Abstract
Inorganic polyphosphate (poly P), a polymer of tens or hundreds of phosphate residues linked by high-energy, ATP-like bonds, is found in all organisms and performs a wide variety of functions. Myxococcus xanthus, a social bacterium that feeds on other bacteria and forms fruiting bodies and spores, depends on poly P for motility, development, and nutritional predation. Two poly P metabolizing enzymes were studied in M. xanthus: poly P kinase 1, which synthesizes poly P reversibly from ATP, and poly P:AMP phosphotransferase, which uses poly P as a donor to also reversibly convert AMP to ADP. The null mutant of ppk1 is defective in social motility, overproduces pilin protein on the cell surface, is delayed in fruiting body formation, produces fewer spores, is delayed in germination, and forms far smaller plaques on a lawn of Klebsiella aerogenes. The pap mutant is also impaired in social motility, but shows only slightly reduced abilities in development and predation.
Inorganic polyphosphate (poly P), a polymer of tens or hundreds of phosphate residues linked by high-energy, anhydride bonds, has been found in every cell: bacterial, fungal, plant, and animal (1, 2). The principal enzyme for poly P synthesis, poly P kinase 1 (PPK1), has been identified in a hundred or more bacterial strains and is responsible for a wide variety of functions (3, 4), including long-term survival, adaptation to nutrient downshift, resistance to stresses, motility, biofilm formation and, in many major pathogens, virulence (5-11). Remarkably, PPK1 is conserved in certain eukaryotic species, including the social slime mold Dictyostelium discoideum, in which poly P participates in the development of fruiting bodies and spores and in predation on bacteria (12). Among other poly P enzymes, poly P:AMP phosphotransferase (PAP), which uses poly P as a donor to convert AMP to ADP has been identified in Acinetobacter johnsonii (13). In Bacillus cereus, mutants are deficient in motility and biofilm formation (14).
Myxococcus xanthus is a Gram-negative, gliding, soil bacterium that exhibits social interactions and undergoes multicellular developmental cycles (15). During the vegetative phase, biofilms of M. xanthus consume other bacteria in a wolf-pack-like manner by producing several lytic enzymes and antibiotics (16). Upon starvation, cells at high density initiate a multicellular developmental cycle in which 105 cells aggregate into a mound-shaped fruiting body that contains thousands of resistant myxospores (16). The spherical myxospores germinate into rod-shape, vegetative cells when reexposed to a nutrient-rich condition.
M. xanthus glides on surfaces by two genetically distinct systems: adventurous motility (A-motility) and social motility (S-motility) (17). A-motility translocates cells by slime secretion, whereas S-motility translocates cells by pilus retraction (18, 19); neither system uses flagellae. Motility is important for vegetative growth and development because it helps aggregate cells into fruiting bodies and movement within the body so that a signaling threshold is reached that is adequate for sporulation (20-22). S-motility requires polarly localized type IV pili (TFP) (23, 24), fibril polysaccharides (25-27), and LPS O-antigen (28). Specific sugar moieties of fibril polysaccharides provide binding targets for the TFP of adjacent cells; when pilus retraction is triggered, the cell is pulled forward (27). The S-motility of M. xanthus is related to twitching motility in Pseudomonas aeruginosa and Neisseria gonorrhoea in that all are TFP-dependent (28). M. xanthus and P. aeruginosa also share highly homologous genes for ppk1 and pap. We have examined the roles of poly P in S-motility, development, and predation with the aid of the phenotypes of the null mutants of the genes.
Materials and Methods
Bacterial Strains, Plasmids, and Growth. Bacterial strains and plasmids are listed in Table 1. M. xanthus strains were grown vegetatively at 30°C in CTT broth (1% casitone/8 mM MgSO4/10 mM Tris·HCl/1 mM KPO4, pH 7.6) or on CTT 1.5% agar plates. For fruiting body development, TPM plates (10 mM Tris·HCl/8 mM MgSO4/1 mM KPO4, pH 7.6) with 1.5% agar were used. Kanamycin was present at 40 μg/ml.
