Abstract
Three genome-wide RNA interference screens were performed in Drosophila S2 cells to dissect the contribution of host processes to Listeria monocytogenes entry, vacuolar escape, and intracellular growth. Among the 116 genes identified, several host pathways previously unrecognized as playing a role in listerial pathogenesis were identified: knockdowns affecting vacuolar trafficking to and from the multivesicular body bypassed the requirement for the essential pore-forming toxin listeriolysin O in mediating escape from phagocytic vacuoles and knockdowns affecting either subunit of serine palmitoyltransferase, a key enzyme in ceramide and sphingolipid biosynthesis, enhanced the toxicity of listeriolysin O expressed in the host cell cytosol, leading to lack of appropriate toxin activity compartmentalization and host cell death. Genome-wide RNA interference screens using Drosophila S2 cells proved to be a powerful approach to dissect host-pathogen interactions.
Keywords: Listeria monocytogenes, listeriolysin O, multivesicular bodies, serine palmitoyltransferase
Infectious diseases caused by intracellular pathogens are responsible for an enormous amount of worldwide morbidity and mortality. These pathogens exploit the basic processes of host cells to establish their intracellular niche (1). Listeria monocytogenes, a facultative intracellular Gram-positive bacterial pathogen, thrives in the cytosol of host cells. The intracellular life cycle of L. monocytogenes has been well defined (2) and can be summarized as follows. Bacteria enter cells by either phagocytosis or bacteria-mediated internalization. Subsequent to internalization, the bacteria produce a cholesterol-dependent pore-forming cytolysin, termed listeriolysin O (LLO), and two phospholipases C (PLCs) that mediate rupture of the resulting phagosome, thereby allowing bacteria access to the rich milieu of the host cytosol. Once in the cytosol, bacteria grow rapidly and exploit a host system of actin-based motility to move intracellularly and spread from cell to cell. Mutants lacking LLO cannot escape from the phagosome, whereas those lacking PLCs are partially defective in escape. In some mammalian epithelial cells, however, a requirement for LLO can be bypassed, and in that case, PLCs are required for vacuolar escape (3, 4). Nevertheless, LLO is absolutely required for pathogenicity and is essential in the vast majority of cells analyzed. However, LLO is a double-edged sword that can kill the host cell if expressed inappropriately. Mutations affecting its acidic pH optimum or in a PEST-like sequence result in inappropriate LLO expression in the cytosol, leading to plasma membrane damage, premature cell death, and severe attenuation in experimental listeriosis (5-7).
Although much has been learned about the cellular microbiology of L. monocytogenes infection, the characterization of host processes contributing to pathogenesis has been hampered by the lack of tools for whole-genome genetic manipulations of the host. Many unanswered questions remain, such as how do bacteria escape from a phagocytic vacuole, and what is the precise role of LLO in mediating this process; how does the host cell process LLO in the cytosolic compartment; and how do cells exert innate immune mechanisms in the cytosol. The development of RNA interference (RNAi) technology and the use of model organisms have emerged as postgenomic approaches to screen for host mutations that affect host-pathogen interactions (8-10).
Recently, Drosophila S2 cells have been established as a suitable host model for bacterial infections (11-13). These macrophage-like cells were shown to behave similarly to mammalian cells in in vitro infection assays using L. monocytogenes. Most importantly, S2 cells are exquisitely sensitive to RNAi and thus conducive for use in genomic analyses (8, 14). In this study, we conducted three genome-wide RNAi-based screens to identify host processes that contribute to L. monocytogenes pathogenesis. Two of these screens focused specifically on host processes involved in the function of the major virulence protein LLO. Many previously uncharacterized host processes involved in various steps of listerial pathogenesis were identified. These RNAi screens may provide insight into the host processes used by other pathogens to subvert their hosts.
Materials and Methods
Bacterial Strains and Cell Cultures. Bacterial strains used in this study were the wild-type L. monocytogenes 10403S; hly in-frame deletion strain DP-L2161 (LLO-minus); the LLOS44A point mutant strain DP-L4057, in which amino acid 44 has been altered; and the hly, plcA, and plcB in-frame deletion strain DP-L2319 (LLO&PLC-minus). Bacterial cultures were grown overnight in brain heart infusion broth at 30°C without shaking. Before infection, bacterial cultures were washed and resuspended in equal volumes of PBS. Drosophila S2 cells were obtained from Invitrogen and cultured in Schneider's Drosophila medium (Invitrogen), supplemented with 10% FBS (Gemini Biological Products, Calabasas, CA).
