Abstract
Numerous cellular mRNAs encoding proteins critical during cell stress, apoptosis, and the cell cycle seem to be translated by means of internal ribosome entry sequences (IRES) when cap-dependent translation is compromised. The underlying molecular mechanisms are largely unknown. Using a HeLa-based cell-free translation system that mirrors the function of cellular IRESs in vitro, we recently demonstrated that translation from the c-myc IRES continues after proteolytic cleavage of eukaryotic translation initiation factor (eIF) 4G. To address the role of eIF4G in cellular IRES-driven translation directly, we immunodepleted eIF4GI from the HeLa cell translation extracts. After efficient depletion of eIF4GI (>90%), both cap-dependent and c-myc IRES-dependent translations are diminished to residual levels (<5%). In striking contrast to cap-dependent translation, c-myc IRES-dependent translation is fully restored by addition of the conserved middle fragment of eIF4GI, harboring the eIF3- and eIF4A-binding sites. p97, an eIF4G-related protein that has been described both as an inhibitor of translation and as a modulator of apoptosis, not only suffices to also rescue c-myc IRES-driven (but not cap-dependent) translation, but it even superinduces IRES-mediated translation 3-fold compared with nondepleted extracts. Interestingly, both p97 and the middle fragment of eIF4GI also rescue translation driven by proapoptotic (p97) and antiapoptotic [X-linked inhibitor of apoptosis (XIAP) and cellular inhibitor of apoptosis 1 (c-IAP1)] IRESs, reflecting a broader role of these polypeptides in cellular IRES-mediated translation and indicating their importance in apoptosis.
For most eukaryotic mRNAs, translation initiation involves binding of the small ribosomal subunit to the capped 5′ end of the mRNA followed by scanning to the start codon. The eukaryotic initiation factor (eIF) 4G plays a key role in cap-mediated ribosome recruitment. The function of eIF4G includes circularization of the mRNA by interaction with the cap-binding protein eIF4E and the poly(A)-binding protein (PABP), delivery of the RNA-helicase eIF4A to the 5′ end of the mRNA, and bridging of mRNA and 40S ribosomal subunit by means of eIF3 (1-3).
Internal ribosomal entry does not require a 5′ 7mGpppN cap-structure (4, 5) and has been described as an alternative mechanism of translation initiation for many viral RNAs and an increasing number of cellular mRNAs (6, 7). Interestingly, many of these cellular mRNAs encode proteins involved in important cellular processes, including development, cell cycle, apoptosis, wound repair, and angiogenesis (8-15).
The mRNA encoding the transcription factor c-myc can be translated by means of an internal ribosomal entry sequence (IRES) in its 5′ UTR (16, 17). c-myc plays a critical role in the control of both cell proliferation/differentiation and apoptosis. Deregulation of c-myc expression is associated with a wide range of hematopoietic neoplasms, and the c-myc IRES has been implicated in the development of multiple myeloma (18, 19). The c-myc IRES is still active during apoptosis or cellular stress when cap-dependent translation is inhibited (20-22).
Internal ribosomal entry is also involved in the translation of pro- and antiapoptotic proteins. The proteins of the inhibitor of apoptosis (IAP) family are potent cellular suppressors and modulate the cellular response to apoptotic stimuli (23). Translation of two members of the IAP-family, X-linked inhibitor of apoptosis (XIAP) and cellular inhibitor of apoptosis 1 (c-IAP1), can be driven by IRES-elements that remain active during cellular stress (10, 21, 24, 25). The eIF4G-related protein p97 (also named DAP5 or NAT-1) was described as a suppressor of translation (26) as well as a modulator of apoptosis (27). The 5′ UTR of the p97 mRNA contains an IRES element that is stimulated by the p97 protein (15).
Viral IRESs employ diverse molecular mechanisms for function and have distinct requirements for canonical initiation factors. Although translation from the hepatitis C virus (HCV) IRES and the cricket paralysis virus (CrPV) IRES are completely independent of eIF4G (28-30), the IRES of encephalo-myocarditis virus (EMCV) binds eIF4G directly (independent of eIF4E) and promotes 40S recruitment by means of this direct RNA-protein interaction (31).
eIF4G is organized into three domains: the N-terminal fragment harbors eIF4E- and PABP-binding sites, the middle fragment interacts with eIF4A and eIF3, and the C-terminal fragment harbors a second eIF4A-binding site as well as a binding site for the Mnk1 kinase. The eIF3-/eIF4A-binding region located in the middle fragment of eIF4GI is highly conserved in many species as well as in the eIF4G-related protein p97 (27). This middle fragment of eIF4G together with eIF4A is sufficient for EMCV IRES-mediated translation initiation (32) and can also stimulate cap-independent translation from RNAs without IRES elements (33).
Although IRES-binding transacting factors (ITAFs) (6) and eIF2α phosphorylation (8, 34) have been implicated in IRES function (7), much remains to be learned about the molecular mechanism of cellular IRES-mediated translation and the role, if any, of eIF4G during internal ribosomal entry. Several cellular IRESs remain active under conditions where eIF4G is cleaved, e.g., during apoptosis and infections (35-37). On the basis of cell culture experiments, eIF4G cleavage products, as well as p97 and its cleavage product p86, have been implicated in the function of several cellular IRESs (15, 21, 38). Here, we demonstrate directly, employing biochemical depletion-readdition experiments, that the conserved eIF3-/eIF4A-binding domain of eIF4GI promotes translation driven by the c-myc IRES as well as pro- and antiapoptotic IRESs in the absence of full-length eIF4GI. These results directly implicate eIF4GI in cellular IRES-mediated translation. We furthermore show that recombinant p97 not only rescues, but even superinduces cellular IRES-mediated translation in eIF4GI-depleted HeLa cell extracts.