Table 1. Myxobacterial strains and plasmids.
| Strain or plasmid | Characteristics | Source |
|---|---|---|
| DK1622 | M. xanthus WT | 21 |
| DKΔppk1 | DK1622 Δppk1 | This work |
| DKΔpap | DK1622 Δpap | This work |
| DKΔppk1Δpap | DK1622Δppk1Δpap | This work |
| pBJ113 | pUC118 containing Kmr and galK; derived from pKG2 | 30 |
| pBJ114 | pUC119 containing Kmr and galK; derived from pKG2 | 30 |
| pBJ113ppk1 | pBJ113 with deletion construction for ppk1; Kmr | This work |
| pBJ114pap | PBJ114 with deletion construction for pap; Kmr | This work |
Kmr, kanamycin resistance.
poly P Determination. Cells were grown in CTT broth containing 25 μCi/ml (1 Ci = 37 GBq) [32P]Pi and collected at several growth stages. Trichloroacetic acid (TCA)-soluble compounds were removed by extraction with 2% cold TCA; the TCA-insoluble pellets were suspended in 50 mM Tris·HCl (pH 7.5) and treated with 300 μg/ml DNase I and RNaseA at 37°C for 30 min followed by 300 μg/ml proteinase K at 37°C for 30 min; EDTA was added to 8 mM. The aqueous phase was saved after following sequential phenol-chloroform and chloroform extractions. poly P was precipitated by addition of three volumes of ethanol and kept at -20°C for 1 h or more. The precipitated poly P was then collected by 20,000 × g centrifugation at 4°C for 20 min, washed with 70% ethanol, dried under vacuum, and resuspended in 50 mM Tris·HCl, pH 7.4/1 mM EDTA. Samples were digested with yeast exopolyPase to determine poly P levels (2) and applied to 20% PAGE to assess the sizes of poly P (29). Spores were freeze-dried and broken by Wig-L-Bug; poly P was then extracted as described above.
Assay for PPK1. A crude lysate was prepared as follows: Cells were harvested by 8,000 × g centrifugation at 4°C for 10 min, suspended in 50 mM Tris·HCl (pH 7.5), treated with 250 μg/ml lysozyme for 1 h at 4°C, and disrupted by sonication three times for 10 s at 4°C. The cell debris was removed by 14,000 × g centrifugation at 4°C for 10 min. The supernatant was designated “crude lysate.” The PPK1 activity was measured in a 25-μl reaction mixture containing 50 mM Tris·HCl (pH 7.5), 2 mM MgCl2, 1.5 mM poly P75 (type 75; in phosphate residues; Sigma), and 1 mM [γ-32P]ATP (20 cpm/pmol ATP). After incubation for 5 min at 37°C, the reaction was stopped, and the [32P]poly P was measured as described in ref. 3. One unit of enzyme is defined as the amount incorporating 1 pmol of phosphate into acid-insoluble poly P per min.
Assay for PAP. This activity was measured in the membrane fraction prepared by further centrifuging the crude lysate at 50,000 × g at 4°C for 1 h. The pellet was suspended in 50 mM Tris·HCl (pH 7.5) and used in the assay. The 10-μl reaction mixture contained 50 mM Hepes·KOH (pH 8.0), 4 mM MgCl2, 40 mM (NH4)2SO4, 1 mM [3H]AMP, and 3 mM poly P65 (type 65; Sigma). After incubation at 37°C for 30 min, 1 μl of the reaction mixture was applied to polyethyleneimine-TLC plates and developed in 0.4 M LiCl and 1 M formic acid. The AMP, ADP, and ATP spots were visualized under UV light, cut out, and quantified in a liquid scintillation counter. One unit of enzyme is defined as the transfer of 1 pmol of phosphate residue to AMP per min.
