Abstract
For gene therapy of inherited diseases, targeted integration/gene repair through homologous recombination (HR) between exogenous and chromosomal DNA would be an ideal strategy to avoid potentially serious problems of random integration such as cellular transformation and gene silencing. Efficient sequence-specific modification of chromosomes by HR would also advance both biological studies and therapeutic applications of a variety of stem cells. Toward these goals, we developed an improved strategy of adenoviral vector (AdV)-mediated HR and examined its ability to correct an insertional mutation in the hypoxanthine phosphoribosyl transferase (Hprt) locus in male mouse ES cells. The efficiency of HR was compared between four types of AdVs that contained various lengths of homologies at the Hprt locus and with various multiplicities of infections. The frequency of HR with helper-dependent AdVs (HD AdVs) with an 18.6-kb homology reached 0.2% per transduced cell at a multiplicity of infection of 10 genomes per cell. Detection of random integration at DNA levels by PCR revealed extremely high efficiency of 5% per cell. We also isolated and characterized chromosomal sites where HD AdVs integrated in a random manner. In contrast to retroviral, lentiviral, and adeno-associated viral vectors, which tend to integrate into genes, the integration sites of AdV was distributed randomly inside and outside genes. These findings suggest that HR mediated by HD AdVs is efficient and relatively safe and might be a new viable option for ex vivo gene therapy as well as a tool for chromosomal manipulation of a variety of stem cells.
Keywords: gene therapy, homologous recombination, hypoxanthine phosphoribosyl transferase
For gene therapy of inherited diseases, integrating vectors, such as retroviral, lentiviral, and adeno-associated virus (AAV) vectors, have been used to introduce a therapeutic gene into random sites on target cell chromosomes (1, 2). However, random integration has potential problems, such as insertional mutagenesis and possible effects on gene expression. Homologous recombination (HR) is an ideal strategy for gene therapy of inherited diseases because it has the potential to repair mutant genes even in the case of dominant mutations. It also can be applied to introduce a gene into a safe and transcriptionally active chromosomal site to obtain a predictable level of expression. HR between electroporated exogenous DNA and chromosomal loci in mammalian cells has been extensively applied in mouse ES cells to make gene knockout mice. However, its application in other cell types, including human ES cells, has been hampered by a lack of a suitable gene delivery method to obtain HR (3). Considering that a variety of embryonic and adult stem cells have been isolated and characterized, development of a general strategy to obtain efficient HR in various cell types would be of paramount importance for biological studies and clinical applications of stem cells. However, HR between exogenous and chromosomal DNA is extremely inefficient, especially in mammalian cells. Many approaches, including chimeric DNA-RNA oligonucleotides, triplex-forming oligonucleotides, and small single- or double-stranded oligonucleotides, have been used in combination with nonviral gene delivery methods for gene repair (4, 5). Nevertheless, these approaches allow correction of only point mutations or small deletions/insertions. AAV vectors have also been exploited as a viral vector for gene repair (6, 7). However, the vector shows even higher frequency of random integration, which might obscure the advantage of targeted integration. Furthermore, currently available AAV vectors are less efficient in transducing certain types of stem cells, such as ES cells and hematopoietic cells, than adenoviral vectors (AdVs) (8, 9).
In general, by choosing appropriate fiber serotypes, AdVs can infect almost any cell type efficiently, and their genome is transferred to and stays in the host nuclei as stable double-stranded linear episomal DNA (10, 11). Importantly, unlike AAV vectors, AdVs are believed to integrate into host chromosomes at relatively low frequencies (1, 2). Previously, AdVs have been used to target extrachromosomal and chromosomal loci in mammalian cells by HR, albeit with limited success (12-14). In the case of adenine phosphoribosyl transferase locus in CHO cells transduced with E3-deleted replication-competent AdVs, the chromosomal integration frequencies were 10-5 to 10-6, in which 6-20% was by HR (14). When self-replicating extrachromosomal plasmid DNA (≈100 copies per cell) was targeted by replication-incompetent E1-deleted (E1D) AdVs in a mouse cell line, the HR frequencies were 1.9-8.3 × 10-4 per cell (12). An E1-deleted AdV was also used to target the Fgr locus in mouse ES cells with the frequencies of 1.9 × 10-7 to 1.2 × 10-5 (13). Helper-dependent adenoviral vectors (HD AdV) were originally developed to overcome host immune responses against E1D AdVs in vivo (reviewed in ref. 15). Because all of the viral genes are removed from the vector genome, they offer additional advantages such as decreased cytotoxicity and expanded cloning capacity, permitting insertion of larger segments of homologous DNA for HR. These features of HD AdVs might be advantageous to obtain highly efficient site-specific integration into host chromosomes through HR for therapeutic purposes.
In this study, we examined the ability of HD AdVs to correct an insertional mutation at the hypoxanthine phosphoribosyl transferase (Hprt) locus in male mouse ES cells. The frequency was 0.02-0.2% per transduced cell by using a HD AdV with an 18.6-kb homology at a relatively low multiplicity of infection (moi) of 10, although the frequency of random integration was much higher and was ≈5%, with 72% of the integration sites being intergenic. These observations suggest that HR mediated by HD AdVs might be applicable to ex vivo gene correction therapy as well as manipulation of chromosomes in a variety of stem cells.
Materials and Methods
Cell Culture. The wild-type mouse ES cell line, AB1, and an HPRT-knockout mouse ES cell line, AB1/HprtRV7.0PGK, which is derived from AB1 and has the 1.6-kb PGK-neo cassette inserted into exon 3 of the Hprt locus (kindly provided by Allan Bradley, Wellcome Trust Sanger Institute, Cambridge, U.K.), were cultured as described in ref. 16. HEK293 cells were maintained in DMEM supplemented with 10% FCS. HEK293Cre66 cells (kindly provided by Gudrun Schiedner and Stefan Kochanek, University of Cologne, Cologne, Germany) were maintained in MEM-α supplemented with 10% FCS.