Materials and Methods
Plasmids. All constructs containing the firefly luciferase reporter are in a pBluescript-KS+ vector (Stratagene). The pT3Luc(pA), the pMycLuc(pA), and pHCV-IRES-Luc plasmids have been described (39). To block 5′ end-dependent translation in addition to using an ApppG cap structure, a synthetic DNA fragment (5′-CCCGGAGCGCCCAGATCTGGGCGCTCCGGGGTAC-3′) was inserted 19 nt downstream from the transcription start site of pMycLuc(pA) to introduce a stable stem loop structure (ΔG = -243 kJ/mol) (pSLMLA). To minimize the interference of this inserted structure with the IRES, a 200-nt segment amplified from the human β-globin ORF (corresponding to nucleotides 254-448, GenBank accession no. AF117710) was inserted between stem loop and c-myc IRES [SL200MLuc(pA)].
DNA fragments corresponding to the IRES sequences (http://ifr31w3.toulouse.inserm.fr/IRESdatabase) were amplified from HeLa cell RNA by RT-PCR by using primers 5′-aaactcgagtgaatgatgtggtaatgtcg-3′ (sense) and 5′-aaaccatggcttctcttgaaaataggacttgtcc-3′ (antisense) for the XIAP IRES (10); 5′-aaaggtaccaatataaactgagataaatccagtaaag-3′ (sense) and 5′aaaccatgggagtaggtgacagtactgtttg-3′ (antisense) for the c-IAP1 IRES (40); as well as 5′-aaactcgagcagcagtgagtcggagctctatgg-3′ (sense) and 5′-aaaccatggtttggcggcttgacaacgaagaatc-3′ (antisense) for the p97 IRES (27). The c-myc IRES sequence in pSL200MLuc(pA) was then replaced by the respective other IRES sequences to obtain pSL200XLuc(pA) (XIAP IRES), and pSL200CLuc(pA) (c-IAP1 IRES). The c-myc IRES sequence in pSL200MLuc(pA98) [a plasmid analogous to pSL200MLuc(pA), but containing a 98nt-pA tail] was replaced by the p97 IRES to obtain SL200PLuc(pA). All constructs were verified by restriction enzyme analysis and DNA sequencing.
The expression plasmids encoding the middle domain and the middle plus C-terminal domain of eIF4GI (pGEX4G935 and pGEX4G1404) have been described (33). To express the p97-ORF (26) in Escherichia coli, the p97 cDNA (IMAGp958L20820Q2, RZPD Deutsches Ressourcenzentrum für Genomforschung, Heidelberg) was cloned into the pET16b-vector (Novagen). The construct was verified by DNA sequencing.
In Vitro Transcription and Translation. In vitro transcription of mRNAs in the presence of either 7mGppG or ApppG, the preparation of HeLa cell extracts, and in vitro translation assays have been described (41). The conditions for the in vitro translation assays were 20% HeLa extracts, 2.1 mM MgOAc, and 70 mM KOAc for cap- and cellular IRES-dependent translation, and 4.5 mM MgOAc and 145 mM KOAc for HCV IRES-driven translation. All experiments were performed in micrococcal nuclease-treated HeLa cell extracts (39). The concentration of exogenous mRNAs was 7 nM for all luciferase reporter mRNAs, except for the HCV IRES-mRNA (1.5 nM). Translation reactions were incubated at 37°C for 30 min.
eIF4GI Depletion from HeLa Extracts. Immunopurified polyclonal rabbit anti-eIF4GI antibodies and magnetic protein A beads (Dynal, Great Neck, NY) were incubated at a ratio of 0.25 μg of antibody per μl of bead slurry for 1 h at room temperature. Beads were washed three times with PBS and incubated at a ratio of 0.25 μl of micrococcal nuclease-pretreated HeLa cell extract per μl of bead slurry for 1 h on ice. Small aliquots of the eIF4GI-depleted supernatant were frozen in liquid nitrogen and stored at -80°C. For mock depletion, HeLa cell extract was incubated at the same ratio with protein A beads, which were not coupled to anti-eIF4GI antibodies.
Recombinant Proteins. His-p97 was expressed in E. coli BL21/DE3 and purified on Ni-NTA Sepharose (Qiagen, Valencia, CA) by standard procedures. GST-4G1404 and GST-4G935 were expressed, purified, and cleaved as described (33, 39). All proteins were dialyzed against 100 mM KCl, 20 mM Hepes (pH7.6), and 1 mM DTT.
Western Blotting and Antibodies. Polyclonal anti-eIF4GI antibodies were raised against the middle plus C-terminal domain of human eIF4GI [GST-4G1404, purified on glutathione beads (Amersham Pharmacia)] in two rabbits. Affinity purification was carried out on CNBr-activated Sepharose beads coupled to GST-4G1404. Polyclonal anti-PABP antibodies and anti-eIF4A antibodies were raised against the respective N-terminal His-tagged full-length proteins [purified on Ni-NTA Sepharose (Qiagen)] in two rabbits, each.