Double-Stranded RNA (dsRNA) Library Screen. The library containing 7,216 dsRNA species targeting Drosophila genes was described by Foley and O'Farrell (9). S2 cells were treated with dsRNA, as described (14). After 5 days, dsRNA-treated S2 cells were infected with three strains of L. monocytogenes: wild-type, LLO-minus, and LLOS44A-producing mutants. Cells were scored visually for both quantity and quality of infection in LLO-minus and LLOS44A-producing strains and crosschecked with the wild-type screen. Infections were performed in 96-well Con A-(Sigma) coated glass-bottom plates. Each well contained a 120-μl reaction: 20 μl of resuspended dsRNA-treated S2 cells and 100 μl of L. monocytogenes strains at a 1:200 dilution of overnight-grown bacteria in Schneider's Drosophila serum media (SDM) supplemented with 50 μM methyl-β-cyclodextrin-cholesterol (Sigma). Cells were incubated for 1 h at 30°C, followed by a wash with SDM. One hundred microliters of fresh SDM serum media containing 10 μg/ml gentamicin (Sigma) was then added and incubated for an additional 6.5 h. When entry defect phenotypes for a set of genes needed to be distinguished from intracellular growth phenotypes, infection was performed for a total of 2 h instead of 7.5 h. In some cases, S2 cell infections were scaled up to 24-well culture dishes containing Con A-coated 12-mm coverslips for easier quantification.
Immunofluorescence. After infection, glass-bottom plates or coverslips with infected S2 cells were washed once with PBS and fixed in 4% paraformaldehyde. Before staining, cells were washed with TNT buffer (200 mM Tris·HCl/150 mM NaCl/0.5% Triton X-100) for 10 min. L. monocytogenes was stained with a rabbit polyclonal anti-Listeria antibody (Difco, 1:2,000 dilution in TNT buffer with 1% BSA) followed by a secondary Alexa 488-labeled anti-rabbit antibody (Molecular Probes, 1:2,000 dilution), and cytosolic F-actin was stained with tetramethylrhodamine B isothiocyanate-phalloidin (Sigma, 1:1,000 dilution). Each antibody was incubated for 30 min followed by three washes with TNT buffer. Coverslips were mounted with Vectashield mounting medium with DAPI (Vector Laboratories), and 96-well plates were covered with 100 μl of DAKO fluorescent mounting medium (DakoCytomation). Samples were viewed at ×600 with a Nikon TE300 inverted microscope.
Bacterial entry (% Infected) was quantified by the number of S2 cells that contained fluorescent bacteria over the total number of S2 cells counted [(no. of infected cells/infected + noninfected S2 cells) × 100] at 2 h after infection. The percentage of escape (% Escape) was determined by the number of S2 cells containing fluorescent bacteria that had spread throughout the cytosol after 7.5 h of infection over the total number of infected S2 cells [(% Escaped/% Infected S2 cells) × 100] (11). Escape of LLO-minus and LLO&PLC-minus bacteria was represented as fold difference of the no-RNAi control (set arbitrarily at 1). Numbers were obtained from the average of at least three experiments. Approximately 500 S2 cells were counted to obtain the percentages of infection, and ≈300 infected S2 cells were evaluated for the percentages of escape.
Infection of Drosophila S2 Cells and Bacterial Growth Curve. Drosophila S2 cells were infected by L. monocytogenes strains, as described (11). Proteasome inhibition was achieved by treating cells with 2.5 μM lactacystin (Calbiochem) 30 min before and throughout infection.
Infection of Bone Marrow-Derived Macrophages (BMDM) with L. monocytogenes. Growth curves in BMDMs were performed as described (15). Briefly, cells were seeded on coverslips and cultured in DMEM containing 20% FBS (HyClone) and 30% L929-cell conditioned medium. BMDMs were infected with L. monocytogenes strains at a multiplicity of infection of 1.5:1 for 30 min. The medium was then replaced, and 50 μg/ml gentamicin was added 1 h postinfection. Serine palmitoyltransferase (SPT) inhibition was achieved by treating cells with 10 μM myriocin (Sigma) the night before experiments and throughout infection. At the specified time after infection, cells were lysed, and the number of bacteria per coverslip (three for each time point) was determined.