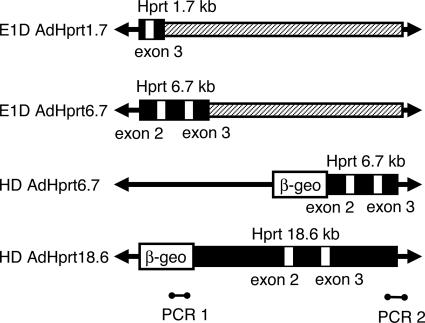
Construction of AdVs. To construct E1D AdHprt1.7 and E1D AdHprt6.7, the 1.7-kb HindIII-EcoRI and the 6.7-kb EcoRI fragment, both encompassing the Hprt exon 3, were subcloned into pΔE1sp1A (17), to make pΔE1Hprt1.7 and pΔE1Hprt6.7, respectively. To construct HD AdHprt6.7, the 6.7-kb EcoRI fragment and the PGKβ-geobpA (18) cassette were subcloned into HD AdV plasmid, pSTKCMVβ-gal (19), to make pHDAdHprt6.7. To construct HD AdHprt18.6, the 18.6-kb NotI fragment encompassing the exons 2 and 3 of the mouse Hprt locus (20) was subcloned into HD AdV plasmid, pNEBITRSRαβ-geo (K.M., unpublished data), to make pHDAdHprt18.6. The schematic structure of these vectors is shown in Fig. 1. A detailed description of these subclonings can be provided on request.
Fig. 1.
Schematic representations of Hprt-targeting AdVs. Locations of Hprt genomic sequence (black box), exons (white box), inverted terminal repeats, and the packaging signal of Ad5 (arrows), adenoviral sequence (striped box),β-geo marker gene, and stuffer DNA fragment (thin line) are shown. The locations of two PCR primer pairs, used to detect HD AdHprt18.6, are shown as PCR 1 and PCR 2.
Preparation of AdVs. pΔE1Hprt1.7 and pΔE1Hprt6.7 were cotransfected into 293 cells with pJM17 (21) and pBHG10 (17) to rescue AdHprt1.7 and AdHprt6.7, respectively. The resultant E1D AdVs were propagated by serial passages on 293 cells, as described in ref. 22. After transfection of PmeI-linearized pHDAdHprt6.7 and pHDAdHprt18.6, the rescued HD AdVs were propagated by serial passages on 293Cre66 cells with the addition of the Ad5E3stLYLR helper virus (F. P. Hernandez and K.M., unpublished data) at each passage, as described in ref. 23. Both E1D and HD vectors were purified on two rounds of CsCl density gradient ultracentrifugation, as described in ref. 22. Infectious vector titers were adjusted with respect to the copy number of vector DNA in transduced ES cells. For this titration, ES cells were infected with serially diluted vectors. Four hours after infection, DNA was extracted from the cells, digested with EcoRV and subjected to quantitative Southern hybridization, and probed with the 1.2-kb EcoRI fragment encompassing the exon 3 of the Hprt locus (data not shown). Throughout this study, the moi is based on the copy number of vector genome per ES cell after infection.
Measurement of Homologous Recombination and Random Integration Frequencies. Because HPRT-proficient cells are hypoxanthine/aminopterin/thymidine (HAT)-resistant, HR events, which restore the Hprt locus in AB1/RV7.0PGK cells, were detected by HAT selection. Because AB1/RV7.0PGK cells already contain the neomycin-resistant gene inserted into exon 3 of the Hprt locus, the parental AB1 cells were used in studies to detect random integration events of the HD AdVs, which encode the β-geo marker gene, by G418 selection. For these assays, the cells were plated into six-well dishes on day 1 and, on day 2, the cells were counted and infected with the vectors at various mois for 1 h at room temperature. In one set of experiments, the cells were treated with 300 μM trichostatin A (TSA) for the first 2 days of infection with the HD AdHprt18.6 vector. On day 3 or 5, the cells were replated in six-well dishes and, on the following day, HAT or G418 selection was initiated. The frequencies of HR or random integration were calculated by dividing the total number of HAT- or neomycin-resistant colonies by the total number of cells plated. The efficiency of colony formation of infected cells was also measured in the absence of selection to normalize the frequencies of integration. Experiments were performed in duplicate and analyzed by t test.
Electroporation. Ten million AB1/RV7.0PGK cells were electroporated with 25 μg of PmeI-linearized pHDAdHprt18.6 plasmid DNA, which has structure similar to the virus genome with inverted terminal repeats attached at both ends, at 230 V and 500 μF, and plated onto a 6-cm dish. The next day, cells were diluted and plated in duplicate. HAT selection was initiated 2 days after electroporation. The efficiency of colony formation was also determined in the absence of selection. Under this condition, the average gene transfer efficiency, measured by using a plasmid DNA encoding the GFP gene and FACS analysis, was ≈20%, and the plating (colony-forming) efficiency after electroporation was 42% (data not shown).
DNA Analysis of ES Clones. To analyze the structure of the Hprt locus after HR, DNA was extracted from HAT-resistant AB1/RV7.0PGK clones, digested with BamHI, and subjected to Southern hybridization with the Hprt exon 3 probe. To analyze whether extra copies of the vector exist in HAT-resistant ES cell clones, which went through HR, DNA from HAT-resistant AB1/RV7.0PGK clones was subjected to PCR analysis with primer pairs amplifying the β-gal sequence (PCR 1 in Fig. 1) and the junction between adenovirus and Hprt sequences of HD AdHprt18.6 (PCR 2 in Fig. 1). The sequences of oligonucleotide primers are as follows: β-gal216, CTGGCTGGAGTGCGATCTTC, and β-gal291R, CGCATCGTAACCGTGCATC, for β-gal PCR; and Ad1295, CTAAAATGGCGCCTGCTATC, and mHprtR, ACTAGAATGATCAGTCAACGGG, for Ad/Hprt PCR. PCR was also used to detect vector DNA directly from infected AB1/RV7.0PGK clones, which were grown in the absence of HAT selection. DNA was extracted and subjected to PCR with the same Ad/Hprt and β-gal primer pairs as above.
