Abstract
Vitamin B6 is an essential metabolite in all organisms. It can act as a coenzyme for numerous metabolic enzymes and has recently been shown to be a potent antioxidant. Plants and microorganisms have a de novo biosynthetic pathway for vitamin B6, but animals must obtain it from dietary sources. In Escherichia coli, it is known that the vitamin is derived from deoxyxylulose 5-phosphate (an intermediate in the nonmevalonate pathway of isoprenoid biosynthesis) and 4-phosphohydroxy-l-threonine. It has been assumed that vitamin B6 is synthesized in the same way in plants, but this hypothesis has never been experimentally proven. Here, we show that, in plants, synthesis of the vitamin takes an entirely different route, which does not involve deoxyxylulose 5-phosphate but instead utilizes intermediates from the pentose phosphate pathway, i.e., ribose 5-phosphate or ribulose 5-phosphate, and from glycolysis, i.e., dihydroxyacetone phosphate or glyceraldehyde 3-phosphate. The revelation is based on the recent discovery that, in bacteria and fungi, a novel pathway is in place that involves two genes (PDX1 and PDX2), neither of which is homologous to any of those involved in the previously doctrined E. coli pathway. We demonstrate that Arabidopsis thaliana has two functional homologs of PDX1 and a single homolog of PDX2. Furthermore, and contrary to what was inferred previously, we show that the pathway appears to be cytosolic and is not localized to the plastid. Last, we report that the single PDX2 homolog is essential for plant viability.
Keywords: Arabidopsis, isoprenoid, pyridoxine, deoxyxylulose 5-phosphate
Vitamin B6 is well renowned in the medical field for being involved in more bodily functions than any other single nutrient. The vitamin exists in various forms, i.e., pyridoxal, pyridoxine, pyridoxamine, and their phosphorylated derivatives. As pyridoxal 5′-phosphate, it is an essential cofactor for numerous metabolic enzymes including amino acid metabolism and antibiotic biosynthesis. Most interestingly, it has recently been found that the vitamin is a potent antioxidant with a particular ability to quench reactive oxygen species such as superoxide and singlet oxygen (1, 2). The de novo biosynthesis of vitamin B6 takes place in microorganisms and plants, but this ability has been lost in animals, making it essential in the human diet.
Despite the vitamin's obvious physiological and pharmaceutical importance, its biosynthesis has predominantly been studied in the Gram-negative bacterium, Escherichia coli, where it is derived from deoxyxylulose 5-phosphate (DXP, an intermediate in the nonmevalonate pathway of isoprenoid biosynthesis) and 4-phosphohydroxy-l-threonine (refs. 3-7 and Scheme 1). Astonishingly, although plants are a major source of vitamin B6 in the human diet, our understanding of the pathway therein is very limited and has been assumed to be derived the same way as in E. coli. There is one preliminary study that reports on its biosynthesis in spinach (8), whereas other, more in-depth studies have addressed the topic based on the formation of 4′-O-methylpyridoxine, a vitamin B6 derivative found in Ginkgo biloba (9, 10). Knowledge of the pathway in plants may not only aid in studies designed for overproduction of the vitamin for beneficial effects, but proteins involved could also provide novel herbicidal targets if proven to be essential for viability of the organism.
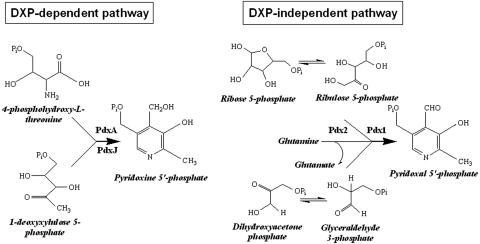
Scheme 1.
Model depicting the DXP-dependent and -independent pathways of vitamin B6 biosynthesis.
Recently, it has become clear from genetic and labeling studies in fungi (2, 11-14) that a pathway alternative to the one in E. coli that does not involve DXP exists for vitamin B6 biosynthesis. Only two genes (PDX1 and PDX2), which show no homology to any of the E. coli genes, appear to be involved in this alternative pathway (2, 15). Furthermore, based on extensive genomic analyses, it has been shown that the pathways of vitamin B6 biosynthesis are autoexclusive in that organisms have the genes for one or the other pathway but not both (2, 15). PDX1 and PDX2 have been predicted to function as a glutamine amidotransferase with PDX2 as the glutaminase domain and PDX1 as the acceptor/synthase domain. Indeed, glutaminase activity has been demonstrated for PDX2 from a number of organisms (16-18). More recently, Burns et al. (19) and our own independent studies (51) have been able to reconstitute vitamin B6 formation from intermediates of glycolysis and the pentose phosphate pathway by using the PDX1 and PDX2 homologs from the Gram-positive bacterium Bacillus subtilis.
Here, we address how vitamin B6 is synthesized in plants by using the model organism Arabidopsis thaliana. We report on the identification of homologs of PDX1 and PDX2 and demonstrate that, contrary to what is often quoted in the plant literature (e.g., refs. 20-22), vitamin B6 biosynthesis does not depend on the isoprenoid precursor DXP. Moreover, we demonstrate biosynthesis of the vitamin in a reconstituted system from A. thaliana. Last, we show that disruption of the single PDX2 homolog is lethal for the plant resulting in arrested embryo development.
