The activation of T lymphocytes is finely calibrated by positive and negative signals to allow a vigorous response to invading pathogens while at the same time avoiding the recognition and destruction of the host. Positive signals are delivered through the antigen-specific T cell receptor (TCR) and integrated with additional costimulatory signals. Counteracting these stimulatory effects, inhibitory receptors on lymphocytes serve to limit the response. Viruses have developed counterattacks to the host immune system, interfering with the process of antigen presentation (1) as well as with chemokine and cytokine signals (2). In a recent issue of PNAS, Cheung et al. (3) revealed that two evolutionarily divergent herpes family viruses, Herpes simplex virus (HSV), a member of the α herpes family, and human cytomegalovirus (hCMV), a member of the β herpesvirus family, target the same cosignaling pathway. Cheung et al. show that a previously identified orphan receptor of the TNFR family, UL144, expressed by human CMV, binds to an inhibitory receptor to down-regulate T cell responses. HSV-1 glycoprotein D (gD) binds to the ligand of this same inhibitory receptor and uses it as an entry mediator (4).
Two major families of cosignaling receptors participate in modulating the T cell response: the CD28 family and the TNFR superfamily. Whereas engagement of CD28 by its B7 family ligands results in positive signals delivered to T cells, additional CD28 family members contribute positively or negatively to T cell activation (5). Several members of the TNFR family also provide activation and/or survival signals to T cells (6). The TNFR family member herpesvirus entry mediator (HVEM) delivers costimulatory signals to T cells upon binding its TNF family ligand LIGHT (for lymphotoxin-like, exhibits inducible expression, and competes with HSV glycoprotein D for HVEM, a receptor expressed by T lymphocytes) (7).
Recently, a new member of the inhibitory family of CD28/Ig superfamily receptors, B and T lymphocyte attenuator (BTLA), was identified (8). BTLA contains immune receptor tyrosine inhibition motifs and upon aggregation recruits tyrosine phosphatases SHP-1 and -2, leading to attenuation of T cell activation (8). BTLA is expressed on activated and anergic T cells, resting B cells, and, at lower levels, on dendritic cells and macrophages (8, 9). Surprisingly, HVEM was discovered to be a ligand for BTLA (10, 11), providing the first example of a functional interaction between a TNFR and an Ig superfamily member. Binding of HVEM to BTLA delivers an inhibitory signal to T cells, whereas BTLA binding does not appear to trigger HVEM (10, 11). Upon binding their trimeric ligands, TNFR family members signal by recruiting trimeric TNFR-associated factors to the receptor trimers (12). If the interaction of BTLA with HVEM imposes a different oligomerization state on HVEM, this may explain why HVEM-BTLA binding leads to BTLA but not HVEM signaling.
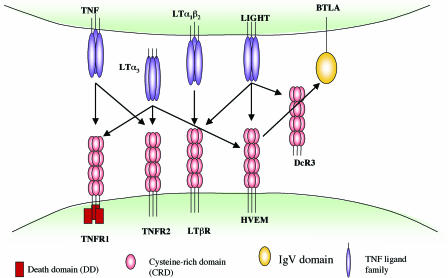
The implications of these findings are that HVEM may act as a molecular switch, delivering positive costimulatory signals upon binding LIGHT or acting as a ligand to deliver inhibitory signals via BTLA. Yet, this picture is further complicated by additional HVEM ligands. In addition to LIGHT, HVEM also binds to the TNF family ligand lymphotoxin-α (LTα3), and LIGHT itself has three receptors, HVEM, LTβ receptor (LTβR), and the decoy receptor DcR3 (Fig. 1) (13). Moreover, as its name suggests, HVEM was initially identified as a receptor for HSV gD (4). It is intriguing that HSV-1 and -2 have coopted HVEM, a molecule that serves both positive and negative regulatory functions in the immune system, as a receptor for cell entry.
Fig. 1.
HVEM and LIGHT interactions within the TNFR and Ig superfamily. TNF-family ligands usually exist as trimers, and the receptors are also trimeric in their ligand-bound form, although higher order oligomers are possible (12). The oligomerization state of HVEM bound to BTLA or LIGHT is unknown. The networked interactions of this cosignaling family suggest that interference with one receptor/ligand interaction may have a domino effect by freeing up other interacting partners.