Results and Discussion
Drosophila S2 Cell RNAi Screens. We conducted RNAi-based screens using a library of 7,216 dsRNA representing ≈50% of the predicted genes in the Drosophila genome. These genes were selected based on their homology with genes from human and/or Caenorhabditis elegans (9). Three different RNAi screens were performed. (i) The wild-type L. monocytogenes strain 10403S was used on the first screen to identify knockdowns that affected bacterial entry, vacuolar escape, and intracellular growth. (ii)An LLO-minus L. monocytogenes strain was used in the second screen designed to identify host knockdowns that bypass the requirement for LLO. We hypothesized that knockdowns in pathways that are normally disrupted or targeted by LLO would allow vacuolar escape of this mutant. (iii) A L. monocytogenes strain harboring a point mutation in the PEST-like sequence (LLOS44A) was used in a third screen. This mutant displayed an increased production of LLO in the cytosol, causing a slight toxicity to the host cell (6, 40). To identify host genes controlling LLO toxicity, we sought RNAi knockdowns that enhanced the cytotoxic phenotype of the LLOS44A-producing strain.
dsRNA-treated Drosophila S2 cells were infected with L. monocytogenes strains in 96-well glass-bottom plates. Infection was followed by treatment with the antibiotic gentamicin to kill extracellular bacteria or bacteria in host cells whose cytoplasmic membrane was compromised. Infected cells were incubated for a total of 7.5 h, fixed, and stained for indirect immunofluorescence microscopy (Fig. 6, which is published as supporting information on the PNAS web site). S2 cells were visualized by staining with tetramethylrhodamine B isothiocyanate-phalloidin, which stains actin and highlights cells, as well as polymerized actin surrounding cytosolic L. monocytogenes. Bacteria were stained by indirect immunofluorescence with Alexa 488. Plates were processed manually, and each well was visually inspected by using an inverted microscope at ×600. Any sample that showed an observable difference in either quantity or quality of infection was flagged, and candidates were then retested in independent experiments to confirm the mutant phenotype. Overall, 116 host genes were identified; knockdown of 89 genes affected the infectious process by the wild-type strain, knockdown of 29 genes allowed escape of LLO-minus strain (10 of which also had phenotypes in the wild-type screen), and 8 knockdowns were candidates for control of LLO toxicity. Genes identified by the screens are listed in Table 2, which is published as supporting information on the PNAS web site. Fig. 1 shows representative examples of phenotypes discussed throughout the text.
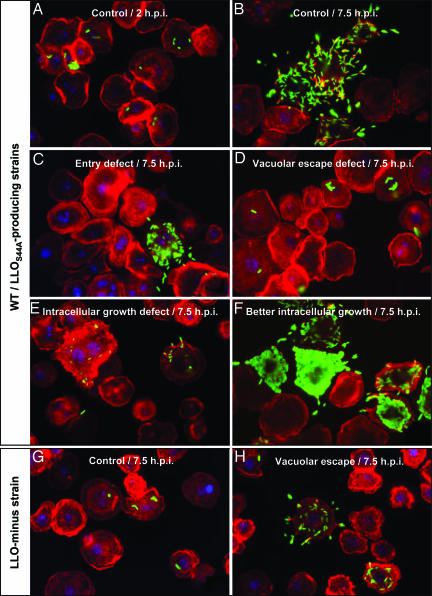
Fig. 1.
Immunofluorescence micrographs of S2 cells infected with L. monocytogenes strains. Bacteria were labeled by using a rabbit polyclonal anti-Listeria antibody followed by secondary labeling with an Alexa 488-coupled anti-rabbit antibody (shown in green). S2 cells and actin were stained in red with tetramethylrhodamine B isothiocyanate-phalloidin, and cell nuclei were stained in blue with DAPI. Phenotypes for S2 cells infected with wild-type bacteria and the LLOS44A-producing strain were very similar; herein, only micrographs of cells infected with wild-type L. monocytogenes are presented (A-F). (A and B) Control cells (no RNAi treatment) at 2 and 7.5 h postinfection (h.p.i.), respectively. (C-H) Micrographs depict RNAi phenotypes at 7.5 h.p.i. (C) Entry defect when chc (clathrin heavy chain) was silenced. (D) Defect in vacuolar escape when vha13 (vATPAse subunit) was silenced; more bacteria are seen as “clumps.” (E) Defect in intracellular growth when peanut (Septin 7 homologue) was silenced. (F) Better intracellular growth when CG5451 was silenced. (G) Control cells infected with LLO-minus bacteria; LLO-minus is defective in vacuolar escape. (H) Vacuolar escape phenotype of the LLO-minus strain when dor (Vps18 homologue) was silenced; here a knockdown bypassed the requirement for LLO in vacuolar escape.