Materials and Methods
Plant Material. When grown in Petri dishes, A. thaliana seeds were surface sterilized and grown on Murashige and Skoog (MS) medium (23) (containing 1% wt/vol sucrose), either with or without vitamin B6 depending on the experiment, and under constant illumination at 22°C. WT (Col-2 or Ws), heterozygous SALK_072168, heterozygous RIKEN GSC 53-2381-1, and heterozygous cla1-1 plants were maintained on soil under 16-h light/8-h dark cycles and genotyped by PCR. Homozygous cla1-1 plants were obtained by allowing seeds from heterozygous plants to germinate in Petri dishes and selecting albino plants for analysis.
Amplification of the Genes. In the case of PDX1.1-1.3, the absence of introns allowed the direct amplification from genomic DNA isolated from A. thaliana ecotype Columbia, whereas PDX2 was amplified from the isolated cDNA.
Yeast Complementation. The yeast strains used in this study were Saccharomyces cerevisiae BQS1037 and MML21, which are knockouts of ScSNZ1 and ScSNO1, respectively, and were kindly provided by Enrique Herrero (Universitat de Valencia, Burjassot, Spain; ref. 24). For the complementation studies, the AtPDX1.1-1.3 genes were cloned into the PstI and NotI sites of pCM262 (24), whereas AtPDX2 was cloned into the NotI site of the same plasmid. Complementation was carried out on synthetic complete medium without vitamin B6 and with the appropriate selection markers. All plasmid constructs were maintained episomally.
Transient Expression and Subcellular Localization. Transient expression of GFP fusion proteins in onion epidermal cells was done according to the method of Scott et al. (25). Expression constructs were delivered into the cells by gold particle-mediated gun bombardment. Analysis of the expression in Arabidopsis protoplasts was performed according to ref. 26 by using 3- to 4-week-old plants maintained on MS plates but with some modifications: Incubation in enzyme solution was performed overnight, in the dark, and without agitation. Cells were allowed to recover 24-48 h before analysis by confocal laser scanning microscopy (Leica Microsystems, Wetzlar, Germany) by using an ArKr laser at 488 nm. GFP fluorescence was recorded between 503 and 550 nm.
Analysis of the Dependence of Vitamin B6 Biosynthesis in Plants on DXP. Fosmidomycin (Molecular Probes) and clomazone (Riedel-de Haën, Seelze, Germany) inhibitor treatments were performed by growing seedlings for 7 days after imbibition on MS medium and then transferring them either to fresh MS medium (controls) or to MS medium containing the respective inhibitor. Vitamin B6 was always omitted from the MS medium. Seedling samples were harvested 96 h after inhibitor addition, frozen in liquid nitrogen, and stored at -80°C until further analysis. The amount of vitamin B6 in these samples and in wt and cla1-1 plants was determined by using the microbiological assay as described in ref. 27, employing S. carlsbergensis American Type Culture Collection 9080. The vitamin was extracted from whole plant material (2 mg) by using 0.02 M H2SO4 as described in ref. 28 with the following modifications: After extraction the solution was neutralized to pH 5.2 and centrifuged, and the supernatant was analyzed. Tissue from at least three independent experiments was used.
Recombinant Expression, Purification, and Biochemical Characterization of A. thaliana PDX1.1-1.3. The PDX genes were cloned into the NdeI/XhoI sites of pET21a (Novagen) in such a way as to allow expression of the proteins with a C-terminal hexa-histidine affinity tag in E. coli BL21(DE3) cells. Expression was induced by addition of 0.1 mM isopropyl-1-β-thio-d-galactopyranoside, followed by growth for 5 h at 30°C. The proteins were purified by Ni-NTA chromatography (Qiagen) by using the nondenaturing protocol described by the manufacturer. The proteins were judged to be >90% homogeneous from an SDS/PAGE analysis. Enzyme assays were carried out in 50 mM Tris·HCl, pH 8, at 37°C containing 40 μM of the isolated protein and 500 μM of the respective substrates, ribose 5-phosphate or ribulose 5-phosphate, and either dihydroxyacetone phosphate or dl-glyceraldehyde 3-phosphate (1 mM) in the presence of 10 mM ammonium sulfate. The progress of the reaction was monitored from the appearance of the absorbance maximum at 414 nm. For the HPLC analysis (System Gold, Beckman), the reaction product was deproteinized by ultrafiltration, followed by precipitation with methanol-chloroform and applied to a C18 reversed phase column (LiChroCART 250-4, RP-18, 5 mM; Merck) in 0.1% trifluoroacetic acid. An isocratic elution, followed by a gradient of 0-50% acetonitrile, was used in all experiments. For the electron impact-MS, the reaction product was treated with alkaline phosphatase, deproteinized, purified by HPLC, and lyophilized. The spectrum was acquired on a Micromass Autospec Ultima instrument by using a source temperature of 200°C and with electrons accelerated at 70 eV.
Analysis of T-DNA Insertion Knockouts of AtPDX2. The pdx2.1 mutant line was identified from the Salk Institute Genomic Analysis Laboratory T-DNA insertion lines (http://signal.salk.edu), and pdx2.2 was identified from the available transposon tagged lines at the RIKEN BioResource Center (http://www.brc.riken.jp/lab/epd/Eng/catalog/seed.shtml). For the histological analysis, siliques were fixed with ethanol and acetic acid (3:1 vol/vol) overnight, followed by rehydration with ethanol and water (3:1 vol/vol). The defect was observed by clearing the siliques in chloral hydrate, glycerol, and water (10:1:2.5, wt/wt/wt) overnight and visualized with a compound microscope equipped with Nomarski optics.
Supporting Information. Additional experimental procedures used in this study are provided in more detail in Supporting Text, which is published as supporting information on the PNAS web site.