The extracellular domain of HVEM, like other TNFR family members, is made up of cysteine-rich repeat domains (CRD) of which human HVEM has four. The LIGHT-binding site has been mapped to CRD2 and CRD3 (14), whereas HVEM binds BTLA through its CRD1 (11). BTLA competes with gD for binding HVEM (10). HSV-1 gD contains a V-like Ig domain core and binds to HVEM through an N-terminal hairpin extension that contacts the CRD1 as well as making additional contact with CRD2 on the face opposite the LIGHT binding site (15). Cheung et al. (3) used site-directed mutagenesis to further delimit these binding sites. They provide evidence that gD and BTLA bind to distinct but overlapping sites on the HVEM CRD1. Although HSV gD and LIGHT bind on opposite sides of HVEM (Fig. 2) and had previously been shown to form a ternary complex with HVEM when components were soluble (10), gD blocks HVEM-Fc binding to membrane-bound LIGHT, likely by steric hindrance (3, 7). Therefore, with membrane-bound LIGHT, gD appears to nullify all interactions of HVEM while using it as an entry mediator. How this will impact on HVEM ligands LIGHT and LTα3, which presumably would then be free to interact with their other cognate receptors (Fig. 1), remains to be determined.
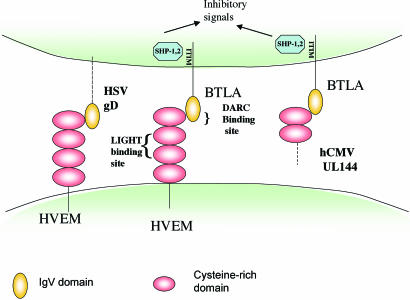
Fig. 2.
Viral interactions with the BTLA/HVEM/LIGHT cosignaling pathway. HSV gD binds to the membrane-distal CRD1 domain of HVEM opposite the LIGHT-binding site and overlapping the binding site of HVEM for BTLA. hCMV UL144 acts as a mimic of HVEM and binds to BTLA to send an inhibitory signal to T cells (3). gD can be expressed as a transmembrane protein in infected cells but would also bind to HVEM as part of the HSV envelope. UL144 is expressed intracellularly in infected cells (16) but as a transmembrane protein could form part of the viral envelope or be shed from infected cells. The binding site for LIGHT on HVEM is opposite its “DARC” side. ITIM, immune receptor tyrosine inhibition motif.
The precise mapping of the BTLA-binding site on HVEM by Cheung et al. (3) proved to be particularly informative. They found that K64, with additional contributions from R62 and E65, is critical for BTLA binding. These residues form a charged ridge on the solvent-exposed surface of HVEM. Accordingly, the BTLA/gD binding site on HVEM opposite the LIGHT-binding site has been coined by Cheung et al. (3) as the “DARC” side of HVEM (gD and BTLA binding site on the TNF receptor HVEM in the cysteine-rich domain 1) (Fig. 2). Insightfully, the authors noted that the key amino acids on HVEM required for binding BTLA were conserved in the hCMV-encoded protein, UL144, previously identified as an orphan receptor of the TNFR family (16). The authors went on to show that a soluble human BTLA-Fc fusion protein bound to cells transfected with UL144 from five different clinical isolates of human CMV. In T cell activation assays, HVEM-Fc and UL144-Fc each inhibited T cell proliferation. Thus, the studies of Cheung et al. identify UL144 as a virally encoded mimic of HVEM that coopts the BTLA inhibitory pathway to inhibit T cell activation.
UL144-Fc inhibits T cell proliferation with greater efficacy than HVEM-Fc despite a lower affinity for BTLA, suggesting that its effects in vivo might be to subvert the immune system via BTLA (3). Indeed, the finding that UL144 from five diverse clinical isolates of CMV maintains BTLA binding despite the sequence variation in the CRD1 argues for the importance of UL144-BTLA interaction in the virulence of hCMV (3). One puzzling aspect of UL144 is that although it is readily detected on transfected cells, when expressed in CMV infected fibroblasts, it appeared to be largely intracellular (16). However, hCMV- and HIV-coinfected individuals have antibodies against UL144, suggesting exposure of the human immune system to this protein (16).