Construction of Mutants. A method based on Wu and Kaiser (30) was used to create deletions in ppk1 and pap in DK1622. The 1,072-bp 5′ flanking region of ppk1 (including the first 81 codons of ppk1 with EcoRI or BamHI sites on 5′ or 3′ end) and the 970-bp 3′ flanking region of ppk1 (including the last 30 codons of ppk1 with BamHI or HindIII sites on the 5′ or 3′ end) were generated by PCR. Both PCR products were subcloned into pBJ113 EcoRI and HindIII sites by three fragment ligations to get pBJ113ppk1. Similarly, pBJ114pap was constructed by sub-cloning the 915-bp 5′ flanking region (containing the first 17 codons) and the 948-bp 3′ flanking region (containing the last 23 codons) of pap into pBJ114. The pBJ113ppk1 and pBJ114pap constructs were electroporated into DK1622, and kanamycin-resistant colonies were selected. By using the galK gene in the plasmids as a counterselectable marker, galactose-resistant colonies were obtained and screened for deletion of ppk1 (DKΔppk1) and of pap (DKΔpap). Subsequently, the strain DKΔppk1Δpap was obtained by making a pap deletion in DKΔppk1.
Assays for Development, Germination, and Motility. For fruiting body formation, exponentially growing cells in CTT broth (Klett unit of 100) were centrifuged at 3,000 × g at room temperature for 15 min, washed with an equal volume of TPM buffer, and suspended in TPM at a concentration of 5 × 109 cells per ml (Klett unit of 1,000). An aliquot of 20 μl was spotted on TPM agar and incubated for several days at 30°C. Pictures were taken at various times to record fruiting body formation.
To prepare myxospores for germination, fruiting bodies were harvested by gentle scraping of the agar plates, suspended in cold distilled water, and sonicated three times for 10 s each to disrupt vegetative cells. Debris was removed by four sequential centrifugations of the pellet at 3,000 × g at 4°C for 15 min to yield a pellet of pure spores, which were counted in a hemocytometer. The spores were frozen in liquid nitrogen and stored at -80°C until used. The spores were suspended and heated at 49°C for 2 h and inoculated into germination medium (31) (0.2% casitone/10 mM Tris·HCl, pH 7.6/8 mM MgSO4/1 mM CaCl2) at a density of 1 × 108 to 5 × 108 spores per ml. Samples were taken at different times and observed under the microscope.
For motility assays, cells were harvested from CTT medium at an exponential stage and concentrated to a Klett unit of 1,000 in fresh CTT medium. Five microliters of concentrated cells was spotted on soft agar plates (CTT with 0.4% agar) for S-motility, and CTT plates solidified with 1.5% agar were used for A-motility. After incubation at 30°C for several days, swarm diameters were measured for each culture. For Western blots, cell-surface pilin was isolated and assayed as described in ref. 32.
Assay of Predation. An overnight culture of Klebsiella aerogenes formed a lawn on a nutrient agar plate (1% peptone/0.5% yeast extract/10 mM Tris·HCl, pH 7.6/1.5% agar). Log-phase M. xanthus cells in CTT medium were collected and diluted to a Klett unit of 1 (≈5 × 106 cells per ml); 5 μl of a culture was spotted on a lawn of K. aerogenes and incubated at 30°C. Plaque sizes were measured at intervals. The same number of cells was spotted on nutrient agar plates without K. aerogenes as a control for measurement of colony size.
Results
PPK1: The Gene and Activity. The deduced protein sequence of single-copy ppk1 in the WT showed a 40% identity with PPK1 of P. aeruginosa and a 33% identity with that of E. coli. PPK1 activity measured in the WT lysate was very low (50 units/mg) when using the assay conditions for P. aeruginosa or E. coli (3). However, by adding poly P, omitting (NH4)2SO4, and altering the pH and concentration of MgCl2, the activity was increased 10-fold to 500 units/mg (see Materials and Methods).
PAP: The Gene and Activity. The pap gene that encodes PAP was first identified in A. johnsonii and later found in other bacteria that include P. aeruginosa (4, 13) and B. cereus (14). The level of activity in M. xanthus increases sharply after cells enter the stationary phase. The activity resides in the membrane fraction and is not solubilized, even after repeated sonication.