Results and Discussion
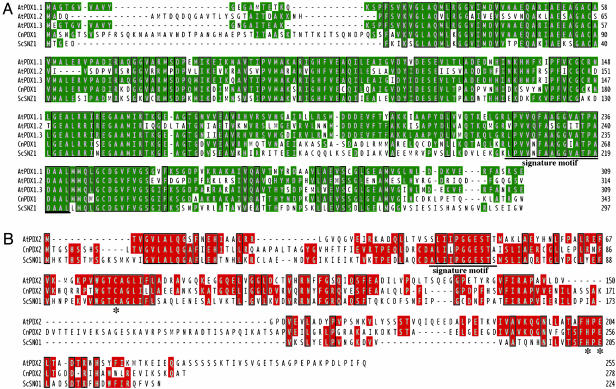
Identification and Functional Characterization of the Arabidopsis PDX1 and PDX2 Genes. Using blast with the C. nicotianae PDX1 and PDX2 sequences as queries (2, 11), three genes encoding putative PDX1s (At2g38230, At3g16050, and At5g01410) and a single gene encoding a putative PDX2 (At5g60540) were identified in the completely sequenced genome of Arabidopsis. For ease of reference, we propose to name the PDX1 homologs AtPDX1.1, AtPDX1.2 and AtPDX1.3, respectively, whereas the homolog of PDX2 will be referred to as AtPDX2. An alignment of the hypothetical A. thaliana PDX1 protein sequences (AtPDX1.1 shows 58% and 87% identity to AtPDX1.2 and AtPDX1.3, respectively) together with those from the fungi C. nicotianae (69%, 44%, and 68% identity with AtPDX1.1-1.3, respectively) and S. cerevisiae (58%, 41%, and 58% identity with AtPDX1.1-1.3, respectively) shows an extremely high degree of conserved sequence throughout the entire length of the genes (Fig. 1A). Whereas AtPDX1.2 is not as strongly conserved as those of the other A. thaliana homologs, it also does not have the complete UPF0019 sequence LPVVNFAAGGVATPADAAL, a characteristic of this protein family (www.ebi.ac.uk). An amino acid sequence alignment of PDX2 also shows a high degree of sequence identity between the plant and fungal homologs (35% and 25% identity between Cercospora nicotianae and S. cerevisiae, respectively) with almost the entire signature sequence of this protein family (UPF0030, [GA]LI[LIV]PGGEST[STA]) being retained (Fig. 1B). Importantly, the characteristic catalytic triad of class I glutamine amidotransferases, which consists of a cysteine, histidine, and glutamate residue in the glutaminase domain, is conserved in PDX2 (Fig. 1B and refs. 29 and 30).
Fig. 1.
The PDX1 and PDX2 genes in Arabidopsis. Shown is the amino acid sequence alignment of the PDX1 (A) and PDX2 (B) homologs identified from Arabidopsis. Amino acids identical in at least two of the sequences are shaded in green (PDX1) and red (PDX2). The signature motifs of the respective protein class are underlined. The asterisk denotes the conservation of the Cys-His-Glu catalytic triad in PDX2. At, A. thaliana; Cn, C. nicotianae; Sc, S. cerevisiae.
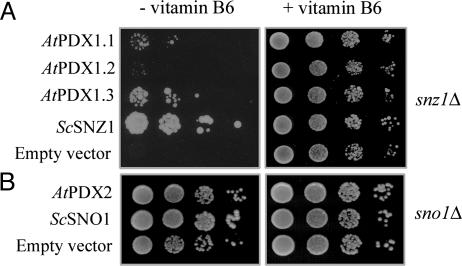
Expressed sequence tags can be found for all of these genes in a variety of tissues (http://mips.gsf.de). Moreover, total RNA isolation and real-time RT-PCR by using specific primers confirmed that all genes are expressed (Fig. 6, which is published as supporting information on the PNAS web site). To determine whether AtPDX1.1-1.3 and AtPDX2 encode functional enzymes, the genes were cloned into pCM262, a yeast expression vector under the control of a tetracycline regulatable promoter system (24, 31), and were tested for their ability to complement the SNZ1 (functional PDX1 homolog) and SNO1 (functional PDX2 homolog) disruption mutants, BQS1037 and MML21, respectively, from S. cerevisiae (24). When grown in minimal medium in the absence of vitamin B6, these mutants have a growth defect that is most pronounced in BQS1037 (24). Complementation of BQS1037 and MML21 with the S. cerevisiae SNZ1 and SNO1 genes, respectively, served as positive controls. In the absence of vitamin B6, the snz1 mutant transformed with either Arabidopsis PDX1.1 or PDX1.3 grew significantly faster than the strain transformed with the empty vector (Fig. 2A), suggesting that these plant homologs can restore vitamin B6 prototrophy in yeast. On the other hand, AtPDX1.2 did not restore prototrophy (Fig. 2A). The sno1 mutant transformed with AtPDX2 showed enhanced growth in comparison to that of cells carrying the empty vector, indicating functional homology of the two genes (Fig. 2B; growth in liquid culture is presented as Fig. 7, which is published as supporting information on the PNAS web site). When pyridoxine was included in the medium, all strains grew equally well (Fig. 2).
Fig. 2.