Analysis of Vector Integration Sites. To determine vector-chromosome junction sequences of randomly integrated AdVs, we performed adaptor-ligated PCR (AL-PCR) as described in ref. 24. AB1 cells were infected with the HD AdHprt18.6 vector, G418-resistant colonies were isolated, and their DNA was partially digested with Sau3AI. Sequences of oligonucleotide primers used for AL-PCR and sequencing are as follows: adaptor 1, ACAGCAGGTCAGTCAAGCAGTA; adaptor 2, AGCAGTAGCAGCAGTTCGATAA; Ad5 280R, CCTAAAACCGCGCGAAAATTGTC; Ad5 228R, CCTAAAACCGCGCGAAAATTGTC; mHprt18.6-3′ first, TTCTATCGCCTTCTTGACGAGT; mHprt18.6-3′ second, TGTACTGAGAGTGCACGTCGAA. The PCR products were gel-extracted and sequenced by using either the Ad5 280R primer or the mHprt18.6-3′ second primer (AB1 PRISM 310 DNA Sequencer, PE Applied Biosystems, Foster City, CA). The sequences of integration sites determined by AL-PCR were blast-searched against the public mouse genome database thorough the National Center for Biotechnology Information. The chromosomal localization of each integration site was blast-searched by using the University of California, Santa Cruz, Genome Bioinformatics Site (May 2004 assembly). A “gene” was defined as a genomic region between transcriptional start and stop boundaries of one of the RefSeq genes, as defined in the investigation of AAV integration sites (25). The bias for or against preferred integration in RefSeq genes was compared with computer-simulated 10,000 random integrations (26% into the genes; ref. 25), and the statistical significance was assessed with the χ2 test.
Results
Effects of Length of Homologies and mois on Homologous Recombination. To clarify whether the length of homology and/or the moi have effects on HR or random integration frequency, we constructed four types of AdVs and examined their ability to correct an insertional mutation in exon 3 of the Hprt locus in male mouse ES cells. HR between the vector and the mutated Hprt locus would restore the HPRT activity and make the cell HAT-resistant. The frequency of random integration was examined by measuring the number of G418-resistant colonies. Each AdV contained a wild-type exon 3 sequence of the mouse Hprt gene with variable lengths of flanking intronic sequences ranging from 1.7 kb to 18.6 kb (Fig. 1). The PGK and SRα promoter-driven β-geo gene were encoded in the HD AdHprt6.7 and the HD AdHprt18.6 vector, respectively. AB1 and AB1/RV7.0PGK mouse ES cells were infected with these vectors at various mois from 10 to 1,000, and the selections were started 4 days after infection. HR and random integration frequencies were calculated by dividing the total number of HAT- and neomycin-resistant colonies by the total number of cells plated, respectively.
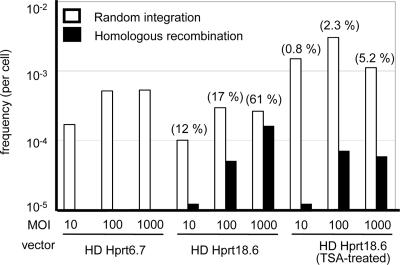
As shown in Fig. 2, HR was most efficiently obtained when using the HD AdHprt18.6 vector. Under this condition, the frequency of HR was nearly 0.0002 per infected cell at an moi of 1,000. In our study, the length of homologous sequence appeared to be critical to achieve HR. In the case of HD AdHprt6.7 and E1D AdHprt1.7, no HR events were detected. The frequency of HR by E1D AdHprt6.7 was 2.0 × 10-7 per cell at an moi of 1,000 (data not shown). The HR frequency by electroporation of the plasmid pHDAdHprt18.6 was 23-fold less than that by vector infection (6.7 × 10-4 per cell vs. 2.4 × 10-5 per cell in experiment 3, Table 1), indicating the significance of AdV-mediated DNA delivery for efficient HR. The structure of the exon 3 region of Hprt locus was analyzed in HAT-resistant ES clones by Southern analysis with the exon 3 probe. All 22 (10 by AdV infection and 12 by electroporation) showed a pattern of faithful HR (data not shown).
Fig. 2.
Frequencies of homologous recombination and random integration. Cells were infected with each targeting vector at an moi of 10, 100, and 1,000. Selection was initiated 4 days after infection. The values were determined by dividing the number of G418 or HAT-resistant colonies by the total number of cells plated. Frequencies of random integration and HR are shown in white bars and black bars, respectively. The numbers enclosed in parentheses indicate the percentage of HR in random integration.
Table 1. Effects of incubation time between infection and start of selection on frequencies of homologous recombination.
| Experiment | Start of selection | Homologous recombination | Random integration |
|---|---|---|---|
| 1 | Day 2 | 5.5 × 10−4 | 1.4 × 10−3 |
| (1.8 × 10−4)* | (7.9 × 10−4) | ||
| Day 4 | ND | 5.0 × 10−4 | |
| (1.2 × 10−4) | (1.8 × 10−4) | ||
| 2 | Day 2 | 2.2 × 10−3 | 3.1 × 10−3 |
| Day 4 | 3.2 × 10−4 | 4.8 × 10−4 | |
| 3 | Day 2 | 6.7 × 10−4 | ND |
| Day 4 | 5.6 × 10−4 | ND | |
| Electroporation† | 2.4 × 10−5 | ND |
Cells were infected with HD AdHprt18.6 vector at an moi of 10. After infection, cells were passaged every 2 days at a density of 105 cells per cm2 into a six-well dish. Two and 4 days after infection, selection was initiated. Frequencies of random integration of HR were calculated by dividing the total number of G418- or HAT-resistant colonies by the total number of cells plated, and these were normalized with plating (colony-forming) efficiency. Experiments were performed in duplicates. ND, not done.