Host Pathways That Affect L. monocytogenes Entry, Vacuolar Escape, and Intracellular Growth. Approximately 42% of host genes identified were in host endocytic and vesicular protein trafficking pathways, suggesting that L. monocytogenes engages these pathways to promote uptake, escape from a vacuole, and grow intracellularly (Fig. 2). Notably, among the host genes identified were two phosphoinositide (PI) phosphatases (CG3573 and myotubularin). PI species have been known to associate with membranes and to play an important role in the recruitment and distribution of numerous proteins involved in vesicular trafficking and signaling events (16, 17). Conversion or hydrolysis of PI by PI kinases and PI phosphatases, respectively, allows their spatial and temporal control. Previously, PI3 kinases were shown to be activated during invasion of host cells by L. monocytogenes (18). We observed that knockdowns in CG3573, a type II inositol 1,4,5-5-phosphatase that hydrolyzes Ins-1,4,5-P3, Ins-1,3,4,5-P4, and PI(4,5)P2 (19); myotubularin, the orthologue of the mammalian myotubular myopathy-related protein 2 (MTMR2), a PI (3)P and PI(3,5)P2 phosphatase; and sbf, a regulating partner of the myotubularin orthologue (MTMR5) (20) led to decreased bacterial entry (≈50% of the no-RNAi-treated control) and less-effective vacuolar escape (≈35% less escape than the no-RNAi-treated control). Knockdowns of CG3573 and sbf also led to reduced intracellular growth. Quantification of defects in entry and vacuolar escape for these knockdowns is shown in Table 1. To our knowledge, these specific PI phosphatases have not been previously associated with host-pathogen interactions. Many trafficking factors and host pathways that affected host entry, phagosomal escape, and intracellular growth are likely not L. monocytogenes-specific. For example, some of the knockdowns that affected L. monocytogenes entry were also needed for entry of Escherichia coli and Staphylococcus aureus (8). Our RNAi screens identified many host processes that could potentially be used by other pathogens.
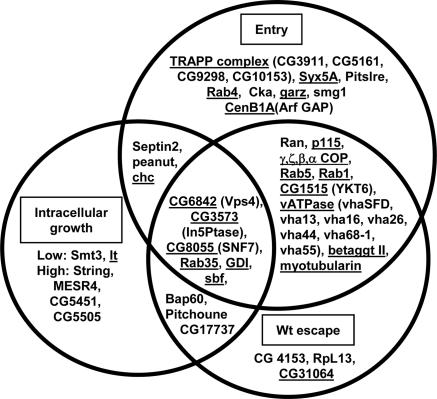
Fig. 2.
RNAi affecting L. monocytogenes entry, escape, and intracellular growth. Diagram illustrating the interconnecting phenotypes of select genes identified in the wild-type L. monocytogenes RNAi screen. Genes are categorized based on phenotypes for entry, vacuolar escape, and intracellular growth. Underlined are genes involved in protein or vesicular trafficking.
Table 1. Entry and vacuolar escape of wild-type L. monocytogenes.
| Gene* | CG no. | Percent infected | Percent escape | Lower intracellular growth |
|---|---|---|---|---|
| No RNAi | 13 ± 1 | 55 ± 2 | − | |
| In5Ptase | 3573 | 6 ± 1 | 35 ± 4 | + |
| Myotubularin | 9115 | 9 ± 1 | 38 ± 3 | − |
| sbf | 6939 | 7 ± 1 | 35 ± 2 | ++ |
Percent infected and Percent escape were obtained as described in Materials and Methods. Lower intracellular growth was scored visually; +, presence of S2 cells with fewer bacteria in the cytosol.
Genes targeted by RNAi.
To identify host genes that limit bacterial replication, we screened for knockdowns that resulted in increased number of intracellular bacteria (Fig. 1F). We identified four knockdowns that led to enhanced L. monocytogenes intracellular growth: MESR4, string, CG5451, and CG5505 (Fig. 1F). Studies in Drosophila suggest that both MESR4 and string may act as antagonists of the Ras/mitogen-activated protein kinase (MAPK) signaling pathway (21, 22). If the phenotype observed is related to the Ras/MAPK pathway, inhibitors of this pathway may decrease L. monocytogenes replication. MAPK are activated upon phosphorylation of both tyrosine and threonine residues by MAPK kinases. Kügler et al. (23) reported that inhibition of tyrosine kinase, using the inhibitor genistein, decreased intracellular growth of L. monocytogenes in J774 macrophages. However, we cannot rule out that the RNAi phenotypes of MESR4 and string might be independent of the Ras/MAPK pathway.