The three A. thaliana PDX1 homologs identified and the single PDX2 homolog were tested for their ability to restore prototrophy to the S. cerevisiae strains BQS1037 and MML21. These strains are deletion knockouts in SNZ1 and SNO1, which are functional PDX1 and PDX2 homologs in S. cerevisiae, respectively. (Left) Growth of 6-day-old BQS1037 (A) and 4-day-old MML21 (B) in the absence of vitamin B6. (Right) Growth of 3-day-old BQS1037 (A) and MML21 (B) in the absence of vitamin B6. Each row represents a serial 10-fold dilution from a starter culture with an OD at 600 nm of 0.5.
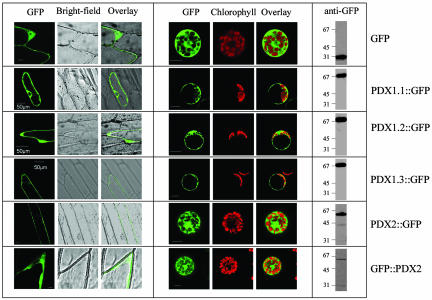
Subcellular Localization of AtPDX1.1-1.3 and AtPDX2. No clear targeting sequence could be identified in AtPDX1.1-1.3 or AtPDX2 from an in silico analysis. We therefore investigated the subcellular location of each of the proteins in vivo by fusing each one to the N terminus of GFP employing the vector pCAMBIA1302 (ref. 32; www.cambia.org) such that the fusions were under control of the CaMV35S promoter. We noted an extension at the C terminus of AtPDX2 when compared with homologous sequences of nonplant origin and, therefore, also investigated a C-terminal GFP fusion construct by using the vector pSH11 (33). Each construct was introduced into onion epidermal cells and protoplasts of Arabidopsis mesophyll cells by particle gun bombardment and polyethylene glycol-mediated chemical transformation, respectively. The subcellular location of the transiently expressed GFP fusion proteins was viewed by confocal microscopy. As expected in the case of GFP alone, a diffuse fluorescence pattern typical of a cytosolic location was found in both the onion epidermal cells and Arabidopsis protoplasts with a tendency to accumulate in the nucleoplasm (Fig. 3). All three PDX1-GFP fusions and both GFP-PDX2 and PDX2-GFP exhibited a similar localization pattern (Fig. 3), implying that they are cytosolic. It is clear from the Arabidopsis protoplast green fluorescence that there is no overlap with the chlorophyll autofluorescence nor did we observe a punctuate pattern typical of mitochondrial or microbody targeting in either the nonphotosynthetic (onion) or photosynthetic (Arabidopsis) tissue types. Western blot analyses of the transfected protoplasts by using a GFP antibody revealed that all of the fusion proteins remained intact (Fig. 3).
Fig. 3.
Subcellular localization of AtPDX1.1-1.3 and AtPDX2. (Left) Transient expression of the indicated GFP fusion proteins in onion epidermal cells. (Center) The transient expression observed in isolated A. thaliana protoplasts. (Scale bars: 10 μm, unless indicated otherwise.) (Right) Western blot of crude extracts is shown from isolated A. thaliana protoplasts expressing the indicated constructs after immunodecoration with anti-GFP, indicating that the fusion proteins were intact.
Vitamin B6 Biosynthesis in Plants Occurs Independent of Deoxyxylulose 5-Phosphate. It is generally assumed in the literature that deoxyxylulose 5-phosphate and 4-phosphohydroxy-l-threonine are the precursors for the formation of vitamin B6 in plants (20-22). Although it is well established that DXP is a precursor for the biosynthesis of isoprenoids through the nonmevalonate (MEP) pathway in prokaryotes (34, 35) and plants (36-40), its role in vitamin B6 biosynthesis has merely been inferred from extrapolation of the pathway as it occurs in E. coli (4, 5). There is only a single, very preliminary report (8) in the literature that claims that DX was used to form vitamin B6 in spinach chloroplasts, but no proof of the nature of the reaction product was provided. Using a microbiological assay, we measured the vitamin B6 content in the Arabidopsis cla-1 mutant. CLA-1 encodes deoxyxylulose 5-phosphate synthase (DXS) (41, 42). Although the vitamin content of the cla-1 mutant plants is somewhat reduced compared with that of wild-type plants (Table 1), it is clear that vitamin B6 biosynthesis can still occur in the absence of this protein. In addition, the percentage of cla-1 seeds that germinate in the absence of exogenous vitamin B6 is almost identical to that of the wild type (data not shown). However, two additional genes, closely related to CLA-1, exist in Arabidopsis (designated DXS2 and DXS3), which may encode DXP synthase but have not yet been functionally analyzed. Thus, the plants were treated with clomazone (Pestanal), an agent that has been shown to specifically inhibit DXP synthase (43). A slight decrease comparable with that in the cla-1 mutant plants was observed in the vitamin B6 content (Table 1). The observed decrease in vitamin B6 could indeed indicate that there are two routes functioning in its biosynthesis in plants (i.e., DXP dependent and DXP independent). However, an analysis of the available genomes has shown that there are no genes homologous to either PdxA or PdxJ, which catalyze formation of the pyridoxine ring in E. coli from DXP and 4-phosphohydroxy-l-threonine, in plants (and indeed most organisms with the exception of the γ-division of proteobacteria; refs. 2 and 15), arguing against the utilization of DXP in vitamin B6 biosynthesis. This conclusion is corroborated by the observation of E. Leistner's group, referred to in ref. 9, that isotopically labeled DX or DXP was not incorporated into the vitamin B6 derivative, 4′-O-methylpyridoxine, in G. biloba. An explanation for the observed decrease in vitamin B6 can thus be that inhibition of DXP synthase depletes tissue of a metabolite that may positively regulate a key protein participating in the DXP-independent route. On the other hand, blocking the second enzyme in the nonmevalonate isoprenoid pathway, i.e., DXP reductoisomerase, would be expected to result in a buildup of DXP and, hence, vitamin B6. When wild-type plants were treated with fosmidomycin, which specifically inhibits DXP reductoisomerase (44, 45), no significant effect on the vitamin B6 content was observed (Table 1). Functionality of both clomazone and fosmidomycin as inhibitors under the conditions used was indicated by the bleaching phenotype observed after 48 h (Fig. 8, which is published as supporting information on the PNAS web site). This data indicates that vitamin B6 biosynthesis occurs in plants independent of DXP.