The numbers enclosed in parentheses indicate the frequencies obtained when the infected cells were plated in the presence of uninfected cells to keep a high cell density (1.2 × 105 cells per cm2).
The selection was started 2 days after electroporation.
Effects of Culture Conditions on Homologous Recombination. We examined the effects of tissue culture conditions, such as treatment with the histone deacetylase inhibitor, TSA, and incubation time between infection and the start of HAT selection, on HR with AdVs. We reasoned that TSA-mediated histone acetylation in ES cells, resulting in relaxed chromatin structure, might promote HR. However, TSA treatment did not improve the HR frequency, probably because Hprt is a housekeeping gene and it already has open chromatin (Fig. 2).
To optimize the selection conditions, we started HAT and G418 selection at different times after infection. The HR and random integration frequencies did not change significantly whether the selections were started 2 days or 4 days after infection (Table 1). The frequencies were 3.2 × 10-4 to 2.2 × 10-3 per cell for HR and 4.8 × 10-4 to 3.1 × 10-3 per cell for random integration. These results suggest that HR and random integration occur within 2 days after infection. Porteus et al. (26) also reported that HR occurs within 2-3 days after gene transfer. In contrast to a previous report that suggested that HAT selection at low cell density reduces the frequency of HR (27), we found that plating infected cells at a high cell density by adding uninfected cells before selection (1.2 × 105 cells per cm2 in a six-well dish) did not improve HR frequency in our system (Table 1, experiment 1).
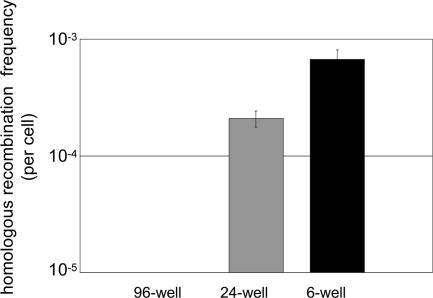
Efficient Homologous Recombination with Reduced Numbers of Cells. Because the number of target cells might be limited in ex vivo stem cell gene therapy, we next investigated whether HD AdV-mediated HR can be efficiently achieved starting with fewer cells. AB1/RV7.0PGK cells were plated in 6-well, 24-well, or 96-well dishes at a density of 2 × 105 cells per cm2 and infected with HD AdHprt18.6 vector. To infect the cells at the same moi of 10, the relative volume of virus-containing solution per cell had to be increased for a smaller well (30, 100, and 200 μl for 96-, 24-and 6-well, respectively). Therefore, the infection efficiency might have been compromised, resulting in a lower HR efficiency. This low infection efficiency might be one of the reasons why HAT-resistant colonies were not formed in a 96-well dish and the efficiency in a 24-well dish was lower than that in a 6-well dish. Nevertheless, we successfully obtained HAT-resistant colonies at a frequency of 2.1 × 10-4 per infected cell in a 24-well dish (25 HAT-resistant colonies of 1.2 × 105 cells infected, on average, Fig. 3). These results clearly indicate gene correction can be achieved relatively efficiently in a smaller number of cells by using AdVs.
Fig. 3.
Homologous recombination in smaller numbers of target cells. AB1/RV7.0PGK cells plated in a 6-well, 24-well, and 96-well at a cell density of 2 × 105 cells per cm2 in duplicate. Each cell plate was infected with HD AdHprt18.6 vector at an moi of 10. Two days after infection, the HAT selection was started. Frequency of HR was determined by dividing the total number of HAT-resistant colonies by the total number of cells plated, which were normalized by colony forming efficiency. Experiments were performed in duplicates.
Random Integration Frequencies of HD AdVs. To measure the random integration frequency of HD AdVs, AB1 cells were infected with the HD AdHprt6.7 or the HD AdHprt18.6 vector at various mois from 10 to 1,000. As shown in Fig. 2, both HD AdVs achieved similar random integration frequencies of 10-4 to 10-3 per cell, which was measured as the frequency of G418-resistant colonies. In contrast to the HR frequency, which showed an moi-dependent increase, the random integration frequency reached a plateau at an moi of 100. As a result, the ratio of HR to random integration was increased at an moi of 1,000 to >50% (Fig. 2).
After treating the cells with TSA, the frequency of random integration apparently increased relative to that of HR (Fig. 2). This result suggests that the nonspecific histone deacetylase activity of TSA promotes global relaxation of higher order chromatin structure, thereby presenting more sites for random integration. It is also possible that the random integration frequencies, measured by G418-resistance, might be underestimated, possibly because gene silencing prevents integrated vectors from expressing the β-geo marker gene. To directly detect vectors at the DNA level, we next performed a PCR that amplified regions close to both the left and the right end of the vector (Fig. 1). DNA was prepared from AB1/RV7.0PGK cells that were transduced with HD AdHprt18.6 by vector infection or by plasmid electroporation and formed colonies without HAT selection. As a result, HD AdHprt18.6 DNA was detected in 4 of the 74 samples (5.4% of colonies; Table 2). On the other hand, no random integration event was detected in ES clones formed after electroporation (0 of 36). These results indicate that the random integration frequency of HD AdVs may be much higher than reported in ref. 28. We also examined whether randomly integrated vector exists in HAT-resistant clones, which had achieved targeted vector integration. We used the same two primer pairs as above, and the vector sequence was detected in 3.1% (1 of 32) of the HAT-resistant clones (Table 2). There was no statistically significant change in the random integration frequency in HAT-resistant colonies compared with nonselected colonies, suggesting that targeted and random integration with AdVs occur independently.