The following antibodies were used for Western blotting: rabbit anti-eIF4GI antibody (1:250 dilution); rabbit anti-eIF4GII antibody (kindly provided by N. Sonenberg, McGill University, Montreal, Canada) (1:1,500 dilution); rabbit anti-p97 antibody (kindly provided by T. Preiss, University of New South Wales, Sydney, Australia) (1:500 dilution); rabbit anti-PABP antibody (1:500 dilution); rabbit anti-eIF4A antibody (1:500 dilution); and rabbit anti-eIF4E antibody (Cell Signaling Technology, Beverly, MA) (1:500 dilution).
Samples were separated by SDS/PAGE and electroblotted onto poly(vinylidene difluoride) (PVDF) membranes (Immobilon, Millipore). Protein signals were detected by using the enhanced chemiluminescent (ECL) procedure.
Results
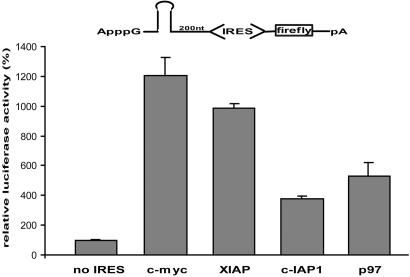
c-myc, XIAP, c-IAP1, and p97 IRES Elements Are Active in HeLa Cell Extracts. ORFs can be translated by 7mGpppN-mediated translation, by means of cap-structure independent but 5′ end-dependent ribosome entry, or by IRES-mediated internal ribosome recruitment. To strictly monitor the last and exclude the former two modes of translation, we designed reporter mRNAs bearing an ApppG instead of the canonical 7mGpppN cap structure at the 5′ end followed by a very stable stem loop structure (ΔG = -243 kJ/mol). These modifications inhibit 5′ end-dependent ribosome entry by 99.5% (data not shown). The IRES elements under study are inserted 200 nt downstream from the hairpin following a spacer element, with an mRNA lacking an IRES insertion serving as a negative control. Reporter mRNAs containing the c-myc, XIAP, c-IAP1, or p97 IRES are translated 4 (c-IAP1) to 12 (c-myc) times more efficiently than the control (Fig. 1).
Fig. 1.
Cellular IRESs are active in HeLa cell extracts. (A) Schematic representation of the IRES-containing reporter mRNAs is shown above the bar graphs. HeLa cell extracts were programmed with ApppG-capped c-myc, XIAP, c-IAP1, or p97 IRES reporter mRNAs bearing a stable hairpin upstream of a spacer sequence and the IRES. A reporter mRNA lacking an IRES element upstream of the firefly luciferase ORF was used as negative control. Luciferase activity obtained with the control mRNA was defined as 100%. Error bars denote the standard deviation from the mean of at least three independent experiments.
Reporter mRNAs bearing an ApppG cap but lacking the stem loop structure allow 5′ end-dependent (but cap-independent) translation initiation, and observed luciferase activity from such IRES constructs reflects the contribution of both 5′ end-dependent and IRES-dependent translation initiation. Whereas the c-myc IRES contributes most of the observed translational activity of a construct lacking the stem loop structure, the IRES contribution increases to close to 100% of the total after the insertion of the stem loop structure (Fig. 5, which is published as supporting information on the PNAS web site).
These results show that translation driven by these cellular IRESs is reconstituted in the HeLa cell extracts, facilitating mechanistic studies without confounding indirect effects that cannot be eliminated in experiments by using living cells.
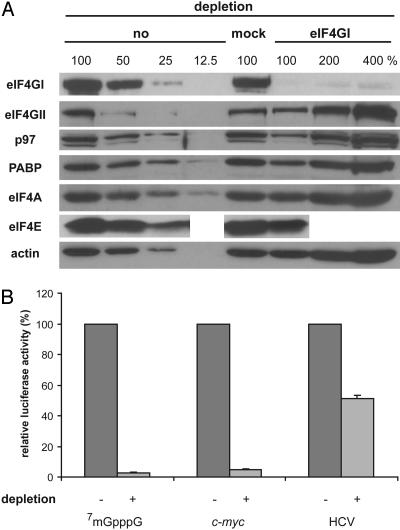
Depletion of eIF4GI from HeLa Cell Extracts Abolishes Cap- and c-myc IRES-Driven Translation. To investigate the role of eIF4GI in cellular IRES-driven translation, we specifically immunodepleted eIF4GI from the cell-free translation extracts. By using immunopurified polyclonal antibodies, >90% of endogenous eIF4GI is depleted from these HeLa cell extracts without a significant reduction of eIF4GII and eIF4A. Approximately 50% of p97, PABP, and eIF4E are codepleted together with eIF4GI (Fig. 2A). Consistent with the crucial role of intact eIF4G in cap-dependent translation and the eIF4G-independent mode of HCV IRES-mediated translation (28, 30), cap-dependent translation is almost completely abolished (to <5%), whereas HCV IRES-driven translation is far less affected (≈2-fold) in eIF4GI-depleted HeLa cell extracts (Fig. 2B). Depletion of eIF4GI inhibits translation from the c-myc IRES as effectively as cap-mediated translation.
Fig. 2.
Immunodepletion of eIF4GI from HeLa cell extracts abolishes cap- and cellular IRES-driven translation. (A) Western blot analysis of eIF4GI, eIF4GII, p97, PABP, eIF4A, eIF4E, and actin in non-, mock-, and eIF4GI-depleted HeLa cell extracts. To facilitate quantitative comparisons, different dilutions are loaded, as indicated. (B) Mock-depleted (-) or eIF4GI-depleted (+) HeLa cell extracts were programmed with a 7mGpppG-capped reporter mRNA, an ApppG-capped c-myc IRES reporter mRNA bearing a stable hairpin upstream of a spacer sequence and the IRES, or an ApppG-capped HCV IRES reporter mRNA, as indicated. Translation of the respective mRNA in mock-depleted HeLa cell extracts was defined as 100%. Error bars denote the standard deviation from the mean of at least three independent experiments.
