Abstract
ISHp608 from Helicobacter pylori is active in Escherichia coli and represents a recently recognised group of insertion sequences. Its transposase and organisation suggest that it transposes using a different mechanism to that of other known transposons. The IS was shown to excise as a circular form, which is accompanied by the formation of a resealed donor plasmid backbone. We also demonstrate that TnpA, which is less than half the length of other transposases, is responsible for this and for ISHp608 transposition. Transposition was shown to be site specific: both insertion and transposon excision require a conserved target, 5′TTAC. Deletion analysis suggested that potential secondary structures at the left and right ends are important for transposition. In vitro TnpA bound both ends, showed a strong preference for a specific single-stranded DNA and introduced a single-strand break on the same strand at each end. Although many of the characteristics of ISHp608 appear similar to rolling-circle transposons, there are differences suggesting that, overall, transposition occurs by a different mechanism. The results have permitted the formulation of several related models.
Keywords: excision, in vitro, in vivo, integration, transposase activity
Introduction
Transposable genetic elements are found in most sequenced genomes and in many organisms for which the entire genome sequence is not yet available. They can be classified into several groups based on the chemistry used in their mobility. Currently, four major groups have been identified. These are DDE transposases, by far the most abundant recognised at present; Y- and S-transposases, related to the tyrosine (Y) and serine (S) site-specific recombinases; and Y2 enzymes, which share many characteristics of the rolling-circle (RC) replicases (RCR; for reviews, see Chandler and Mahillon, 2002; Curcio and Derbyshire, 2003; Cornet and Chandler, 2004). Recently, analysis of a group of insertion sequences isolated from Helicobacter pylori revealed another type of enzyme that does not appear to carry characteristic features of these groups and whose catalytic properties are at present unknown (Kersulyte et al, 1998, 2000, 2002; Chandler and Mahillon, 2002). It is related to the predicted transposase of the insertion sequence IS200 of Salmonella typhimurium (Lam and Roth, 1983).
IS200-like sequences are present in a wide range of bacteria including Gram-positive and Gram-negative eubacteria and archae (Devalckenaere et al, 1999; Beuzon et al, 2004). Some, such as IS605, carry two open reading frames (orfs), tnpA and tnpB. Using IS1541 from Yersinia pestis (Odaert et al, 1998) as an enquiry sequence confirmed that tnpA from IS605, isolated from H. pylori, was a member of this group. A more recent search of the public databases using tnpA and tnpB from IS605 as query sequences revealed a large number of related elements in over 65 bacterial species (B Ton-Hoang and M Chandler, unpublished results). These exhibit various combinations and configurations of the tnpA and tnpB reading frames. Some are listed in Figure 1. Members of one large group (Figure 1Ai), including IS200 (Lam and Roth, 1983) and IS1541 (Odaert et al, 1998; Beuzon et al, 2004), carry a single orf, tnpA. For those that carry two orfs, these may be divergent (Figure 1Aii: e.g. IS605, IS1523) or expressed in the same direction with tnpA upstream of tnpB. In these latter cases, the orfs may be separate (Figure 1Aiii: e.g. IS8301, ISH1–8) or partially overlapping (Figure 1Aiv: e.g. ISHp608). In addition, isolated copies of tnpB derivatives can also be found. Some of these are thought to be IS elements (e.g. IS891, IS1341, http://www-is.biotoul.fr) while for others, such as the virulence gene gipA of the Salmonella phage Gifsy1 (Stanley et al, 2000), no mobility functions have been suggested. tnpB is also sometimes associated with another transposase which is a member of the S-transposases (e.g. IS607; Kersulyte et al, 2000; see also ISfinder: www-is.biotoul.fr).
Figure 1.

The IS200/IS605 family. (A) Genetic organization. (i) IS200/IS1541 group with tnpA alone: examples include IS200 (Salmonella typhimurium, E. coli), IS1541 (Yersinia pseudotuberculosis, Yersinia pestis), IS1469 (Clostridium perifringens) and ISW1 (Wolbachia sp). (ii) IS605/IS606 group with tnpA and tnpB in a divergent orientation: IS605, IS606 (Helicobacter pylori); ISLjo5 (Lactobacillus johnsonii). (iii) IS8301 group with nonoverlapping tnpA and tnpB orfs in the same direction: IS8301 (Deinococcus radiodurans), ISH-1 (Halobacterium), ISEfa4 (Enterococcus faecium), IS1253 (Dichelobacter nodosus). (iv) ISHp608 with overlapping tnpA and tnpB: ISHp608 (H. pylori). LE and RE are represented by dark and light grey boxes, respectively. orfs are indicated as boxes with arrowheads (showing the direction of translation). tnpA is shown in dark grey and tnpB in light grey. Note that the expression of the overlapping tnpA and tnpB frames in ISHp608 may be translationally coupled. (B) Comparison of different TnpA proteins. Sequence alignment was performed using Multalin (Corpet, 1988). Sequences corresponding to aa 5–130 of TnpA are shown. The consensus was generated using IS605 TnpA genes (see www-is.biotoul.fr). Residues with 100% conservation over the entire family are shown in black on a grey background.