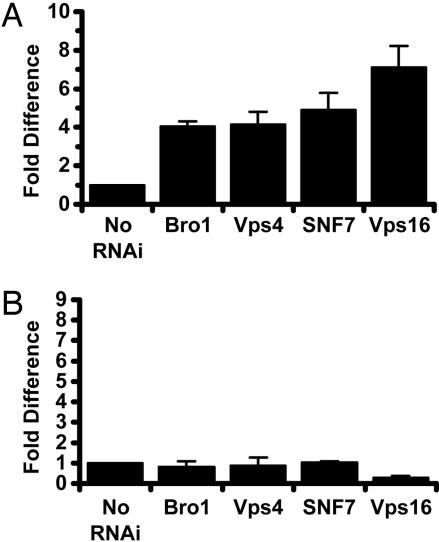
RNAi Screen for Host Knockdowns That Bypassed a Need for LLO in Vacuolar Escape. To identify L. monocytogenes-specific pathways, we relied on the use of bacterial strains with mutations affecting specific steps in the pathogenesis of L. monocytogenes. Although LLO-minus mutants are generally incapable of phagosomal escape (Fig. 1G), we previously noted that LLO-minus mutants escape, albeit at reduced efficiency, from the phagosomes of human epithelial cell lines such as HeLa cells (3, 4). This escape phenotype could be because of one or more mutations leading to the loss or aberrant function of host processes, differences in expression patterns, or maturation rate of HeLa cell phagosomes. We hypothesized that screening for knockdowns that bypass the need for LLO in the escape of S2 cell vacuoles might shed light on host processes or the vesicular trafficking stages targeted by LLO. Many of the host knockdowns that led to this phenotype were in components of vesicular trafficking complexes that control trafficking to and from multivesicular bodies (MVB)/late endosomes (Table 2 and Fig. 1H). We identified components of the Class E/ESCRT (endosomal sorting complex required for transport) complexes (TSG101, SNF7, Vps4, and Bro1) and most of the components of the Class C/B vacuolar sorting complexes involved in the docking and tethering of MVBs to lysosomes (Vps16, Vps18, Vps33, Vps39, and Vps41) (24, 25). Knockdowns of select vacuolar sorting proteins increased the escape of the LLO-minus strain by 4- to 7-fold when compared with the no-RNAi treatment control (Fig. 3A). These data suggest that blocking the later stages of vesicular trafficking bypassed the requirement for LLO. One trivial explanation for these results was that the phagosomes in these knockdowns were distorted or fragile (25). We then evaluated vacuolar escape of L. monocytogenes mutants lacking both LLO and PLCs (LLO&PLC-minus strain) in both control and knockdown mutants. This triple mutant was not able to escape, suggesting that the bypass mutations were LLO-specific (Fig. 3B). Knockdowns of vesicular and trafficking proteins likely led to vacuolar conditions that resembled those produced by LLO.
Fig. 3.
RNAi knockdowns that bypass vacuolar escape defect of LLO-minus L. monocytogenes. (A) Vacuolar escape of LLO-minus bacteria. In no-RNAi-treated S2 cells, LLO-minus bacteria escaped vacuoles at an average of 2 ± 0.4%. Vacuolar escape of knockdown mutants was represented as fold difference of the no-RNAi-treated control (set at the arbitrary unit of 1). Vacuolar escape was calculated as described in Materials and Methods. (B) Vacuolar escape of LLO&PLC-minus bacteria. LLO&PLC-minus mutant escaped at an average of 0.7 ± 0.1%.
Results of the LLO bypass screen provide a view that may shed light on the natural functions of LLO. To date, not much is known about the vesicular trafficking stage that is targeted by L. monocytogenes. Previous research suggested that L. monocytogenes modulated vesicular trafficking by targeting Rab5a, a small GTPase needed for directing membrane fusion events at the early endosome stage (26, 27). However, these observations do not correlate well with the optimal function of LLO. The pH optimum of LLO is 5.5, consistent with the properties of late endosomes (7). We propose that LLO inserts into a maturing phagosome, the MVB/late endosome, thereby aborting further maturation and promoting the activity of PLCs. This model is supported by our observations that knockdowns affecting seven of the eight vacuolar ATPase subunits, and presumably vacuolar pH were blocked in escape of wild-type L. monocytogenes (Table 2 and Fig. 1D). The bypass of LLO does not occur in cells treated with dsRNA against both vacuolar ATPase and vacuolar sorting proteins (data not shown), thus underscoring the need for vacuolar maturation past the early endosome stage before LLO-mediated vacuolar escape at the MVB stage, where conditions are optimal for its pore-forming toxin LLO. Results from this RNAi screen also correlate well with previous data that showed the requirement for vacuolar acidification before escape from macrophage vacuoles (7, 28). Recent data also showed that L. monocytogenes escaped from macrophage endosomes labeled with Rab7, a late endosome marker, but were devoid of Rab5 (29). The MVB seems to be a critical stage in vacuolar maturation that is targeted by other pathogens as well. Just as HIV, which interacts with MVB components to bud out via the plasma membrane or the exosome pathway (24), and Anthrax toxin, which uses the MVB to promote intoxication (30), LLO may be adapted for function in the MVB.