These results show that eIF4GI is critical for c-myc IRES translation and that the endogenous eIF4GII cannot replace eIF4GI in this system. Moreover, although c-myc IRES translation does not require intact eIF4GI (39), this result indicates that it absolutely requires a factor that has been depleted, possibly a cleavage fragment of eIF4GI. We did not expect that the eIF4GI depletion would affect HCV IRES translation (a specific 2-fold effect), because the HCV IRES functions without initiation factors of the eIF4 group (28, 30). This result was not pursued further; it suggests that a factor contributing to HCV IRES function was (co) depleted.
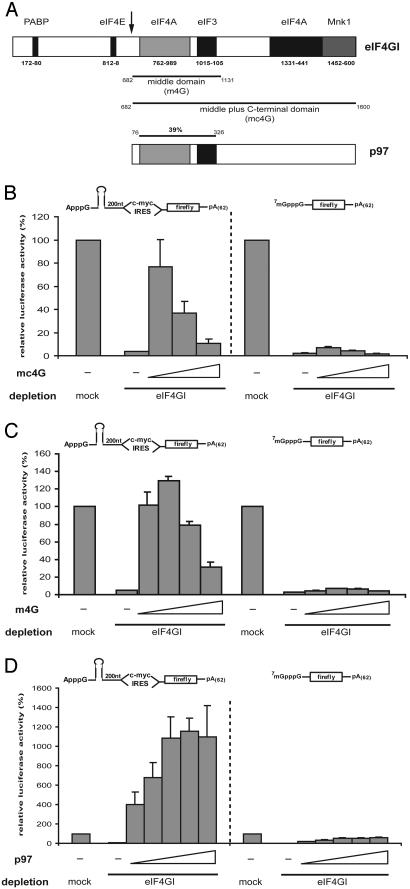
The Conserved Middle Domain of eIF4GI Mediates c-myc IRES Translation. During polioviral infection, protease 2A cleaves an N-terminal fragment from the C-terminal two-thirds of eIF4GI (Fig. 3A). This cleavage separates the binding sites for eIF4E and PABP from the rest of the molecule. Similar eIF4G cleavage fragments are generated during apoptosis by caspase 3 (36). We have recently shown that treatment of the HeLa cell extracts with coxackieviral protease 2A cleaves eIF4GI and eIF4GII and stimulates c-myc IRES-driven translation in vitro (39). This experiment does not distinguish between causative effects of cleaving eIF4GI, eIF4GII, or a different protein that could stimulate the IRES.
Fig. 3.
The middle domain of eIF4GI or p97 mediates c-myc IRES-driven translation in eIF4GI-depleted HeLa cell extracts. (A) Schematic representations of eIF4GI and p97. The coxsackieviral protease 2A cleavage site is indicated by the arrow. The C-terminal two-thirds (mc4G) and the middle fragment (m4G) of eIF4GI are indicated beneath the representation of eIF4GI. The conserved eIF3-/eIF4A-binding region (39% identity) is indicated above the representation of p97. (B-D) Mock- or eIF4GI-depleted HeLa cell extracts were programmed with the indicated mRNAs in the presence of dialysis buffer (-), mc4G (2 pmol, 6 pmol, and 10 pmol, respectively), m4G (2 pmol, 6 pmol, 12 pmol, and 20 pmol, respectively), or p97 (0.5 pmol, 1 pmol, 2.5 pmol, 5 pmol, and 10 pmol, respectively). A schematic representation of the reporter mRNAs is shown above the bar graphs. Translation of the respective mRNA in mock-depleted HeLa cell extracts was defined as 100%. Error bars denote the standard deviation from the mean of at least three independent experiments.
To test the involvement of eIF4GI in c-myc IRES-driven translation directly, a recombinant polypeptide corresponding to the middle and C-terminal domain of eIF4GI (mc4G) was added to eIF4GI-depleted HeLa cell extracts. Addition of only 2 pmol of mc4G (corresponding to ≈70% of endogenous eIF4GI; Fig. 6A, which is published as supporting information on the PNAS web site) stimulates c-myc IRES-dependent translation nearly 20-fold, reaching almost 80% of translation in mock-depleted HeLa cell extracts (Fig. 3B). By contrast, cap-dependent translation is stimulated marginally at best by addition of up to 10 pmol of mc4G.
The C-terminal domain of eIF4GI has been shown to have a modulating effect on cap-dependent translation (42), whereas the middle domain alone (m4G) is sufficient for EMCV IRES-driven translation initiation (32). We therefore tested whether m4G mediates c-myc IRES-dependent translation. Addition of 2 pmol of m4G (≈70% of endogenous eIF4GI) fully (100%) rescues c-myc IRES-driven, but not cap-dependent translation in eIF4GI-depleted HeLa cell extracts (Fig. 3C). Maximal stimulation of c-myc IRES-driven translation (to ≈130%) is observed after addition of 6 pmol of m4G (≈200% of endogenous eIF4GI). In contrast, addition of even up to 20 pmol of m4G fails to reconstitute cap-dependent translation, as expected.