Table 2. Frequency of random integration measured by PCR.
| HAT selection | no, n (%) | yes, n (%) |
|---|---|---|
| No. of colonies analyzed | 74 | 32 |
| No. of positives, % | ||
| β-gal PCR | 4 (5.4%) | 1 (3.1%) |
| Ad/Hprt PCR | 2 (2.7%)* | 1 (3.1%)* |
AB1/RV7.0PGK cells were infected with HD AdHprt18.6 at an moi of 10 and plated to form colonies in the presence or absence of HAT selection. DNA was extracted from these colonies and subjected to PCR analysis by using two primer pairs (Fig. 1) to directly detect vector DNA.
These colonies, positive for Ad/Hprt PCR, were also positive for β-gal PCR.
Integration Sites of HD AdVs. In view of the unexpectedly high frequency of random integration, if AdVs favored integration at chromosomal regions near or within active genes, as reported for retroviral, lentiviral, and AAV vectors, it could potentially have deleterious effects. Therefore, we determined the nucleotide sequences of the AdHprt18.6 vector integration sites. Because of the large genome size of AdV, the plasmid rescue strategy, which was used for cloning of AAV integration sites, could not be used (29, 30). Instead, we used AL-PCR (24). In most of the cases, only one of two vector ends could be determined. By aligning the sequences of AL-PCR products with the mouse genome sequence, it was found that HD AdVs have a tendency to integrate not in genes (5 of 18 integrants, 28%) but in intergenic regions (13 of 18 integrants, 72%) (Table 3). All five intragenic integrations (A7, A8, A11, B4, and C2) were into introns, and at least four of them (A7, A8, A11, and B4) are expressed in ES cells (Stanford SOURCE web site). One of the integrations (C7) was into the B2 repeats, and, therefore, we could not determine the chromosomal location. Also, in most of the cases, small deletions of integrated vector DNA were found at the end of inverted terminal repeats, ranging from 1 to 99 bp (Table 3). Furthermore, as in the case of AAV vectors, small insertions or microhomologies between the vector end and the integration sites were often found (Table 3). In three cases, both ends were determined (Table 4). In one case (A5), the vector right end was found in an intergenic region on chromosome 15/D3 whereas another end was in the L1 repeat. Because the closest L1 repeat to the right end integration site was at least 20 kb away, we concluded that either a >20-kb deletion or a translocation occurred in this clone. In two other cases (A11 and B4), both vector ends were located close to each other. However, in one case (A11), a chromosomal region that is originally 1.5 kb upstream was inverted and inserted 100 bp upstream of the integration site. In another case (B4), inversion of chromosomal DNA at the integration site, which is at least 125 bp in size, was found. Inversion, translocation, insertion, and deletion were all observed at the integration sites of other DNA viruses, such as AAV and hepatitis B virus, suggesting that the mechanisms involved in random chromosomal integration of these DNA viruses are similar (25, 29-33). According to Nakai et al. (25), computer simulated frequency of 100,000 random integration into mouse RefSeq sequences is 26.0% (October 2003 assembly). By using the χ2 analysis, we concluded that the observed frequency of random integration in our study (28%) is not significantly different from the expected frequency. Therefore, random insertion with HD AdV does not show any sequence-specific of gene-specific preferences and is thus truly random unlike retrovirus and AAV integration patterns.
Table 3. Chromosomal integration sites of HD AdVs.
| ID | End | Chromosome | Nearest gene | Vector/chromosome,* bp | Vector deletion,† bp |
|---|---|---|---|---|---|
| 2 | Left | 6/D2 | 2.5 kb upstream of Slc25a26 | 4 insertion | 4 |
| 3 | Left | 12/D1 | 5.3 kb upstream of Psen 1 | 7/8 homology | 9 |
| 6 | Left | 4/B2 | 60.6 kb upstream of Smc211 | 30 insertion | 1 |
| 9 | Left | 4/B3 | 178 kb downstream of Rgs3 | 3 homology | 0 |
| A3 | Right | 13/D2.3 | 5.5 kb downstream of Parp8 | 6 insertion | 4 |
| A5 | Left | L1 repeat | NA | 2 homology | 9 |
| Right | 15/D3 | 174 kb upstream of Bai 1 | 14 homology | 2 | |
| A7 | Left | 11/A5 | NudC domain containing 2‡ | 4 insertion | 99 |
| A8 | Right | 2/E1 | MAP kinase activating death domain‡ | 3 insertion | 1 |
| A11 | Both | 4/C7 | Fas-associated factor 1‡ | 5 homology | 2/2 |
| B3 | Left | 4/B1 | 5.1 kb upstream of Tgfbr1 | 2 homology | 2 |
| B4 | Both | 4/E2 | Brain acyl-CoA hydrolase‡ | 9 insertion§ | 1/1 |
| B6 | Right | 11/E1 | 91.3 kb downstream of Axin2 | 32 insertion | 22 |
| B9 | Left | 19/C3 | 8.4 kb upstream of Fbxw4 | 5/6 homology | 8 |
| B11 | Left | 10/C2 | 536 kb upstream of Tmpo | 4 insertion | 11 |
| C1 | Right | 16/A1 | 58.5 kb downstream of Adcy9 | 13 insertion | 8 |
| C2 | Left | 9/A5.1 | Poliovirus receptor-related 1‡ | No | 2 |
| C3 | Left | 7/F3 | 17.1 kb upstream of Atxn21 | 3 homology | 1 |
| C6 | Left | 7/B5 | 1.7 kb downstream of Snrpa | 3 insertion | 14 |
| C7 | Right | B2 repeat | NA | 4 homology | 0 |
| C12 | Left | 1/H3 | 9.2 kb downstream of Fcr13 | 2 homology | 9 |
AB1/RV7.0PGK cells were infected with HD AdHprt18.6 and plated to form colonies under G418 selection. DNA was extracted from the colonies, and vectors/chromosome junctions were determined by AL-PCR. NA, not applicable.