Screen for Host Processes That Enhance LLO Toxicity. We wished to identify host factors that controlled the potentially toxic effects of LLO secreted into the host cytosol. As an intracellular pathogen, L. monocytogenes has evolved mechanisms to avoid killing its host cell. We have noted (5) that LLO contains a PEST-like sequence essential for effective compartmentalization of its activity. Mutations in the PEST-like sequence result in bacteria that escape normally from phagocytic vacuoles but led to premature permeabilization of the host cell cytoplasmic membrane subsequent cell death. A single mutation in the PEST-like sequence (S44A) has a subtle but clearly reproducible phenotype in S2 cells that manifests as a decrease in intracellular bacteria because of the influx of gentamicin resulting from the introduction of pores into the host cell cytoplasmic membrane (Fig. 4).
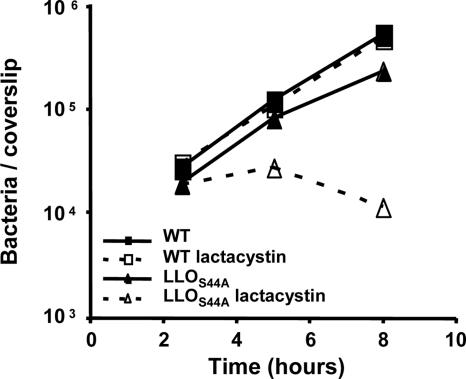
Fig. 4.
Intracellular growth of wild-type and LLOS44A-producing strains in Drosophila S2 cells upon inhibition of the proteasome. Inhibition of the proteasome with 2.5 μM lactacystin (dashed line) increased toxicity caused by the LLOS44A-producing strain (triangle). Growth of wild-type bacteria (square) was not affected by the addition of lactacystin. Gentamicin was added to the medium 1 h postinfection to kill extracellular bacteria and bacteria in host cells whose plasma membrane was compromised. Decrease in intracellular growth of bacteria is correlated with the degree of LLO toxicity.
There is evidence that LLO secreted by cytosolic L. monocytogenes is degraded by the host proteasome (31). Therefore, we reasoned that host knockdowns in pathways controlling LLO toxicity would increase the toxicity displayed by the strain expressing LLOS44A. In agreement with this hypothesis, pharmacological inhibition of the host proteasome with lactacystin synergized with the LLOS44A-producing strain in mammalian cells (40) as well as in Drosophila cells (Fig. 4). Our RNAi screen also confirmed the involvement of the host proteasome (Table 2). However, identification of proteosomal subunits during the screen has been made difficult by the toxicity of the interference itself, which led to a decrease in cell viability (32).
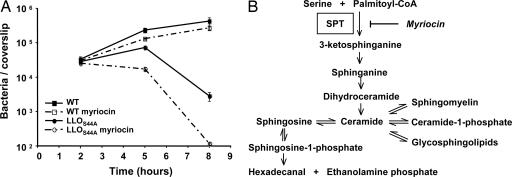
The most interesting result revealed by this screen was the identification of both subunits of SPT, both of which enhanced the toxicity of the LLOS44A-producing strain. SPT catalyzes a critical step in the biosynthesis of sphingolipids (Fig. 5B) (33), suggesting that sphingolipids play a role in controlling LLO toxicity. SPT is specifically inhibited by the drug myriocin (34). Therefore, we were able to validate a role for sphingolipid biosynthesis by blocking SPT in murine bone marrow-derived macrophages. As shown in Fig. 5A, inhibition of SPT increased toxicity caused by bacteria producing LLOS44A. The use of the LLOS44A-producing strain was critical to reveal this pathway, because inhibition of sphingolipid biosynthesis had a negligible effect on the wild-type strain grown in these in vitro conditions.
Fig. 5.
Effect of SPT inhibition on bacterial intracellular growth in murine bone marrow-derived macrophages. (A) Growth of wild-type L. monocytogenes (square) and LLOS44A-producing strains (circle) in murine bone marrow-derived macrophages, upon inhibition of SPT with 10 μM myriocin (dashed lines). Inhibition of SPT increased toxicity of the LLOS44A-producing strain leading to the increased influx of gentamicin into the cytosol and subsequent killing of intracellular bacteria. (B) Mammalian sphingolipid and catabolism pathways. Myriocin is an inhibitor of SPT.