Addition of purified eIF4F from rabbit reticulocyte lysate fully restores both cap- and c-myc IRES-driven translation in eIF4GI-depleted HeLa cell extracts, indicating further the specificity of the eIF4GI depletion procedure (Fig. 7, which is published as supporting information on the PNAS web site).
These results and the analysis of a bicistronic reporter system (Fig. 8, which is published as supporting information on the PNAS web site) reveal that the middle fragment of eIF4GI harboring the eIF3/eIF4A-binding regions represents a critical initiation factor for c-myc IRES-mediated translation.
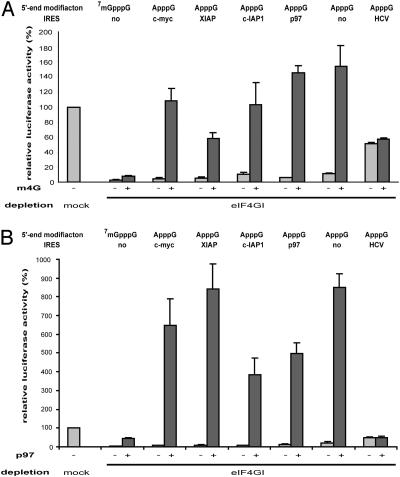
Multiple Cellular IRESs for Translation of Pro- and Antiapoptotic Proteins Require m4G as a Critical Initiation Factor. Several cellular IRESs have been reported to be active during apoptosis (20, 21, 25), when mammalian eIF4GI is cleaved and cap-dependent translation is impaired (36, 37). The central part of caspase-cleaved eIF4GI resembles m4G but includes the eIF4E-binding site located within the N-terminal third of eIF4GI (36). To explore whether m4G is also a critical factor for IRES-mediated translation of pro- and antiapoptotic proteins, we tested the role of m4G for the IRES-elements of the mRNAs encoding XIAP, c-IAP1, and p97 (Dap5/NAT-1).
Like c-myc, translation mediated by any of these IRES elements is severely inhibited in eIF4GI-depleted HeLa extracts (to 5-10%) reflecting a more general importance of eIF4GI in cellular IRES-driven translation (- lanes in Fig. 4A). Addition of 6 pmol of m4G (200% of endogenous eIF4GI) strongly stimulates the translation of all cellular IRESs tested (10-fold for XIAP and c-IAP1, 25-fold for p97), resulting in a complete rescue of p97 IRES and c-IAP1 IRES-driven translation. Translation mediated by the XIAP IRES is restored to ≈60% by 6 pmol of m4G, whereas addition of up to 20 pmol of m4G does not have an additional effect (data not shown).
Fig. 4.
Role of m4G and p97 in translation driven by pro- and antiapoptotic IRESs. Mock- or eIF4GI-depleted HeLa cell extracts were programmed with the indicated mRNAs in the presence of dialysis buffer (-), 6 pmol of m4G (A), or 5 pmol of p97 (B). Messenger RNAs bearing a cellular IRES element are furnished with the strong stem loop structure at their 5′ end (Fig. 1) to ensure that bona fide IRES activity is monitored, whereas mRNAs without cellular IRES element lack this structure to permit translation initiated from the 5′ end. The HCV-IRES control construct was used as published (39) and also lacks the stem loop structure. Translation of the respective mRNAs in mock-depleted HeLa cell extracts was defined as 100%. Error bars denote the standard deviation from the mean of at least three independent experiments.
It has been reported that translation of uncapped mRNAs is stimulated by m4G (33). Therefore, we also tested the translation of an mRNA lacking an IRES and the 7mGpppG cap structure. The relative degree of stimulation of translation in eIF4GI-depleted HeLa cell extracts of this mRNA is comparable with that of mRNAs bearing a cellular IRES (Fig. 4A). In contrast, HCV IRES-driven translation is not affected by addition of m4G, demonstrating the specificity of the observed effects.
When added to mock-depleted HeLa cell extracts, m4GI also stimulates the translation from cellular IRESs and ApppG-capped mRNA (Fig. 9A, which is published as supporting information on the PNAS web site). As expected, it inhibits 7mGpppG-capped mRNA translation by means of the endogenous, full-length eIF4G, and it does not affect translation from the HCV-IRES. Depletion of eIF4GI or addition of m4G does not alter reporter mRNA stability in the HeLa cell extracts (Fig. 10, which is published as supporting information on the PNAS web site).
An Effective Second eIF4GI-Independent Mode of Cellular IRES Translation by Means of p97. The N-terminal part of the eIF4G-related protein p97 is highly similar [39% identity, 63% similarity (27)] to m4G, and its role in translation is subject to intensive investigations (21, 26, 38). Thus, we asked whether p97 could substitute for m4G in eIF4GI-depleted extracts. Strikingly, addition of only 0.5 pmol of recombinant p97 (20% of endogenous p97; Fig. 6B) to eIF4GI-depleted HeLa cell extracts not only rescues c-myc IRES-dependent translation, but even super-induces it 4-fold compared with mock-depleted extracts (Fig. 3D). Maximal stimulation of c-myc IRES-driven translation is observed after addition of 5 pmol of p97 (≈200% of endogenous p97). By contrast, cap-dependent translation is elevated modestly (to ≈40%) (Fig. 3D), and HCV IRES-dependent translation is not affected by addition of p97 to eIF4GI-depleted HeLa cell extracts (Fig. 4B).