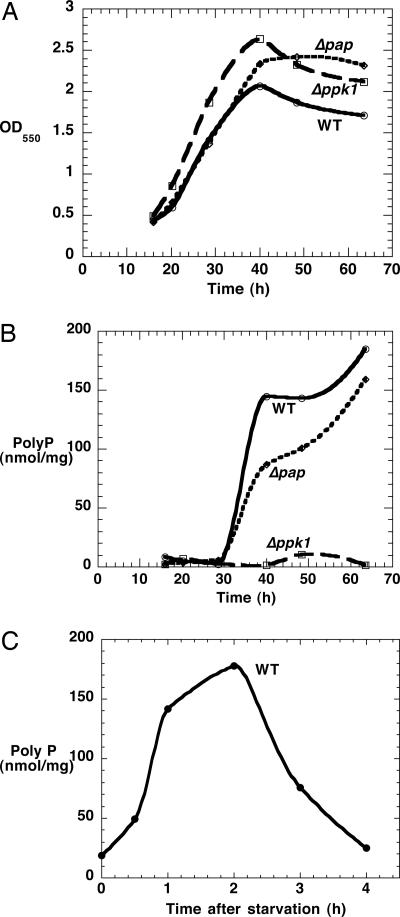
Enzyme and poly P Levels in WT and Mutants. All strains grew at similar rates in the CTT medium (Fig. 1A). Among the mutant strains, the PPK1 activity was nearly absent in Δppk1 and the double mutant Δppk1Δpap (each were 30 units/mg, ≈6% of WT). PAP activity was undetectable in the Δpap and in the double mutant (data not shown).
Fig. 1.
Growth and poly P accumulation of WT and mutants. (A) Growth in CTT medium. (B) poly P levels in CTT medium. (C) poly P accumulation upon starvation. Mid-log cells in CTT medium were washed with TPM buffer and resuspended in an equal volume of TPM buffer.
The poly P (as measured by exopolyPase digestion) in the WT appeared abruptly when cells entered stationary phase and remained at a high level (Fig. 1B); it was <5% in Δppk1 mutant and slightly reduced in the Δpap mutant. The chain lengths of poly P from WT and Δpap mutant are similar and distributed in a wide range, from ≈20-750 phosphate residues (data not shown). The WT cells also accumulated large amounts of poly P within 2 h when WT log-phase cells were transferred from the nutrient-rich, CTT broth to the starvation TPM buffer (Fig. 1C), a condition that triggers the developmental cycle. poly P was then removed in the next 2 h, indicating that the accumulated poly P is used at an early developmental stage. WT and Δpap mutant spores contain 2 nmol/mg poly P, whereas the Δppk1 mutant has no detectable poly P in spores.
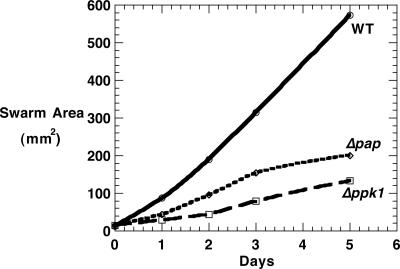
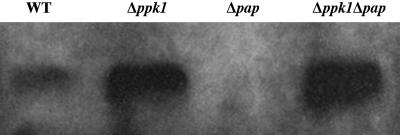
Motility Defects in Mutants. When assayed on soft agar (0.4% CTT), the swarm expansion rate that is provided mostly by S-motility of Δppk1 was only 20% that of WT, whereas that of Δpap was reduced to 46% after 3 days and 37% after 5 days compared with WT (Fig. 2). Inasmuch as S-motility is TFP-dependent, the cell-surface PilA (pilin protein) released from cells by shearing them was assayed by immunoblotting with anti-PilA antibody. The levels were elevated in the Δppk1 and Δppk1Δpap mutants, a phenotype characterized as hyperpiliation (Fig. 3). The A-motility, measured on hard agar, showed no significant difference between the mutants and WT.
Fig. 2.
M. xanthus swarm sizes on soft agar.
Fig. 3.
Detection of cell-surface pilin protein. Pilin was sheared from 108 cells, separated by 15% SDS/PAGE, electroblotted, and probed with a polyclonal antibody against PilA. The apparent molecular mass of PilA is ≈25 kDa.