The sphingolipid biosynthesis pathway leads to formation of many important plasma membrane compounds (Fig. 5B). Sphingomyelin is a major constituent of lipid rafts. Two studies in which LLO was added extracellularly report the association of LLO with lipid rafts (35, 36). However, if lipid rafts were the preferential sites of LLO insertion inside the cells, we would expect that inhibition of sphingomyelin synthesis would rescue instead of enhance the toxic effect of LLO. The sphingolipid biosynthesis pathway also leads to synthesis of ceramide or sphingosine-1-phosphate, known to behave as lipid-signaling molecules mediating apoptosis and cell survival, respectively (37). One can hypothesize that such molecules could signal host cells to respond to LLO toxicity. Further studies will be required to determine the precise role played by sphingolipid metabolism in controlling LLO toxicity. At the moment, we speculate that either (i) changing plasma membrane composition by preventing synthesis of specific plasma membrane components would favor LLO activity, or (ii) intermediates of this pathway would act as signaling molecules that trigger LLO elimination and/or membrane repair. Interestingly, recent studies have shown that cells lacking SPT are more sensitive to Diphtheria toxin (38) and Epsilon toxin of Clostridium perfringens (39).
Intracellular pathogens exploit and manipulate the basic processes of host cells to establish and maintain their intracellular niche. By combining bacterial mutations with host knockdowns, the results of this study provide insights into the pathogenic mechanisms used by L. monocytogenes. We now have a model by which LLO mediates escape of L. monocytogenes from a phagosome by acting before phagolysosome fusion. We have also identified SPT, a key enzyme in sphingolipid metabolism, as necessary to control the damaging effects of LLO in the cytosol. Last, we have identified host genes that apparently control bacterial intracellular replication. These and other factors or host pathways identified in our screens may be conserved in mammalian systems and thus can further shed light on host-pathogen interactions for other intracellular pathogens.
Supplementary Material
Acknowledgments
The dsRNA library used in this study was produced from a collaboration among Graeme Davis, Ben Eaton, Edan Foley, Patrick O'Farrell, Steve Rogers, Nico Stuurman, and Ronald D. Vale at the University of California, San Francisco. The library construction was made possible by a gift from the Sandler family. We thank Kirkwood Land and Jess Leber for critical review of the manuscript. We thank Nicole Meyer-Morse and Aimee Geissler for help in the isolation of bone marrow-derived macrophages. This work was supported by the National Institutes of Health Grants AI27655 and AI29619 (to D.A.P.), National Research Service Award Fellowship AI51896 (to L.W.C.), and the Howard Hughes Medical Institute (to R.D.V.).
Abbreviations: RNAi, RNA interference; dsRNA, double-stranded RNA; LLO, listeriolysin O; PLC, phospholipase C; MVB, multivesicular bodies; SPT, serine palmitoyltransferase; PI, phosphoinositide; MAPK, mitogen-activated protein kinase.
References
- 1.Alonso, A. & Garcia-del Portillo, F. (2004) Int. Microbiol. 7, 181-191. [PubMed] [Google Scholar]
- 2.Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., González-Zorn, B., Wehland, J. & Kreft, J. (2001) Clin. Microbiol. Rev. 14, 584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marquis, H., Doshi, V. & Portnoy, D. A. (1995) Infect. Immun. 63, 4531-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundling, A., Gonzalez, M. D. & Higgins, D. E. (2003) J. Bacteriol. 185, 6295-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decatur, A. L. & Portnoy, D. A. (2000) Science 290, 992-995. [DOI] [PubMed] [Google Scholar]
- 6.Glomski, I. J., Decatur, A. L. & Portnoy, D. A. (2003) Infect. Immun. 71, 6754-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glomski, I. J., Gedde, M. M., Tsang, A. W., Swanson, J. A. & Portnoy, D. A. (2002) J. Cell Biol. 156, 1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rämet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R. A. (2002) Nature 416, 644-648. [DOI] [PubMed] [Google Scholar]