To investigate the effects of p97 on pro- and antiapoptotic IRESs, we tested its ability to restore translation mediated by p97, XIAP, and c-IAP1 IRESs in eIF4GI-depleted HeLa cell extracts. Addition of 5 pmol of recombinant p97 to eIF4GI-depleted HeLa extracts mediates a strong stimulation of cellular IRES-driven translation ranging from 40-fold (c-IAP1 IRES) to 100-fold (c-myc-IRES) (Fig. 4B). p97 rescues translation mediated by pro- and antiapoptotic IRESs in eIF4GI-depleted extracts and even superinduces translation 4- to 8.5-fold above translation of mock-depleted HeLa extracts. Similar to m4G (Fig. 4A), p97 also exerts its effect on an ApppG-capped mRNA lacking an IRES. Depletion of eIF4GI or addition of p97 does not alter reporter mRNA stability in HeLa cell extracts (Fig. 10). Addition of p97 to mock-depleted HeLa cell extracts exerts similar effects on all tested mRNAs (Fig. 9B).
To exclude the possibility that the stimulatory effect of p97 and m4GI could result from “nonspecific recruitment” of ribosomes to the 200-nt spacer region located between the stem loop and the IRES, we tested also the effect of p97 and m4GI, respectively, on translation of the c-myc IRES reporter mRNA lacking this spacer region. Stimulation of c-myc IRES-driven translation is similar to using reporter mRNAs with or without the 200-nt spacer region (Fig. 11, which is published as supporting information on the PNAS web site, and Fig. 4). These results show that recombinant p97 and m4G can both mediate the translation of the same spectrum of tested mRNAs, suggesting that p97 can provide a second, eIF4GI-independent mode of cellular IRES translation.
Discussion
Numerous eukaryotic mRNAs involved in important physiological processes have the remarkable ability of being translated by means of internal ribosomal entry. Although their 7mGpppN-cap structure allows them to be translated by the eIF4E-mediated mechanism, the IRES endows them with a second alternative that may be physiologically relevant, particularly when cap-dependent translation is compromised. The molecular mechanism of this alternative mode of translation for cellular mRNAs is poorly understood, and little is known about the role of eIF4GI, a factor that plays a central role in cap-dependent translation.
To investigate the role of eIF4GI in cellular IRES-driven translation, we depleted eIF4GI from a cell-free translation system that mirrors the function of cellular IRESs (Fig. 1 and ref. 39). The loss of cellular IRES-driven translation after effective immunodepletion of eIF4GI and the recovery of translation after readdition of recombinant m4G (Fig. 4A) and purified eIF4F (Fig. 7) demonstrate the specificity of the depletion procedure and the importance of eIF4GI for cellular IRES-function (Fig. 2). eIF4GII has been described as a functional homologue of eIF4GI (43). Surprisingly, eIF4GII is not able to substitute for eIF4GI in the cell-free system, because translation is severely inhibited, whereas eIF4GII levels are not reduced after eIF4GI-depletion (Fig. 2). Because eIF4GII is abundant in HeLa cell extracts (Fig. 12 A, which is published as supporting information on the PNAS web site), codepletion of a factor required for eIF4GII (but not m4G or p97) function could explain this unexpected observation. Alternatively, eIF4GII may not be functional with the cellular IRESs tested here. The latter hypothesis is supported by the fact that the middle fragment of eIF4GII (m4GII) only marginally stimulates c-myc IRES-driven translation in nondepleted HeLa cell extracts and m4GII supplementation fails to restore c-myc IRES-driven translation in eIF4GI-depleted HeLa cell extracts (Fig. 12B).
Viral IRESs employ a diversity of mechanisms with grossly different eIF4G requirements (e.g., hepatitis A virus and EMCV versus HCV and cricket paralysis virus (28-31, 44, 45)). Regarding the four different cellular IRESs that we have tested here, the mechanistic similarities among them prevail: they are all inhibited by eIF4GI depletion, their translation is rescued by the middle domain of eIF4GI, and p97 can substitute for m4G, activating their translation very strongly.
The function of p97 in cellular IRES-driven translation is poorly understood, and conflicting data about the effects of p97 in translation have been reported. Initially, p97 was described as a general inhibitor of translation in vivo (26). Overexpression of p97 resulted in an inhibition of cap- and EMCV IRES-dependent translation of reporter mRNAs by 50% and of endogenous protein synthesis by 20%. In contrast, Nevins et al. (21) did not observe any effect of p97 on EMCV IRES-driven (or c-myc- or XIAP IRES-driven) translation, but detected a >20-fold stimulation of p97 IRES function (21). On the other hand, inhibition of cap-dependent translation by p97 was reported by Henis-Korenblit et al. (38), and, according to their data, p97 does not affect p97 IRES-driven translation. In nondepleted rabbit reticulocyte lysate, c-myc IRES-driven translation was not stimulated by p97 (46).
Here, we show that recombinant p97 strongly stimulates the translation mediated by the c-myc, p97, XIAP, and c-IAP1 IRESs, but not cap- or HCV IRES-dependent translation (Fig. 4). The strong stimulation by recombinant p97 is surprising and interesting, particularly because the amount of recombinant p97 added (5 pmol) is only ≈2-fold higher than the level of endogenous p97 in the HeLa cell extract. It seems as if the recombinant p97 is more active than the endogenous protein. It has been reported that the C terminus of p97 is cleaved during apoptosis and p97 is processed to its active form p86 (15, 21, 38). One may speculate that posttranslational modifications that suppress p97 activity may occur at the C terminus of p97 expressed in eukaryotic cells but not in E. coli. As a consequence, endogenous p97 is activated when the C terminus is removed, and the recombinant p97 from E. coli would be constitutively active. Because the effect of recombinant p97 is strong and is observed at relatively low concentrations of the protein, we consider it unlikely to represent a biochemical artifact.