Table 1. Vitamin B6 content of WT, cla 1-1, and inhibitor-treated A. thaliana plants.
| Sample | Vitamin B6, μg/gram of fresh weight |
|---|---|
| WT* | 3.6 ± 0.3 |
| cla 1-1* | 2.8 ± 0.2 |
| WT† | 5.6 ± 0.2 |
| Clomazone†‡ | 3.6 ± 0.3 |
| Fosmidomycin†§ | 5.2 ± 0.1 |
Wassilewskija ecotype.
Columbia ecotype.
Treated with 10−4 M clomazone for 96 h.
Treated with 4 × 10−4 M fosmidomycin for 96 h.
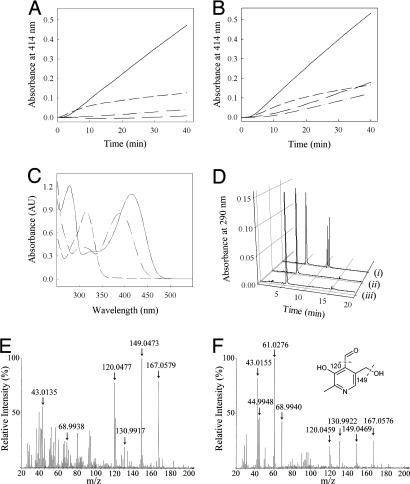
A Pathway of Vitamin B6 Biosynthesis in Plants. In yeast, the nonmevalonate pathway of isoprenoid biosynthesis does not exist (46). Moreover, the extensive studies of Spenser and colleagues (13, 14) indicate that in yeast, vitamin B6 biosynthesis proceeds by a route distinct from that in E. coli in that a pentose, which is not DXP, and a triose are the sugar precursors for formation of the vitamin in this organism. In the bacterial model (B. subtilis), it has recently been established by Burns et al. (19), and indeed by our own independent studies (51), that YaaD (the PDX1 homolog) can accept either ribose 5-phosphate or ribulose 5-phosphate as the pentose sugar and either dihydroxyacetone phosphate or glyceraldehyde 3-phosphate as the triose sugar to form vitamin B6 in the presence of YaaE (the PDX2 homolog) and glutamine. Here, we demonstrate the reconstitution of vitamin B6 biosynthesis from a higher plant. Recombinant AtPDX1.1 and AtPDX1.3 are able to catalyze the formation of the vitamin from either one of the above-mentioned pentose or triose sugars in the presence of ammonium sulfate as the nitrogen source and without addition of AtPDX2 (Fig. 4 A and B). No reaction was observed with glutamine under these conditions. Although the rate observed is substantially higher with ribose 5-phosphate and glyceraldehyde 3-phosphate in the presence of either PDX1 protein than with the other sugar molecules (Fig. 4 A and B), AtPDX1.3 appears to have a greater ability to accept dihydroxyacetone phosphate than AtPDX1.1. Thus far, we have not been successful in isolating a functional recombinant version of AtPDX2 based on the measurement of its ability to hydrolyze glutamine (16), but once achieved, the influence of AtPDX2 on AtPDX1 activity will be examined. No activity was observed with AtPDX1.2 under any of these conditions, indicating that it is not a functional homolog. UV-visible spectrophotometric analysis of the reaction product by treatment with sodium borohydride or sodium hydroxide, results in shifts in the absorption maxima (from 414 nm to 317 nm and 390 nm, respectively) that are characteristic of pyridoxal 5′-phosphate (PLP) (Fig. 4C). The identity of the reaction product under all conditions was confirmed by a HPLC analysis with commercially available standards (PLP, pyridoxal, and pyridoxine) and always resulted in coelution with pyridoxal 5′-phosphate (Fig. 4D). Furthermore, the reaction product was dephosphorylated and submitted to electron impact-mass spectrometry. Although the molecular ion observed agrees very well with the calculated mass of the compound (168.0582), fragment masses corresponding to the loss of H2O (149.0469) and the consecutive loss of the aldehyde group (120.0459), characteristic of pyridoxal, can be clearly observed (Fig. 4 E and F). Because the reaction product of the functional AtPDX1 is PLP, it can be concluded that the alternative vitamin B6 pathway observed in bacteria is also in operation in plants.
Fig. 4.