The size of microhomologies or insertions at the vector/chromosome junctions is shown. Microhomologies indicate the length of perfect matches, except clone 2 and B9, which showed 7 of 8 bp and 5 of 6 bp were shared between the vector and the chromosome, respectively.
The number of nucleotides that were deleted at the left or right end of vector inverted terminal repeat is shown.
All the intragenic integrations are into introns.
An identical 9-bp stretch of DNA was inserted at both ends in an inverted manner.
Table 4. Host chromosomal changes at AdV integration sites.
| Clone ID | End | Chromosomal location | Gene | Host chromosome |
|---|---|---|---|---|
| A5 | Left | Unknown (L1 repeat) | L1 repeat | Multiple potential targets (>20 kb deletion or translocation) |
| Right | 15/D3 | Intergenic | ||
| A11 | Left | 4/C7 | Faf1, intron | 283-bp inversion/insertion |
| Right | 4/C7 | Faf1, intron | 21-bp deletion | |
| B4 | Left | 4/E2 | Bach, intron | At least 125-bp inversion |
| Right | 4/E2 | Bach, intron | No deletion |
Discussion
Although integrating vectors, such as retroviral vectors, have the advantage of stable chromosomal integration, the problem of insertional mutagenesis has been highlighted in their clinical use (34). HR would be the most ideal strategy for gene therapy of inherited diseases. It overcomes various undesirable problems that might be caused by random integration. However, HR in mammalian cells is generally very inefficient. AdVs, especially HD AdVs, have several potential advantages for HR. Therefore, establishing an AdV-mediated HR strategy would be important for gene therapy of inherited diseases. Although we have previously tested the ability of an E1-deleted replication-incompetent AdV to target the single-copy chromosomal Fgr locus in mammalian cells by HR, the efficiencies were low at 10-5 to 10-7 per infected cell (13). Here, we demonstrated that HR can be achieved at higher frequencies in mouse ES cells by using HD AdVs.
Our study showed that although no HR event was observed when E1DAdHprt1.7 or HD AdHprt6.7 AdVs were used, the frequency with HD AdHprt18.6 vector was almost 2 × 10-4 per cell (Fig. 2). The frequency of HR was directly proportional to the moi of AdV used. In comparison with gene knockout studies, our experimental design of measuring the restored function of the HPRT gene is a more stringent assay for HR, because only faithful HR is detected. We conclude that the high cloning capacity of HD AdVs could be critical to achieve successful HR. In fact, a previous study reported that the HR frequency in mouse ES cells obtained by electroporation depended highly on homology length up to 14.6 kb DNA and then reached a plateau (20). It would be interesting to know the minimum length of homology required to achieve efficient HR with AdVs.
One of the critical advantages of using HD AdVs is their high HR efficiency. Even at an moi of 10 (i.e., 10 vector genomes per cell), HD AdHprt18.6 vector achieved HR at frequency of 2.2 × 10-3 (Table 1). Although, it cannot be directly compared, much higher mois (moi of 20,000) were reported to be required for AAV vectors than for HD AdVs to achieve HR of 3 to 7 × 10-3 per cell (7). We also showed that it was possible to achieve HR even in a small number of cells by using HD AdVs (Fig. 3). In contrast to electroporation, which required at least 107 cells to obtain HAT-resistant colonies at a frequency of 2.4 × 10-5 per transduced cell, HD AdV required only 3 × 105 cells to achieve HR, and it was 23-fold more efficient per cell. Because of difficulties in obtaining and growing a sufficient number of some rare stem cells, feasibility of manipulating chromosomes of a small number of cells is a big advantage of viral vector-mediated HR.
Surprisingly, our PCR analysis of random integration at DNA levels revealed that the random integration frequency mediated by HD AdHprt18.6 was 0.05 per transduced cell (i.e., one in every 20 cells), suggesting that the frequency of random integration of AdVs is possibly as high as that of AAV vectors. We previously reported that both E1D and HD AdVs integrate into chromosomes in a variety of mammalian cell lines at frequencies between 10-2 and 10-5 (28). However, because the frequencies were measured as the frequency to produce neomycin-resistant colonies, these numbers might also be an underestimation. If random integration occurred at active gene loci at such a high frequency, it is more likely to induce cellular mutagenesis, as is the case in retroviral vectors. Previous reports showed that 59%, 75-80%, 61%, 57%, and 60% of integrations were into genes with AAV, HIV, murine leukemia virus, avian sarcoma-leukosis virus vectors, and human T lymphotropic virus type I, respectively (24, 25, 35). Our analysis of integration sites showed that HD AdVs have a tendency to integrate not in genes (5 of 18, 28%) but in intergenic regions (13 of 18, 72%), suggesting that HD AdVs integrate into random sites in host chromosomes, in contrast to retroviral, lentiviral, and AAV vectors. Although more integration sites should be characterized, HD AdVs might be a safer option compared with other viral vectors, even as an integrating vector.
Interestingly, in contrast with the HR frequency, which showed an moi-dependent increase, the random integration frequency reached a plateau at an moi of 100 (Fig. 2). Because HD AdVs do not encode proteins to catalyze integration, such as an integrase, the mechanism of random integration of HD AdV might be similar to that of AAV vector and duck hepatitis B virus, one in which viral genomes integrate at spontaneous double-strand breaks, presumably by the nonhomologous end-joining pathway (29, 36). The characteristics of AdV/chromosome junctions, such as microhomologies and small deletions/insertions, support this hypothesis. However, the difference in preferential chromosomal regions of integration (intragenic vs. intergenic) between AAV and Ad vectors suggests that proteins involved in chromosomal integration are not exactly the same. The limited number of preexisting chromosomal breaks (37) might be the reason why the frequency of HD AdV-mediated random integration reached maximum levels at an moi of 100. Interestingly, the double-strand break frequency of 0.05 per cell, measured by the number of γ-H2AX foci in confluent culture of MRC-5 cells (37), coincides with the frequency of random integration of AdVs we obtained.