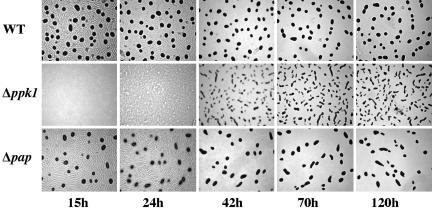
Developmental Defects in Mutants. The Δppk1 mutant was delayed in aggregation and altered in fruiting body morphology (Fig. 4). Fruiting bodies of the WT appeared within 15 h after starvation and reached a limit by 24 h. The Δppk1 mutant did not move in typical ripples and aggregated into irregularly shaped fruiting bodies at 70 h. The Δpap mutant was not delayed, but it formed fewer fruiting bodies. The number of spores formed by Δppk1 was 32% of WT, and that of Δpap was 38%. The shape of the aggregates in Δppk1 is consistent with a defect in sporulation. The cells appear loosely packed in the elongated fruiting bodies, and, as a result, fewer cells exceed the C-signal threshold required for sporulation.
Fig. 4.
Development of Δppk1 and Δpap mutants. Cells were spotted on 1.5% TPM agar plates, incubated at 30°C, and monitored over a period of 5 days.
Spores were germinated by heating at 49°C for 2 h and then transferred into fresh germination medium and examined under a phase-contrast, light microscope. After 20 h, WT and Δpap spores had all become rod-shaped. By contrast, Δppk1 spores were delayed in germination and remained spherical even after 20 h. Only after 33 h were large aggregations of spores observed along with some rod-shaped vegetative cells at the edges of spore clusters; conversion to rod-shaped cells was complete only after 46 h. All WT and mutants spores are viable and can form colonies on CTT plates (data not shown).
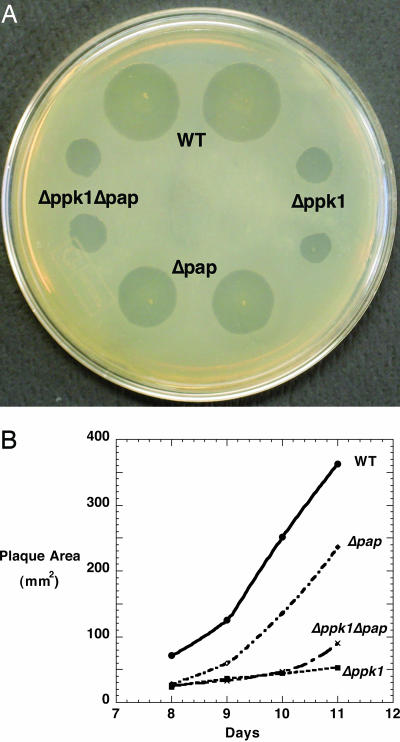
Defective Predation in Mutants. With K. aerogenes as prey in a predation assay, WT produced plaques on a lawn of bacteria on nutrient agar, which increased in size over several days (Fig. 5). The Δppk1 mutant also formed plaques but with an area only 15% of the WT even after 11 days; the Δpap plaques reached 35-65% the size of WT. The double mutant Δppk1Δpap behaved like Δppk1. The reduced plaque size of the mutants was not due to the growth and motility on the nutrient agar plate, given that WT and mutants produced swarms of the same size in the absence of K. aerogenes.
Fig. 5.
Predation on K. aerogenes. Overnight K. aerogenes cultures were plated on nutrient agar. WT and mutants (2.5 × 104 cells) were then spotted on these plates and incubated at 30°C. (A) Plaques on K. aerogenes lawn after 10 days. (B) Sizes of plaques during an 11-day period.
Direct contact of M. xanthus with its bacterial prey was essential for predation. No plaques were observed when the predator and prey were separated by a 0.22-μm membrane filter (Millipore), even when M. xanthus was applied to the bacterial lawn at 100 times the level (data not shown). Thus, the Δppk1 and Δpap mutants appear to be deficient in some cell-surface component essential for feeding on K. aerogenes.
Discussion
poly P is a unique biomolecule (2), a highly ionized reservoir of energy and phosphate, an anionic chelator of metal cations (33), and, as a polyanion sometimes complexed with an actin filament (34), can be localized and dynamic in responses to environmental stresses and strengiencies (5-8). Major bacterial pathogens rely on poly P for virulence (9-11), and eukaryotes, as in the case of D. discoideum, employ poly P in predation and the developmental cycle (12).