The data reported here are consistent with a simple model for the function of the examined cellular IRESs: as in cap-dependent translation, eIF4G serves as an adaptor for the recruitment of the small ribosomal subunit (2). Although eIF4E and PABP promote highly efficient eIF4G binding to support cap-dependent translation, the ability of a noncapped mRNA (or a capped mRNA when eIF4E-mediated translation is disabled) to be translated depends on its ability to (directly or indirectly) bind eIF4G. The inefficient but detectable cap-independent translation of an mRNA without an IRES and the more efficient IRES-mediated translation may not differ fundamentally in terms of mechanism, but quantitatively in terms of their ability to recruit eIF4G (and/or appropriate proteolytic fragments of it). In this context, it is not surprising that the PABP- and eIF4E-binding N terminus of eIF4G and the C terminus that can modulate the function of eIF4G in cap-dependent translation are dispensable for the IRES-driven mechanism. The ability of p97 to take over this function supports the notion that the conserved property of binding eIF3 and eIF4A represents an important functional parameter.
This simple model predicts a continuum of cellular IRES activities as a function of their abilities to recruit eIF4G and/or p97, starting from just above the background of an uncapped mRNA to the most efficient cellular IRESs. Such a model is fully supported by the finding that m4G can mediate the translation of the downstream cistron of a bicistronic mRNA in vivo when tethered to the intercistronic region (47).
In summary, this work uncovers a common denominator of the function of several cellular IRESs: the role of eIF4G, its proteolytic cleavage products, and the related protein p97. In addition to their mechanistic implications, the findings highlight the role of eIF4G cleavage and p97 in apoptosis.
Supplementary Material
Acknowledgments
We thank our colleagues N. Sonenberg and T. Preiss for their generous gift of antibodies. We thank K. Beckmann (EMBL, Heidelberg) for purified eIF4F and K. Beckmann, K. Duncan, B. Galy, and R. Thermann for helpful discussions. P.H. was funded by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft. C.T. was funded by postdoctoral fellowships from Fritz-Thyssen-Stiftung and the Deutsche Forschungsgemeinschaft. M.W.H. gratefully acknowledges funding from the Gottfried-Wilhelm-Leibniz Prize by the Deutsche Forschungsgemeinschaft.
Abbreviations: eIF, eukaryotic translation initiation factor; PABP, poly(A)-binding protein; IRES, internal ribosome entry sequence; IAP, inhibitor of apoptosis; XIAP, X-linked inhibitor of apoptosis; c-IAP1, cellular inhibitor of apoptosis 1; HCV, hepatitis C virus; EMCV, encephalomyocarditis virus; mc4G, middle plus C-terminal domain of eIF4GI; m4G, middle domain of eIF4GI.
References
- 1.Gingras, A. C., Raught, B. & Sonenberg, N. (1999) Annu. Rev. Biochem. 68, 913-963. [DOI] [PubMed] [Google Scholar]
- 2.Hentze, M. W. (1997) Science 275, 500-501. [DOI] [PubMed] [Google Scholar]
- 3.Imataka, H., Gradi, A. & Sonenberg, N. (1998) EMBO J. 17, 7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier, J. & Sonenberg, N. (1988) Nature 334, 320-325. [DOI] [PubMed] [Google Scholar]
- 5.Jang, S. K., Krausslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C. & Wimmer, E. (1988) J. Virol. 62, 2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellen, C. U. & Sarnow, P. (2001) Genes Dev. 15, 1593-1612. [DOI] [PubMed] [Google Scholar]
- 7.Holcik, M. & Sonenberg, N. (2005) Nat. Rev. Mol. Cell Biol. 6, 318-327. [DOI] [PubMed] [Google Scholar]
- 8.Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A. A., Koromilas, A. E., Zhou, L., Snider, M. D., Scheuner, D., Kaufman, R. J. & Hatzoglou, M. (2003) Cell 113, 519-531. [DOI] [PubMed] [Google Scholar]
- 9.Coldwell, M. J., Mitchell, S. A., Stoneley, M., MacFarlane, M. & Willis, A. E. (2000) Oncogene 19, 899-905. [DOI] [PubMed] [Google Scholar]
- 10.Holcik, M., Lefebvre, C., Yeh, C., Chow, T. & Korneluk, R. G. (1999) Nat. Cell Biol. 1, 190-192. [DOI] [PubMed] [Google Scholar]
- 11.Vagner, S., Gensac, M. C., Maret, A., Bayard, F., Amalric, F., Prats, H. & Prats, A. C. (1995) Mol. Cell. Biol. 15, 35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein, I., Itin, A., Einat, P., Skaliter, R., Grossman, Z. & Keshet, E. (1998) Mol. Cell. Biol. 18, 3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis, S., Bruynooghe, Y., Denecker, G., Van Huffel, S., Tinton, S. & Beyaert, R. (2000) Mol. Cell. 5, 597-605. [DOI] [PubMed] [Google Scholar]
- 14.Pyronnet, S., Pradayrol, L. & Sonenberg, N. (2000) Mol. Cell 5, 607-616. [DOI] [PubMed] [Google Scholar]