A key issue that remains to be resolved is determining the net effect of these competing interactions when the different receptors and ligands are present simultaneously. Does HVEM acts as a molecular switch such that in the presence of costimulation through LIGHT, HVEM-LIGHT interactions override the inhibitory signals provided by HVEM-BTLA interactions, as suggested by Cheung et al.? Indeed, the affinity of soluble LIGHT for HVEM is 10-fold greater than the affinity of HVEM-Fc to membrane-bound BTLA (3). However, affinity measurements between BTLA and HVEM are different depending on which receptor is soluble and which is membrane-bound (3). How these interactions will play out in vivo will depend on the concentration, valency, and accessibility of the ligands and receptors on the surface of interacting immune cells. Moreover, the relative kinetics of expression of LIGHT versus BTLA on activated T cells during an immune response in vivo has not been determined, raising the possibility that the appearance of different HVEM ligands may play out as a “first past the post” phenomenon. The use of gene-targeted mice where various elements of this pathway are deficient in particular immune cell subsets or in antigen-specific T cells as well as kinetic analysis of the receptors and ligands during in vivo immune responses may help to unravel this complex biological system.
Together, the studies of Cheung et al. highlight the ability of HVEM to use two distinct surfaces to interact with its diverse ligands. The finding that two evolutionarily divergent herpesviruses target the same cosignaling pathway underscores the importance of LIGHT/HVEM/BTLA interactions in immune regulation. These findings remind us that viruses have a lot to teach us about the immune system.
See companion article on page 13218 in issue 37 of volume 102.
References
- 1.Tortorella, D., Gewurz, B. E., Furman, M. H., Schust, D. J. & Ploegh, H. L. (2000) Annu. Rev. Immunol. 18, 861-926. [DOI] [PubMed] [Google Scholar]
- 2.Alcami, A. (2003) Nat. Rev. Immunol. 3, 36-50. [DOI] [PubMed] [Google Scholar]
- 3.Cheung, T. C., Humphreys, I. R., Potter, K. G., Norris, P. S., Shumway, H. M., Tran, B. R., Patterson, G., Jean-Jacques, R., Yoon, M., Spear, P. G., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 13218-13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427-436. [DOI] [PubMed] [Google Scholar]
- 5.Greenwald, R. J., Freeman, G. J. & Sharpe, A. H. (2005) Annu. Rev. Immunol. 23, 515-548. [DOI] [PubMed] [Google Scholar]
- 6.Watts, T. H. (2005) Annu. Rev. Immunol. 23, 23-28. [DOI] [PubMed] [Google Scholar]
- 7.Mauri, D. N., Ebner, R., Montgomery, R. I., Kochel, K. D., Cheung, T. C., Yu, G. L., Ruben, S., Murphy, M., Eisenberg, R. J., Cohen, G. H., et al. (1998) Immunity 8, 21-30. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe, N., Gavrieli, M., Sedy, J. R., Yang, J., Fallarino, F., Loftin, S. K., Hurchla, M. A., Zimmerman, N., Sim, J., Zang, X., et al. (2003) Nat. Immunol. 4, 670-679. [DOI] [PubMed] [Google Scholar]
- 9.Hurchla, M. A., Sedy, J. R., Gavrieli, M., Drake, C. G., Murphy, T. L. & Murphy, K. M. (2005) J. Immunol. 174, 3377-3385. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez, L. C., Loyet, K. M., Calemine-Fenaux, J., Chauhan, V., Wranik, B., Ouyang, W. & Eaton, D. L. (2005) Proc. Natl. Acad. Sci. USA 102, 1116-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sedy, J. R., Gavrieli, M., Potter, K. G., Hurchla, M. A., Lindsley, R. C., Hildner, K., Scheu, S., Pfeffer, K., Ware, C. F., Murphy, T. L. & Murphy, K. M. (2005) Nat. Immunol. 6, 90-98. [DOI] [PubMed] [Google Scholar]
- 12.Zhang, G. (2004) Curr. Opin. Struct. Biol. 14, 154-160. [DOI] [PubMed] [Google Scholar]
- 13.Ware, C. F. (2005) Annu. Rev. Immunol. 23, 787-819. [DOI] [PubMed] [Google Scholar]
- 14.Rooney, I. A., Butrovich, K. D., Glass, A. A., Borboroglu, S., Benedict, C. A., Whitbeck, J. C., Cohen, G. H., Eisenberg, R. J. & Ware, C. F. (2000) J. Biol. Chem. 275, 14307-14315. [DOI] [PubMed] [Google Scholar]
- 15.Carfi, A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169-179. [DOI] [PubMed] [Google Scholar]
- 16.Benedict, C. A., Butrovich, K. D., Lurain, N. S., Corbeil, J., Rooney, I., Schneider, P., Tschopp, J. & Ware, C. F. (1999) J. Immunol. 162, 6967-6970. [PubMed] [Google Scholar]