M. xanthus is a soil bacterium remarkable for its location and the social interactions that promote its feeding on bacterial prey. When starved, M. xanthus aggregates into fruiting bodies with resistant spores. The question of how poly P is used in the variety of myxobacterial functions prompted this study of the levels and distribution of poly P and the defects in mutants that lack one or another of the genes essential for poly P functions. Particular attention has been given to the roles of two poly P enzymes: PPK1 responsible for the synthesis of poly P from ATP and PAP that uses poly P to generate ADP from AMP.
The S-motility of M. xanthus depends on a number of features, among them the participation of the polarly localized TFP in which pilin and fibrils that contain fibril exopolysaccharides (EPS) are major components. The PPK1 mutant, which is strikingly defective in S-motility, accumulates an excessive amount of the cell-surface pilin; this hyperpiliation phenotype is likely due to an insufficient amount of fibril EPS (27) and perhaps other fibril components. Biogenesis of EPS involves genes eps and eas (35), a DnaK homolog (encoded by sglK) (36), chemotaxis homologues (dif) genes (37), and some uncharacterized dsp genes. EPS-deficient mutants fail to bind calcofluor white dye (38) and are defective in cellular agglutination, S-motility, and fruiting body formation (25, 26), phenotypes similar to those in the PPK1 mutant. Especially notable is that the S-motility of M. xanthus resembles the twitching motility of P. aeruginosa, which also depends on PPK1, mutants of which produce less alginate, a major component of P. aeruginosa EPS (K. Ishige and A.K., unpublished data).
With regard to the predation of the M. xanthus on bacterial colony or lawn, we have observed the need for direct contact with its prey. When separated by a membrane filter, M. xanthus is ineffective and thus unable to concentrate its arsenal of lytic weapons.
Another feature of M. xanthus is that its response to starvation may depend on a set of signaling genes: asg, bsg, csg, dsg, and esg. The protein encoded by bsg is homologous to the bacterial Lon protease and is essential for expression of early developmental genes (39, 40). Of particular interest is the role of poly P in the activation of Lon when an E. coli culture is switched from a rich medium to a minimal medium (6, 7). poly P accumulates in large amounts as an alarmone signal of the lack of amino acids. poly P binds Lon and increases its activity by >20-fold to generate amino acids from idled ribosomal proteins, thus providing a crucial supply to restart protein synthesis (6, 7). The large and transient accumulationofpolyPin M. xanthus 2 h after being starved in the minimal TPM buffer may well involve an activation of the Lon protease as observed in a down-shifted E. coli culture.
Acknowledgments
We thank Dr. A. Dale Kaiser (Stanford University) and his group for advice and M. xanthus strains, plasmids, and antibody. This work was supported by the National Institute of General Medical Sciences.
Abbreviations: poly P, inorganic polyphosphate; PPK1, polyphosphate kinase 1; PAP, poly P:AMP phosphotransferase; A-motility, adventurous motility; S-motility, social motility; TFP, type IV pili; EPS, exopolysaccharide.
Data deposition: The sequences of M. xanthus pap and ppk1 reported in this paper have been deposited in the GenBank database (accession nos. DQ091855 and DQ091856).