Although it would be necessary to examine the efficiency of HR at other loci and the effect of DNA polymorphisms on HR, we have shown that HD AdVs could be a promising tool for gene therapy of inherited diseases. There are several advantages of using HD AdVs for HR over other gene delivery methods. In comparison with nonviral methods, because efficiency of DNA delivery is high, the same strategy can be applied to a wider variety of cells relatively easily, and it is easily scaled down for transducing a smaller number of cells. In addition, in comparison with AAV vectors, a higher cloning capacity of HD AdVs allows a larger DNA segment to be inserted to provide greater overlap homology with target chromosomal sites. Finally, unwanted random chromosomal integration through non-HR by HD AdVs tends to be located at intergenic regions. However, because the efficiency of HR is still an order of 10-3, this strategy is applicable only to ex vivo gene therapy. Furthermore, because the frequency of random integration is much higher, an appropriate strategy for negative selection, such as the one with the herpes simplex virus thymidine kinase gene, has to be used. Recently, chimeric nucleases, which combine a zinc finger DNA binding domain and an endonuclease domain, have accomplished to cleave DNA in a site-specific manner (38). It was reported that chimeric nucleases could stimulate HR in human somatic cells by several thousand-fold (26, 39). Performing HR by designing HD AdV, which also encodes chimeric nucleases that can stimulate the introduction of a double-strand break at a target locus, may lead to more effective gene targeting with AdVs. An AAV vector has been used to correct dominant mutations in mesenchymal stem cells from patients of osteogenesis imperfecta (6). Because AdVs have wider tropisms than AAVs, this disease and other inherited diseases, especially X-linked severe combined immunodeficiency, hereditary tyrosinemia type I, and Fanconi anemia, might be strong candidates for HD AdV-mediated stem cell gene therapy. These diseases are ideal candidates, because even a small starting pool of corrected cells can be selected for and amplified in vivo and so extensive screening for HR events might not be necessary. Although hematopoietic cells are relatively refractory to AAV vector, HD AdVs with a chimeric fiber can efficiently transduce these cell types (40, 41).
Our strategy also provides a way to manipulate a chromosome of a variety of target cells effectively by taking advantage of highly efficient gene transfer by AdVs. This AdV-mediated HR would have a variety of applications in biological studies such as production of gene knockout and knockin cell lines. Recently, various types of stem cells have been isolated. With the rapid development of regenerative medicine, the necessity for developing a strategy to manipulate cellular chromosome of these cells has been increased. The technology to obtain HR by HD AdVs presented in this study is an important step toward efficient chromosomal manipulation and wider clinical applications of stem cells.
Acknowledgments
We thank Dr. Allan Bradley for helpful discussions and providing the ES cell lines; Dr. Kirk Thomas (University of Utah, Salt Lake City) for providing the Hprt 18.6-kb fragment; Drs. Stefan Kochanek and Gudrun Schiedner for providing 293cre66 cells; Felicia Hernandez, Airi Harui, and John Rudy for excellent technical help; Drs. Masami Muramatsu and Hiroyuki Nakai for helpful discussions and critical reading of the manuscript; Dr. Masaharu Isobe for helpful discussion on the AL-PCR protocol; and members of K.M.'s laboratory at University of California, Los Angeles and at Saitama Medical School for helpful discussions. This work was supported by National Institutes of Health Grant AI-42214, a grant for the Promotion of the Advancement of Education and Research in Graduate Schools by the Ministry of Education, Culture, Sports, Science and Technology, Japan, and a research grant by the Sankyo Foundation of Life Science.
Abbreviations: AAV, adeno-associated virus; AdV, adenoviral vector; Al-PCR, adaptor-ligated PCR; HAT, hypoxanthine/aminopterin/thymidine; HD, helper-dependent; Hprt, hypoxanthine phosphoribosyl transferase; HR, homologous recombination; moi, multiplicities of infection; TSA, trichostatin A.