- 9.Foley, E. & O'Farrell, P. H. (2004) PLoS Biol. 2, E203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sifri, C. D., Begun, J. & Ausubel, F. M. (2005) Trends Microbiol. 13, 119-127. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, L. W. & Portnoy, D. A. (2003) Cell. Microbiol. 5, 875-885. [DOI] [PubMed] [Google Scholar]
- 12.Mansfield, B. E., Dionne, M. S., Schneider, D. S. & Freitag, N. E. (2003) Cell Microbiol. 5, 901-911. [DOI] [PubMed] [Google Scholar]
- 13.Elwell, C. & Engel, J. N. (2005) Cell. Microbiol. 7, 725-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemens, J. C., Worby, C. A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B. A. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auerbuch, V., Brockstedt, D. G., Meyer-Morse, N., O'Riordan, M. & Portnoy, D. A. (2004) J. Exp. Med. 200, 527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odorizzi, G., Babst, M. & Emr, S. D. (2000) Trends Biochem. Sci. 25, 229-235. [DOI] [PubMed] [Google Scholar]
- 17.Botelho, R. J., Scott, C. C. & Grinstein, S. (2004) Curr. Top. Microbiol. Immunol. 282, 1-30. [DOI] [PubMed] [Google Scholar]
- 18.Ireton, K., Payrastre, B., Chap, H., Ogawa, W., Sakaue, H., Kasuga, M. & Cossart, P. (1996) Science 274, 780-782. [DOI] [PubMed] [Google Scholar]
- 19.Schmid, A. C., Wise, H. M., Mitchell, C. A., Nussbaum, R. & Woscholski, R. (2004) FEBS Lett. 576, 9-13. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S. A., Vacratsis, P. O., Firestein, R., Cleary, M. L. & Dixon, J. E. (2003) Proc. Natl. Acad. Sci. USA 100, 4492-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, A. M. & Rubin, G. M. (2000) Genetics 156, 1219-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebay, I., Chen, F., Hsiao, F., Kolodziej, P. A., Kuang, B. H., Laverty, T., Suh, C., Voas, M., Williams, A. & Rubin, G. M. (2000) Genetics 154, 695-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kügler, S., Schuller, S. & Goebel, W. (1997) FEMS Microbiol. Lett. 157, 131-136. [DOI] [PubMed] [Google Scholar]
- 24.Morita, E. & Sundquist, W. I. (2004) Annu. Rev. Cell. Dev. Biol. 20, 395-425. [DOI] [PubMed] [Google Scholar]
- 25.Mullins, C. & Bonifacino, J. S. (2001) BioEssays 23, 333-343. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez-Dominguez, C. & Stahl, P. D. (1999) J. Biol. Chem. 274, 11459-11462. [DOI] [PubMed] [Google Scholar]
- 27.Prada-Delgado, A., Carrasco-Marin, E., Pena-Macarro, C., Del Cerro-Vadillo, E., Fresno-Escudero, M., Leyva-Cobian, F. & Alvarez-Dominguez, C. (2005) Traffic 6, 252-265. [DOI] [PubMed] [Google Scholar]
- 28.Beauregard, K. E., Lee, K. D., Collier, R. J. & Swanson, J. A. (1997) J. Exp. Med. 186, 1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry, R., Shaughnessy, L., Loessner, M. J., Alberti-Segui, C., Higgins, D. E. & Swanson, J. A. (2005) Cell. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 30.Abrami, L., Lindsay, M., Parton, R. G., Leppla, S. H. & van der Goot, F. G. (2004) J. Cell Biol. 166, 645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villanueva, M. S., Sijts, A. J. & Pamer, E. G. (1995) J. Immunol. 155, 5227-5233. [PubMed] [Google Scholar]
- 32.Wójcik, C. & DeMartino, G. N. (2002) J. Biol. Chem. 277, 6188-6197. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins, G. M. (2003) Cell Mol. Life Sci. 60, 701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyake, Y., Kozutsumi, Y., Nakamura, S., Fujita, T. & Kawasaki, T. (1995) Biochem. Biophys. Res. Commun. 211, 396-403. [DOI] [PubMed] [Google Scholar]
- 35.Coconnier, M. H., Dlissi, E., Robard, M., Laboisse, C. L., Gaillard, J. L. & Servin, A. L. (1998) Infect. Immun. 66, 3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gekara, N. O. & Weiss, S. (2004) Biochem. Soc. Trans. 32, 712-714. [DOI] [PubMed] [Google Scholar]
- 37.Pyne, S. & Pyne, N. (2000) Pharmacol. Ther. 88, 115-131. [DOI] [PubMed] [Google Scholar]
- 38.Spilsberg, B., Hanada, K. & Sandvig, K. (2005) Biochem. Biophys. Res. Commun. 329, 465-473. [DOI] [PubMed] [Google Scholar]
- 39.Shimamoto, S., Tamai, E., Matsushita, O., Minami, J., Okabe, A. & Miyata, S. (2005) Microbiol. Immunol. 49, 245-253. [DOI] [PubMed] [Google Scholar]
- 40.Schnupf, P., Portnoy, D. D. & Decatur, A. L. (2005) Cell. Microbiol., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.