- 15.Henis-Korenblit, S., Strumpf, N. L., Goldstaub, D. & Kimchi, A. (2000) Mol. Cell. Biol. 20, 496-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanbru, C., Lafon, I., Audigier, S., Gensac, M. C., Vagner, S., Huez, G. & Prats, A. C. (1997) J. Biol. Chem. 272, 32061-32066. [DOI] [PubMed] [Google Scholar]
- 17.Stoneley, M., Paulin, F. E., Le Quesne, J. P., Chappell, S. A. & Willis, A. E. (1998) Oncogene 16, 423-428. [DOI] [PubMed] [Google Scholar]
- 18.Chappell, S. A., LeQuesne, J. P., Paulin, F. E., deSchoolmeester, M. L., Stoneley, M., Soutar, R. L., Ralston, S. H., Helfrich, M. H. & Willis, A. E. (2000) Oncogene 19, 4437-4440. [DOI] [PubMed] [Google Scholar]
- 19.Paulin, F. E., West, M. J., Sullivan, N. F., Whitney, R. L., Lyne, L. & Willis, A. E. (1996) Oncogene 13, 505-513. [PubMed] [Google Scholar]
- 20.Stoneley, M., Chappell, S. A., Jopling, C. L., Dickens, M., MacFarlane, M. & Willis, A. E. (2000) Mol. Cell. Biol. 20, 1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevins, T. A., Harder, Z. M., Korneluk, R. G. & Holcik, M. (2003) J. Biol. Chem. 278, 3572-3579. [DOI] [PubMed] [Google Scholar]
- 22.Subkhankulova, T., Mitchell, S. A. & Willis, A. E. (2001) Biochem. J. 359, 183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liston, P., Young, S. S., Mackenzie, A. E. & Korneluk, R. G. (1997) Apoptosis 2, 423-441. [DOI] [PubMed] [Google Scholar]
- 24.Warnakulasuriyarachchi, D., Cerquozzi, S., Cheung, H. H. & Holcik, M. (2004) J. Biol. Chem. 279, 17148-17157. [DOI] [PubMed] [Google Scholar]
- 25.Van Eden, M. E., Byrd, M. P., Sherrill, K. W. & Lloyd, R. E. (2004) RNA 10, 469-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imataka, H., Olsen, H. S. & Sonenberg, N. (1997) EMBO J. 16, 817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy-Strumpf, N., Deiss, L. P., Berissi, H. & Kimchi, A. (1997) Mol. Cell. Biol. 17, 1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieft, J. S., Zhou, K., Jubin, R. & Doudna, J. A. (2001) RNA 7, 194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova, T. V. & Hellen, C. U. (2003) Genes Dev. 17, 181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova, T. V., Shatsky, I. N., Fletcher, S. P., Jackson, R. J. & Hellen, C. U. (1998) Genes Dev. 12, 67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova, T. V., Hellen, C. U. & Shatsky, I. N. (1996) Mol. Cell. Biol. 16, 6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova, T. V., Shatsky, I. N. & Hellen, C. U. (1996) Mol. Cell. Biol. 16, 6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Gregorio, E., Preiss, T. & Hentze, M. W. (1998) RNA 4, 828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerlitz, G., Jagus, R. & Elroy-Stein, O. (2002) Eur. J. Biochem. 269, 2810-2819. [DOI] [PubMed] [Google Scholar]
- 35.Liebig, H. D., Ziegler, E., Yan, R., Hartmuth, K., Klump, H., Kowalski, H., Blaas, D., Sommergruber, W., Frasel, L., Lamphear, B., et al. (1993) Biochemistry 32, 7581-7588. [DOI] [PubMed] [Google Scholar]
- 36.Bushell, M., Poncet, D., Marissen, W. E., Flotow, H., Lloyd, R. E., Clemens, M. J. & Morley, S. J. (2000) Cell Death Differ. 7, 628-636. [DOI] [PubMed] [Google Scholar]
- 37.Marissen, W. E. & Lloyd, R. E. (1998) Mol. Cell. Biol. 18, 7565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henis-Korenblit, S., Shani, G., Sines, T., Marash, L., Shohat, G. & Kimchi, A. (2002) Proc. Natl. Acad. Sci. USA 99, 5400-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thoma, C., Bergamini, G., Galy, B., Hundsdoerfer, P. & Hentze, M. W. (2004) Mol. Cell 15, 925-935. [DOI] [PubMed] [Google Scholar]
- 40.Warnakulasuriyarachchi, D., Ungureanu, N. H. & Holcik, M. (2003) Cell Death Differ. 10, 899-904. [DOI] [PubMed] [Google Scholar]
- 41.Bergamini, G., Preiss, T. & Hentze, M. W. (2000) RNA 6, 1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morino, S., Imataka, H., Svitkin, Y. V., Pestova, T. V. & Sonenberg, N. (2000) Mol. Cell. Biol. 20, 468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gradi, A., Imataka, H., Svitkin, Y. V., Rom, E., Raught, B., Morino, S. & Sonenberg, N. (1998) Mol. Cell. Biol. 18, 334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ali, I. K., McKendrick, L., Morley, S. J. & Jackson, R. J. (2001) J. Virol. 75, 7854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borman, A. M. & Kean, K. M. (1997) Virology 237, 129-136. [DOI] [PubMed] [Google Scholar]
- 46.Evans, J. R., Mitchell, S. A., Spriggs, K. A., Ostrowski, J., Bomsztyk, K., Ostarek, D. & Willis, A. E. (2003) Oncogene 22, 8012-8020. [DOI] [PubMed] [Google Scholar]
- 47.De Gregorio, E., Preiss, T. & Hentze, M. W. (1999) EMBO J. 18, 4865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.