Biochemistry of vitamin B6 biosynthesis. (A and B) Initial rates showing vitamin B6 formation in the presence of ribose 5-phosphate (0.5 mM) and dl-glyceraldehyde 3-phosphate (1 mM) (solid line); ribulose 5-phosphate (0.5 mM) and dl-glyceraldehyde 3-phosphate (1 mM) (short dashed line); ribose-5-phosphate (0.5 mM) and dihydroxyacetone phosphate (0.5 mM) (long dashed line); ribulose 5-phosphate (0.5 mM) and dihydroxyacetone phosphate (1 mM) (dotted and dashed line) as measured in 50 mM Tris·HCl, pH 8.0, containing 10 mM ammonium sulfate at 37°C, in the presence of AtPDX1.1 (40 μM) (A) and At- PDX1.3 (40 μM) (B). (C) UV-visible spectrum of the reaction product in 50 mM Tris, pH 8.0 (solid line); in the presence of sodium hydroxide (long dashed line); and in the presence of sodium borohydride (short dashed line). (D) HPLC analysis of (i) available standards that elute in the order PLP, pyridoxal, and pyridoxine; (ii) the reaction product (synthesized from ribulose 5-phosphate and dihydroxyacetone phosphate); and (iii) coelution of PLP and the reaction product. (E and F) Electron impact-MS of commercial pyridoxal (E) and the dephosphorylated reaction product (F). (F Inset) Characteristic fragmentation ions of pyridoxal. Note the masses due to the internal standard perfluorotributyl-n-amine can also be observed, e.g., 130.9922.
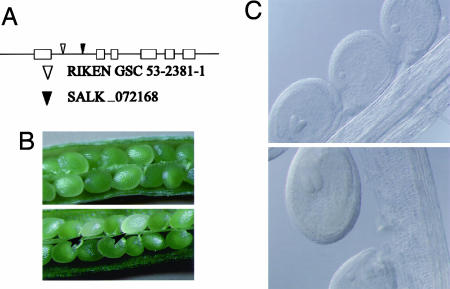
AtPDX2 Is Essential for Plant Viability. Because AtPDX2 is a single-copy gene, various collections of T-DNA and transposon insertion mutants available were searched for a disruption in At5g60540, two of which are described here. SALK_072168 is a T-DNA insertion line (47), whereas 53-2381-1 is a transposon-tagged line from RIKEN GSC envisaged to have one insertion in the gene (48, 49). In the case of SALK_072168, genomic DNA blots revealed a complex insertion of three T-DNA's at the same locus (data not shown). PCR analysis and sequencing established that the disruption in both lines occurs in the first intron as indicated in Fig. 5A. For convenience, we will name these insertion alleles pdx2.1 (SALK_072168) and pdx2.2 (53-2381-1), respectively. Based on a PCR analysis, no homozygous plants could be identified in either the pdx2.1 or pdx2.2 collection of seeds, suggesting that seed development is impaired. A comparison of wild-type siliques with those of heterozygous PDX2.1/pdx2.1 and PDX2.2/pdx2.2 revealed that the heterozygous plants consistently had a population of albino seeds (Fig. 5B). A statistical analysis of the heterozygous PDX2.1/pdx2.1 and PDX2.2/pdx2.2 seeds revealed that ≈25% (±4.0 and 1.3, respectively) showed the albino phenotype, thereby representing the homozygous population. As the albino phenotype could be either due to the loss of pigment(s) or developmental arrest, a histological analysis was carried out. In cleared whole-mount specimens, pdx2.1 and pdx2.2 seeds were developmentally delayed compared with their wild-type or heterozygous siblings. The developmental arrest in all seeds analyzed occurred at the globular stage of embryo development (Fig. 5C). Progress to the heart stage was never observed in pdx2.1 or pdx2.2 even though they were monitored from the globular to the cotyledon stage of embryo development. As far as could be judged, the heterozygous plants developed normally from seedling stage to maturity. The arrest in development observed in pdx2.1 and pdx2.2 can be explained by the assumption that during the early stages of embryogenesis the surrounding maternal tissues supply vitamin B6 among other nutrients, but that the globular to heart transition may be a critical stage in the conversion from heterotrophy to autotrophy. In this context, many of the metabolic enzymes likely to be required at this stage have pyridoxal 5′-phosphate as an essential cofactor, e.g., tryptophan synthase and ornithine decarboxylase involved in tryptophan and spermine and spermidine biosynthesis, respectively, to name but two. It is noted that an allele of pdx2.1 and pdx2.2 has also been described in the Seedgenes database (www.seedgenes.org) where it is reported that a T-DNA insertion in exon 2 of At5g60540 results in arrest of embryo development.
Fig. 5.
Phenotype of the AtPDX2 knockout. (A) Exon-intron structure of PDX2 from A. thaliana indicating the location of the insertions of RIKEN GSC 53-2381-1 (▿) and SALK_072168 (▾). (B) Wild-type immature silique showing normal seeds (Upper), in comparison with one from a heterozygous plant PDX2.1/pdx2.1 (Lower), showing the albino phenotype (indicated arrowheads). (C) Cleared whole-mounted seeds from PDX2.1/pdx2.1 plants, WT or heterozygote at the heart (Upper) and torpedo (Lower) stages and pdx2.1 embryos arrested at the globular stage. The pictures shown are of pdx2.1 (SALK_072168), but analysis of pdx2.2 (RIKEN GSC 53-2381-1) indicated the same phenotype.
We have now initialized experiments to rescue the homozygous phenotype by enriching complete MS growth medium with vitamin B6. However, it is notoriously difficult to rescue embryos arrested at such an early stage of development and may rely on the generation of embryos that can at least reach the globular to heart stage transition (50).
Concluding Remarks. In this report, we show that, contrary to what is generally tacitly assumed, vitamin B6 biosynthesis in plants does not appear to depend on DXP. Instead, the vitamin is synthesized from the pentose phosphate pathway intermediates, ribose 5-phosphate or ribulose 5-phosphate, and either glyceraldehyde 3-phosphate or dihydroxyacetone phosphate (Scheme 1). Moreover, it appears that this pathway is cytosolic, rather than plastidial as was inferred in ref. 8. Of the genes that characterize this previously undescribed pathway, two functional homologs of PDX1 and a single copy of PDX2 are found in Arabidopsis; the latter is essential for plant viability. To summarize, light has been shed onto how this essential vitamin can be synthesized in plants.
