) Cell 59, 637-647. [DOI] [PubMed] [Google Scholar]
- 2.Grewal, S. I. & Klar, A. J. (1996) Cell 86, 95-101. [DOI] [PubMed] [Google Scholar]
- 3.Grewal, S. I. & Elgin, S. C. (2002) Curr. Opin. Genet. Dev. 12, 178-187. [DOI] [PubMed] [Google Scholar]
- 4.Grunstein, M. (1998) Cell 93, 325-328. [DOI] [PubMed] [Google Scholar]
- 5.Rusche, L. N., Kirchmaier, A. L. & Rine, J. (2003) Annu. Rev. Biochem. 72, 481-516. [DOI] [PubMed] [Google Scholar]
- 6.Gartenberg, M. R. (2000) Curr. Opin. Microbiol. 3, 132-137. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad, K. & Henikoff, S. (2002) Mol. Cell 9, 1191-1200. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad, K. & Henikoff, S. (2002) Cell 111, 281-284. [DOI] [PubMed] [Google Scholar]
- 9.Jackson, V. & Chalkley, R. (1981) Cell 23, 121-134. [DOI] [PubMed] [Google Scholar]
- 10.Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. (2004) Cell 116, 51-61. [DOI] [PubMed] [Google Scholar]
- 11.Verreault, A. (2000) Genes Dev. 14, 1430-1438. [PubMed] [Google Scholar]
- 12.Smith, S. & Stillman, B. (1989) Cell 58, 15-25. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman, P. D., Kobayashi, R., Kessler, N. & Stillman, B. (1995) Cell 81, 1105-1114. [DOI] [PubMed] [Google Scholar]
- 14.Gaillard, P. H., Martini, E. M., Kaufman, P. D., Stillman, B., Moustacchi, E. & Almouzni, G. (1996) Cell 86, 887-896. [DOI] [PubMed] [Google Scholar]
- 15.Shibahara, K. & Stillman, B. (1999) Cell 96, 575-585. [DOI] [PubMed] [Google Scholar]
- 16.Murzina, N., Verreault, A., Laue, E. & Stillman, B. (1999) Mol. Cell 4, 529-540. [DOI] [PubMed] [Google Scholar]
- 17.Ye, X., Franco, A. A., Santos, H., Nelson, D. M., Kaufman, P. D. & Adams, P. D. (2003) Mol. Cell 11, 341-351. [DOI] [PubMed] [Google Scholar]
- 18.Hoek, M. & Stillman, B. (2003) Proc. Natl. Acad. Sci. USA 100, 12183-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tchenio, T., Casella, J. F. & Heidmann, T. (2001) Mol. Cell. Biol. 21, 1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nabatiyan, A. & Krude, T. (2004) Mol. Cell. Biol. 24, 2853-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman, P. D., Kobayashi, R. & Stillman, B. (1997) Genes Dev. 11, 345-357. [DOI] [PubMed] [Google Scholar]
- 22.Enomoto, S., McCune-Zierath, P. D., Gerami-Nejad, M., Sanders, M. A. & Berman, J. (1997) Genes Dev. 11, 358-370. [DOI] [PubMed] [Google Scholar]
- 23.Enomoto, S. & Berman, J. (1998) Genes Dev. 12, 219-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, J. S., Caputo, E. & Boeke, J. D. (1999) Mol. Cell. Biol. 19, 3184-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monson, E. K., de Bruin, D. & Zakian, V. A. (1997) Proc. Natl. Acad. Sci. USA 94, 13081-13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutton, A., Bucaria, J., Osley, M. A. & Sternglanz, R. (2001) Genetics 158, 587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman, P. D., Cohen, J. L. & Osley, M. A. (1998) Mol. Cell. Biol. 18, 4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ai, X. & Parthun, M. R. (2004) Mol. Cell 14, 195-205. [DOI] [PubMed] [Google Scholar]
- 29.Poveda, A., Pamblanco, M., Tafrov, S., Tordera, V., Sternglanz, R. & Sendra, R. (2004) J. Biol. Chem. 279, 16033-16043. [DOI] [PubMed] [Google Scholar]
- 30.Le, S., Davis, C., Konopka, J. B. & Sternglanz, R. (1997) Yeast 13, 1029-1042. [DOI] [PubMed] [Google Scholar]
- 31.Tyler, J. K., Adams, C. R., Chen, S. R., Kobayashi, R., Kamakaka, R. T. & Kadonaga, J. T. (1999) Nature 402, 555-560. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Z., Shibahara, K. & Stillman, B. (2000) Nature 408, 221-225. [DOI] [PubMed] [Google Scholar]
- 33.Krawitz, D. C., Kama, T. & Kaufman, P. D. (2002) Mol. Cell. Biol. 22, 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrenhofer-Murray, A. E., Kamakaka, R. T. & Rine, J. (1999) Genetics 153, 1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas, B. J. & Rothstein, R. (1989) Genetics 123, 725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrenhofer-Murray, A. E., Rivier, D. H. & Rine, J. (1997) Genetics 145, 923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, C. P., Slaughter, C. A. & DeMartino, G. N. (1992) J. Biol. Chem. 267, 10515-10523. [PubMed] [Google Scholar]