References
- 1.Kornberg, A. (1995) J. Bacteriol. 177, 491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg, A., Rao, N. N. & Ault-Riché, D. (1999) Annu. Rev. Biochem. 68, 89-125. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, K. & Kornberg, A. (1990) J. Biol. Chem. 265, 11734-11739. [PubMed] [Google Scholar]
- 4.Zhang, H., Ishige, K. & Kornberg, A. (2002) Proc. Natl. Acad. Sci. USA 99, 16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao, N. N. & Kornberg, A. (1996) J. Bacteriol. 178, 1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda, A., Tanaka, S., Ikeda, T., Kato, J., Takiguchi, N. & Ohtake, H. (1999) Proc. Natl. Acad. Sci. USA 96, 14264-14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuroda, A., Nomura, K., Ohtomo, R., Kato, J., Ikeda, T., Takiguchi, N., Ohtake, H. & Kornberg, A. (2001) Science 293, 705-708. [DOI] [PubMed] [Google Scholar]
- 8.Shiba, T., Tsutsumi, K., Yano, H., Ihara, Y., Kameda, A., Tanaka, K., Takahashi, H., Munekata, M., Rao, N. N. & Kornberg, A. (1997) Proc. Natl. Acad. Sci. USA 94, 11210-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rashid, M. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid, M. H., Rao, N. N. & Kornberg, A. (2000) J. Bacteriol. 182, 225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashid, M. H., Rumbaugh, K., Passador, L., Davies, D. G., Hamood, A. N., Iglewski, B. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, H., Gomez-Garcia, M. R., Brown, M. R. W. & Kornberg, A. (2005) Proc. Natl. Acad. Sci. USA 102, 2731-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiba, T., Itoh, H., Kameda, A., Kobayashi, K., Kawazoe, Y. & Noduchi, T. (2005) J. Bacteriol. 187, 1859-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi, X., Rao, N. N. & Kornberg, A. (2004) Proc. Natl. Acad. Sci. USA 101, 17061-17065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dworkin, M. (1996) Microbiol. Rev. 60, 70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg, E. (1984) Myxobacteria: Development and Cell Interactions (Springer, New York).
- 17.Hodgkin, J. & Kaiser, D. (1979) Mol. Genet. 171, 177-191. [Google Scholar]
- 18.Bradley, D. E. (1972) J. Gen. Microbiol. 72, 303-319. [DOI] [PubMed] [Google Scholar]
- 19.Sun, H., Zusman, D. & Shi, W. (2000) Curr. Biol. 10, 1143-1146. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S. K. & Kaiser, D. (1990) Genes Dev. 4, 896-905. [DOI] [PubMed] [Google Scholar]
- 21.Sager, B. & Kaiser, D. (1993) Genes Dev. 7, 1645-1653. [DOI] [PubMed] [Google Scholar]
- 22.Sager, B. & Kaiser, D. (1993) Proc. Natl. Acad. Sci. USA 90, 3690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser, D. (1979) Proc. Natl. Acad. Sci. USA 76, 5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, S. S. & Kaiser, D. (1995) Mol. Microbiol. 18, 547-558. [DOI] [PubMed] [Google Scholar]
- 25.Shimkets, L. J. (1986) J. Bacteriol. 166, 837-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimkets, L. J. (1986) J. Bacteriol. 166, 842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y., Sun, H., Ma, X., Lu, A., Lux, R., Zusman, D. & Shi, W. (2003) Proc. Natl. Acad. Sci. USA 100, 5543-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowden, M. G. & Kaplan, H. B. (1998) Mol. Microbiol. 30, 275-284. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa, N., DeRisi, J. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, S. S. & Kaiser, D. (1996) J. Bacteriol. 178, 5817-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otani, M., Inouye, M. & Inouye, S. (1995) J. Bacteriol. 177, 4261-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, S. S. & Kaiser, D. (1997) J. Bacteriol. 179, 7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown, M. R. W. & Kornberg, A. (2004) Proc. Natl. Acad. Sci. USA 101, 16085-16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez-Garcia, M. R. & Kornberg, A. (2004) Proc. Natl. Acad. Sci. USA 101, 15876-15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, A., Cho, K., Black, W. P., Duan, X., Lux, R., Yang, Z., Kaplan, H. B., Zusman, D. R. & Shi, W. (2005) Mol. Microbiol. 55, 206-220. [DOI] [PubMed] [Google Scholar]
- 36.Weimer, R. M., Creighton, C., Stassinopoulos, A., Youderian, P. & Hartzell, P. L. (1998) J. Bacteriol. 180, 5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancero, H., Brofft, J. E., Downard, J., Birren, B. W., Nusbaum, C., Naylor, J., Shi, W. & Shimkets, L. J. (2002) J. Bacteriol. 184, 1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramaswamy, S., Dworkin, M. & Downard, J. (1997) J. Bacteriol. 179, 2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gill, R. E. & Cull, M. G. (1986) J. Bacteriol. 168, 341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill, R. E., Karlok, M. & Benton, D. (1993) J. Bacteriol. 175, 4538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]