Supplementary Material
Acknowledgments
We dedicate this study to Ian Spenser in appreciation of his poineering work on vitamin B6 biosynthesis. We thank Dr. Enrique Herrero for providing the yeast strains, the pCM262 plasmid and the positive controls used in the yeast complementation experiments; Dr. Cristophe Laloi for his general help and advice with Arabidopsis; Andreas Fürholz and Oliver Laule (Swiss Federal Institute of Technology) for their kind gift of the cla1-1 seeds; the RIKEN BioResource center for supplying line 53-2381-1 for analysis and likewise the Salk Institute for providing line 072168; Philippe Roy for performing the HPLC analysis; Oswald Greter and Bernhard Stump for acquiring the Electron Impact-MS; and Dr. Joanna Wyrzykowska for help with the histological analysis. This work was supported by ETH Zürich Grant 0094/41h2703.5 and Swiss National Science Foundation Grant 3100A0-107975/1.
Abbreviations: DXP, deoxyxylulose 5-phosphate; MS, Murashige and Skoog; PLP, pyridoxal 5′-phosphate; T-DNA, transferred DNA.
References
- 1.Jain, S. K. & Lim, G. (2001) Free Radical Biol. Med. 30, 232-237. [DOI] [PubMed] [Google Scholar]
- 2.Ehrenshaft, M., Bilski, P., Li, M. Y., Chignell, C. F. & Daub, M. E. (1999) Proc. Natl. Acad. Sci. USA 96, 9374-9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cane, D. E., Hsiung, Y., Cornish, J. A., Robinson, J. K. & Spenser, I. D. (1998) J. Am. Chem. Soc. 120, 1936-1937. [Google Scholar]
- 4.Cane, D. E., Du, S., Robinson, J. K., Hsiung, Y. & Spenser, I. D. (1999) J. Am. Chem. Soc. 121, 7722-7723. [Google Scholar]
- 5.Hill, R. E., Himmeldirk, K., Kennedy, I. A., Paulowski, R. M., Sayer, B. G., Wolf, E. & Spenser, I. D. (1996) J. Biol. Chem. 271, 30426-30435. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy, I. A., Hill, R. E., Paulowski, R. M., Sayer, B. G. & Spenser, I. D. (1995) J. Am. Chem. Soc. 117, 1661-1662. [Google Scholar]
- 7.Laber, B., Maurer, W., Scharf, S., Stepusin, K. & Schmidt, F. S. (1999) FEBS Lett. 449, 45-48. [DOI] [PubMed] [Google Scholar]
- 8.Julliard, J.-H. (1992) C.R. Seances Acad. Sci. III 314, 285-290. [Google Scholar]
- 9.Drewke, C. & Leistner, E. (2001) in Vitamins and Hormones, ed. Litwack, G. (Academic, San Diego) Vol. 63, pp. 121-155. [DOI] [PubMed] [Google Scholar]
- 10.Fiehe, K., Arenz, A., Drewke, C., Hemscheidt, T., Williamson, R. T. & Leistner, E. (2000) J. Nat. Prod. 63, 185-189. [DOI] [PubMed] [Google Scholar]
- 11.Ehrenshaft, M. & Daub, M. E. (2001) J. Bacteriol. 183, 3383-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osmani, A. H., May, G. S. & Osmani, S. A. (1999) J. Biol. Chem. 274, 23565-23569. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, R. N., Hemscheidt, T., Sayer, B. G. & Spenser, I. D. (2001) J. Am. Chem. Soc. 123, 11353-11359. [DOI] [PubMed] [Google Scholar]
- 14.Zeidler, J., Gupta, R. N., Sayer, B. G. & Spenser, I. D. (2003) J. Org. Chem. 68, 3486-3493. [DOI] [PubMed] [Google Scholar]
- 15.Mittenhuber, G. (2001) J. Mol. Microbiol. Biotechnol. 3, 1-20. [PubMed] [Google Scholar]
- 16.Belitsky, B. R. (2004) J. Bacteriol. 186, 1191-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, Y. X., Sueda, S., Nikawa, J. & Kondo, H. (2004) Eur. J. Biochem. 271, 745-752. [DOI] [PubMed] [Google Scholar]
- 18.Wrenger, C., Eschbach, M.-L., Müller, I. B., Warnecke, D. & Walter, R. D. (2005) J. Biol. Chem. 280, 5242-5248. [DOI] [PubMed] [Google Scholar]
- 19.Burns, K. E., Xiang, Y., Kinsland, C. L., McLafferty, F. W. & Begley, T. P. (2005) J. Am. Chem. Soc. 127, 3682-3683. [DOI] [PubMed] [Google Scholar]
- 20.Estévez, J. M., Cantero, A., Reindl, A., Reichler, S. & Leon, P. (2001) J. Biol. Chem. 276, 22901-22909. [DOI] [PubMed] [Google Scholar]
- 21.Laule, O., Fürholz, A., Chang, H.-S., Zhu, T., Wang, X., Heifetz, P. B., Gruissem, W. & Lange, B. M. (2003) Proc. Natl. Acad. Sci. USA 100, 6866-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guevara-Garcia, A., San Roman, C., Arroyo, A., Cortés, M. E., Gutiérrez-Nava, M. L. & Leon, P. (2005) Plant Cell 17, 628-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murashige, T. & Skoog, F. (1962) Physiol. Plant 15, 493-497. [Google Scholar]