- 38.Dubiel, W., Pratt, G., Ferrell, K. & Rechsteiner, M. (1992) J. Biol. Chem. 267, 22369-22377. [PubMed] [Google Scholar]
- 39.Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M. & Seraphin, B. (2001) Methods 24, 218-229. [DOI] [PubMed] [Google Scholar]
- 40.Sharp, J. A., Fouts, E. T., Krawitz, D. C. & Kaufman, P. D. (2001) Curr. Biol. 11, 463-473. [DOI] [PubMed] [Google Scholar]
- 41.Gottschling, D. E., Aparicio, O. M., Billington, B. L. & Zakian, V. A. (1990) Cell 63, 751-762. [DOI] [PubMed] [Google Scholar]
- 42.Aparicio, O. M., Billington, B. L. & Gottschling, D. E. (1991) Cell 66, 1279-1287. [DOI] [PubMed] [Google Scholar]
- 43.Laney, J. D. & Hochstrasser, M. (2003) Genes Dev. 17, 2259-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- 45.Sutton, A., Shia, W.-J., Band, D., Kaufman, P. D., Osada, S., Workman, J. L. & Sternglanz, R. (2003) J. Biol. Chem. 278, 16887-16892. [DOI] [PubMed] [Google Scholar]
- 46.Meijsing, S. H. & Ehrenhofer-Murray, A. E. (2001) Genes Dev. 15, 3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motamedi, M. R., Verdel, A., Colmenares, S. U., Gerber, S. A., Gygi, S. P. & Moazed, D. (2004) Cell 119, 789-802. [DOI] [PubMed] [Google Scholar]
- 48.Noma, K., Sugiyama, T., Cam, H., Verdel, A., Zofall, M., Jia, S., Moazed, D. & Grewal, S. I. (2004) Nat. Genet. 36, 1174-1180. [DOI] [PubMed] [Google Scholar]
- 49.Kolodner, R. D. & Marsischky, G. T. (1999) Curr. Opin. Genet. Dev. 9, 89-96. [DOI] [PubMed] [Google Scholar]
- 50.Scholes, D. T., Banerjee, M., Bowen, B. & Curcio, M. J. (2001) Genetics 159, 1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian, Z., Huang, H., Hong, J. Y., Burck, C. L., Johnston, S. D., Berman, J., Carol, A. & Liebman, S. W. (1998) Mol. Cell. Biol. 18, 4783-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belotserkovskaya, R., Oh, S., Bondarenko, V. A., Orphanides, G., Studitsky, V. M. & Reinberg, D. (2003) Science 301, 1090-1093. [DOI] [PubMed] [Google Scholar]
- 53.Belotserkovskaya, R. & Reinberg, D. (2004) Curr. Opin. Genet. Dev. 14, 139-146. [DOI] [PubMed] [Google Scholar]
- 54.Schlesinger, M. B. & Formosa, T. (2000) Genetics 155, 1593-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daganzo, S. M., Erzberger, J. P., Lam, W. M., Skordalakes, E., Zhang, R., Franco, A. A., Brill, S. J., Adams, P. D., Berger, J. M. & Kaufman, P. D. (2003) Curr. Biol. 13, 2148-2158. [DOI] [PubMed] [Google Scholar]
- 57.Mello, J. A., Sillje, H. H., Roche, D. M., Kirschner, D. B., Nigg, E. A. & Almouzni, G. (2002) EMBO Rep. 3, 329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emili, A., Schieltz, D. M., Yates, J. R., 3rd, & Hartwell, L. H. (2001) Mol. Cell 7, 13-20. [DOI] [PubMed] [Google Scholar]
- 59.Hu, F., Alcasabas, A. A. & Elledge, S. J. (2001) Genes Dev. 15, 1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellison, V. & Stillman, B. (2001) Cell 106, 655-660. [DOI] [PubMed] [Google Scholar]
- 61.Franco, A. A., Lam, W. M., Burgers, P. M. & Kaufman, P. D. (2005) Genes Dev. 19, 1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osada, S., Sutton, A., Muster, N., Brown, C. E., Yates, J. R., 3rd, Sternglanz, R. & Workman, J. L. (2001) Genes Dev. 15, 3155-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ray-Gallet, D., Quivy, J. P., Scamps, C., Martini, E. M., Lipinski, M. & Almouzni, G. (2002) Mol. Cell 9, 1091-1100. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, R., Poustovoitov, M. V., Ye, X., Santos, H. A., Chen, W., Daganzo, S. M., Erzberger, J. P., Serebriiskii, I. G., Canutescu, A. A., Dunbrack, R. L., et al. (2005) Dev. Cell 8, 19-30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.