References
- 1.Thomas, C. E., Ehrhardt, A. & Kay, M. A. (2003) Nat. Rev. Genet. 4, 346-358. [DOI] [PubMed] [Google Scholar]
- 2.Verma, I. M. & Weitzman, M. D. (2004) Annu. Rev. Biochem. 74, 711-738. [DOI] [PubMed] [Google Scholar]
- 3.Zwaka, T. P. & Thomson, J. A. (2003) Nat. Biotechnol. 21, 319-321. [DOI] [PubMed] [Google Scholar]
- 4.Kmiec, E. B. (2003) J. Clin. Invest. 112, 632-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seidman, M. M. & Glazer, P. M. (2003) J. Clin. Invest. 112, 487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamberlain, J. R., Schwarze, U., Wang, P. R., Hirata, R. K., Hankenson, K. D., Pace, J. M., Underwood, R. A., Song, K. M., Sussman, M., Byers, P. H., et al. (2004) Science 303, 1198-1201. [DOI] [PubMed] [Google Scholar]
- 7.Hirata, R., Chamberlain, J., Dong, R. & Russell, D. W. (2002) Nat. Biotechnol. 20, 735-738. [DOI] [PubMed] [Google Scholar]
- 8.Smith-Arica, J. R., Thomson, A. J., Ansell, R., Chiorini, J., Davidson, B. & McWhir, J. (2003) Cloning Stem Cells 5, 51-62. [DOI] [PubMed] [Google Scholar]
- 9.Zhong, L., Li, W., Yang, Z., Qing, K., Tan, M., Hansen, J., Li, Y., Chen, L., Chan, R. J., Bischof, D., et al. (2004) Hum. Gene Ther. 15, 1207-1218. [DOI] [PubMed] [Google Scholar]
- 10.Havenga, M. J., Lemckert, A. A., Ophorst, O. J., van Meijer, M., Germeraad, W. T., Grimbergen, J., van Den Doel, M. A., Vogels, R., van Deutekom, J., Janson, A. A., et al. (2002) J. Virol. 76, 4612-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConnell, M. J. & Imperiale, M. J. (2004) Hum. Gene Ther. 15, 1022-1033. [DOI] [PubMed] [Google Scholar]
- 12.Fujita, A., Sakagami, K., Kanegae, Y., Saito, I. & Kobayashi, I. (1995) J. Virol. 69, 6180-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitani, K., Wakamiya, M., Hasty, P., Graham, F. L., Bradley, A. & Caskey, C. T. (1995) Somatic Cell Mol. Genet. 21, 221-231. [DOI] [PubMed] [Google Scholar]
- 14.Wang, Q. & Taylor, M. W. (1993) Mol. Cell. Biol. 13, 918-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer, D. J. & Ng, P. (2005) Hum. Gene Ther. 16, 1-16. [DOI] [PubMed] [Google Scholar]
- 16.Ramirez-Solis, R., Davis, A. C. & Bradley, A. (1993) Methods Enzymol. 225, 855-878. [DOI] [PubMed] [Google Scholar]
- 17.Bett, A. J., Haddara, W., Prevec, L. & Graham, F. L. (1994) Proc. Natl. Acad. Sci. USA 91, 8802-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedrich, G. & Soriano, P. (1991) Genes Dev. 5, 1513-1523. [DOI] [PubMed] [Google Scholar]
- 19.Sato, M., Suzuki, S., Kubo, S. & Mitani, K. (2002) Gene Ther. 9, 472-476. [DOI] [PubMed] [Google Scholar]
- 20.Deng, C. & Capecchi, M. R. (1992) Mol. Cell. Biol. 12, 3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrory, W. J., Bautista, D. S. & Graham, F. L. (1988) Virology 163, 614-617. [DOI] [PubMed] [Google Scholar]
- 22.Ng, P. & Graham, F. L. (2002) Methods Mol. Med. 69, 389-414. [DOI] [PubMed] [Google Scholar]
- 23.Ng, P., Parks, R. J. & Graham, F. L. (2002) Methods Mol. Med. 69, 371-388. [DOI] [PubMed] [Google Scholar]
- 24.Ozawa, T., Itoyama, T., Sadamori, N., Yamada, Y., Hata, T., Tomonaga, M. & Isobe, M. (2004) J. Hum. Genet. 49, 154-165. [DOI] [PubMed] [Google Scholar]
- 25.Nakai, H., Wu, X., Fuess, S., Storm, T. A., Munroe, D., Montini, E., Burgess, S. M., Grompe, M. & Kay, M. A. (2005) J. Virol. 79, 3606-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porteus, M. H. & Baltimore, D. (2003) Science 300, 763. [DOI] [PubMed] [Google Scholar]
- 27.Templeton, N. S., Roberts, D. D. & Safer, B. (1997) Gene Ther. 4, 700-709. [DOI] [PubMed] [Google Scholar]
- 28.Harui, A., Suzuki, S., Kochanek, S. & Mitani, K. (1999) J. Virol. 73, 6141-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, D. G., Petek, L. M. & Russell, D. W. (2004) Nat. Genet. 36, 767-773. [DOI] [PubMed] [Google Scholar]
- 30.Nakai, H., Montini, E., Fuess, S., Storm, T. A., Grompe, M. & Kay, M. A. (2003) Nat. Genet. 34, 297-302. [DOI] [PubMed] [Google Scholar]
- 31.Miller, D. G., Rutledge, E. A. & Russell, D. W. (2002) Nat. Genet. 30, 147-148. [DOI] [PubMed] [Google Scholar]
- 32.Mizusawa, H., Taira, M., Yaginuma, K., Kobayashi, M., Yoshida, E. & Koike, K. (1985) Proc. Natl. Acad. Sci. USA 82, 208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokino, T., Fukushige, S., Nakamura, T., Nagaya, T., Murotsu, T., Shiga, K., Aoki, N. & Matsubara, K. (1987) J. Virol. 61, 3848-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C. S., Pawliuk, R., Morillon, E., et al. (2003) Science 302, 415-419. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell, R. S., Beitzel, B. F., Schroder, A. R., Shinn, P., Chen, H., Berry, C. C., Ecker, J. R. & Bushman, F. D. (2004) PLoS Biol. 2, 1127-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bill, C. A. & Summers, J. (2004) Proc. Natl. Acad. Sci. USA 101, 11135-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothkamm, K. & Lobrich, M. (2003) Proc. Natl. Acad. Sci. USA 100, 5057-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, J., Bibikova, M., Whitby, F. G., Reddy, A. R., Chandrasegaran, S. & Carroll, D. (2000) Nucleic Acids Res. 28, 3361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urnov, F. D., Miller, J. C., Lee, Y. L., Beausejour, C. M., Rock, J. M., Augustus, S., Jamieson, A. C., Porteus, M. H., Gregory, P. D. & Holmes, M. C. (2005) Nature 435, 646-651. [DOI] [PubMed] [Google Scholar]
- 40.Balamotis, M. A., Huang, K. & Mitani, K. (2004) Virology 324, 229-237. [DOI] [PubMed] [Google Scholar]
- 41.Shayakhmetov, D. M., Li, Z. Y., Gaggar, A., Gharwan, H., Ternovoi, V., Sandig, V. & Lieber, A. (2004) J. Virol. 78, 10009-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]