- 24.Rodriquez-Navarro, S., Llorente, B., Rodriguez-Manzaneque, M. T., Ramne, A., Uber, G., Marchesan, D., Dujon, B., Herrero, E., Sunnerhagen, P. & Perez-Ortin, J. E. (2002) Yeast 19, 1261-1276. [DOI] [PubMed] [Google Scholar]
- 25.Scott, A., Wyatt, S., Tsou, P.-L., Robertson, D. & Allen, N. S. (1999) Biotechniques 26, 1125-1132. [DOI] [PubMed] [Google Scholar]
- 26.Jin, J. B., Kim, Y. A., Kim, S. J., Lee, S. H., Kim, D. H., Cheong, G.-W. & Hwang, I. (2001) Plant Cell 13, 1511-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakai, A., Kita, M., Katsuragi, T. & Tani, Y. (2002) J. Biosci. Bioeng. 93, 334-337. [DOI] [PubMed] [Google Scholar]
- 28.Kall, M. A. (2003) Food Chem. 82, 315-327. [Google Scholar]
- 29.Zalkin, H. & Smith, J. L. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72, 87-144. [DOI] [PubMed] [Google Scholar]
- 30.Bauer, J. A., Bennett, E. M., Begley, T. P. & Ealick, S. E. (2004) J. Biol. Chem. 279, 2704-2711. [DOI] [PubMed] [Google Scholar]
- 31.Gari, E., Piedrafita, L., Aldea, M. & Herrero, E. (1997) Yeast 13, 837-848. [DOI] [PubMed] [Google Scholar]
- 32.Hajdukiewicz, P., Svab, Z. & Maliga, P. (1994) Plant Mol. Biol. 25, 989-994. [DOI] [PubMed] [Google Scholar]
- 33.Strassner, J., Schaller, F., Frick, U. B., Howe, G. A., Weiler, E. W., Amrhein, N., Macheroux, P. & Schaller, A. (2002) Plant J. 32, 585-601. [DOI] [PubMed] [Google Scholar]
- 34.Rohmer, M., Knani, M., Simonin, P., Sutter, B. & Sahm, H. (1993) Biochem. J. 295, 517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohmer, M., Seemann, M., Horbach, S., Bringer-Meyer, S. & Sahm, H. (1996) J. Am. Chem. Soc. 118, 2564-2566. [Google Scholar]
- 36.Schwarz, M. K. (1994) Ph.D. thesis (Eidgenössische Technische Hochschule, Zürich).
- 37.Eisenreich, W., Menhard, B., Hylands, P. J., Zenk, M. H. & Bacher, A. (1996) Proc. Natl. Acad. Sci. USA 93, 6431-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lichtenthaler, H. K., Schwender, J., Disch, A. & Rohmer, M. (1997) FEBS Lett. 400, 271-274. [DOI] [PubMed] [Google Scholar]
- 39.Eisenreich, W., Sagner, S., Zenk, M. H. & Bacher, A. (1997) Tetrahedron Lett. 38, 3889-3892. [Google Scholar]
- 40.Zeidler, J. G., Lichtenthaler, H. K., May, H. U. & Lichtenthaler, F. W. (1997) Z. Naturforsch. C, J. Biosci. 52, 15-23. [Google Scholar]
- 41.Estévez, J. M., Cantero, A., Romero, C., Kawaide, H., Jiménez, L. F., Kuzuyama, T., Seto, H., Kamiya, Y. & Leon, P. (2000) Plant Physiol. 124, 95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandel, M. A., Feldmann, K. A., Herrera-Estrella, L., Rocha-Sosa, M. & Leon, P. (1996) Plant J. 9, 649-658. [DOI] [PubMed] [Google Scholar]
- 43.Zeidler, J., Schwender, J., Mueller, C. & Lichtenthaler, H. K. (2000) Biochem. Soc. Trans. 28, 796-798. [PubMed] [Google Scholar]
- 44.Zeidler, J. G., Schwender, J., Mueller, C., Wiesner, J., Weidemeyer, C., Beck, E., Jomaa, H. & Lichtenthaler, H. K. (1998) Z. Naturforsch. 53, 980-986. [Google Scholar]
- 45.Schwender, J., Mueller, C., Zeidler, J. & Lichtenthaler, H. K. (1999) FEBS Lett. 455, 140-144. [DOI] [PubMed] [Google Scholar]
- 46.Eisenreich, W., Bacher, A., Arigoni, D. & Rohdich, F. (2004) Cell. Mol. Life Sci. 61, 1401-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 48.Ito, T., Motohashi, R., Kuromori, T., Mizukado, S., Sakurai, T., Kanahara, H., Seki, M. & Shinozaki, K. (2002) Plant Physiol. 129, 1695-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuromori, T., Hirayama, T., Kiyosue, Y., Takabe, H., Mizukado, S., Sakurai, T., Akiyama, K., Kamiya, A., Ito, T. & Shinozaki, K. (2004) Plant J. 37, 897-905. [DOI] [PubMed] [Google Scholar]
- 50.Patton, D. A., Schetter, A. L., Franzmann, L. H., Nelson, K., Ward, E. R. & Meinke, D. W. (1998) Plant Physiol. 116, 935-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raschle, T., Amrhein, N. & Fitzpatrick, T. B. (July 19, 2005) J. Biol. Chem. ( 10.1074/jbc.M501356200). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.