The group of H. pylori ISs includes IS605, IS606 and ISHp608 (Kersulyte et al, 1998, 2000, 2002). In contrast to IS200 (Haack, 1995), the H. pylori elements transpose in Escherichia coli at reasonable frequencies in a standard ‘mating-out' assay using a derivative of the conjugative F plasmid as a target (Kersulyte et al, 1998, 2000, 2002). IS605 and ISHp608 insert 3′ to conserved AT-rich penta- or tetranucleotides 5′TTTAA or TTAAC (IS605) and TTAC (ISHp608). They do not carry terminal inverted repeats and their insertion does not result in duplication of target sequences. Instead, their left (LE) and right (RE) ends include significant potential secondary structure elements (see below and Figure 5B) reminiscent of the replication origin region of single-stranded (ss) bacteriophages, plasmids with an RC replication mechanism and members of the IS91 family of insertion sequences (see Espinosa et al, 1995; Khan, 1997). The sequence of TnpA, a predicted protein of only 155 aa, is remarkably conserved from element to element (Figure 1B). Interruption of ISHp608 tnpA eliminated transposition activity in E. coli, while interruption of tnpB had no observable effect (Kersulyte et al, 2002).
Figure 5.
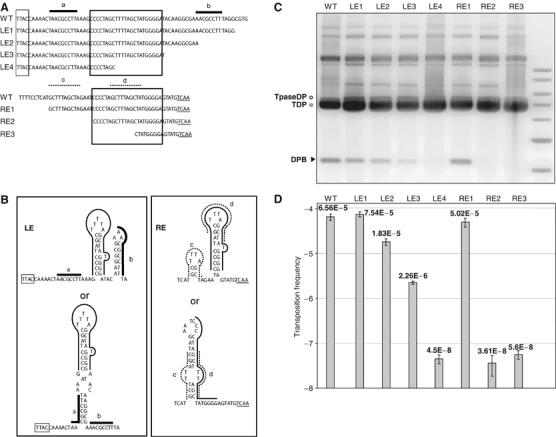
Deletion analysis of LE and RE. (A) Sequences of LE, RE and deletions used for the analysis. The 22–23 bp palindromic (box) and two different smaller directly repeated sequences a-b (LE) and c-d (RE) are indicated (overlined). (B) Potential secondary structures in LE and RE of ISHp608. Secondary structure predictions were obtained with ‘MFolds' (www.bioinfo.rpi.edu). The tetranucleotides TTAC (boxed) and TCAA (underlined) at the tip of RE are indicated. For each sequence, two alternative structures with similar ΔG are shown. Positions of repeated sequences are indicated. (C) Excision activity of truncated substrates: separation of plasmid DNA from alkaline lysates on 0.8% agarose gel. Lane 1: wild-type (WT) pBS102; lanes 2–5: LE1, LE2, LE3 and LE4; lanes 6–8: RE1, RE2 and RE3. The positions of pBS107 transposase donor TpaseDP (o) plasmid, TDP (*) and DPB (arrowhead) are indicated. (D) Frequency of transposition measured in mating-out assay. Error bars are based on n⩾4 individual measurements. The columns correspond to the plasmid lanes of part C.
We present here an analysis of ISHp608 transposition. We demonstrate that the element excises from a donor plasmid molecule, probably in a circular form, and that the donor backbone is precisely resealed. A correlation between excision and transposition was also observed, suggesting that this form may be an intermediate in the ISHp608 transposition pathway. Moreover, the 5′ flanking target 5′TTAC was not only required for integration (Kersulyte et al, 2002) but was essential for ISHp608 excision and transposition. The sequence requirements at LE and RE for transposition were explored using a series of deletion derivatives. In vitro TnpA bound and cleaved each transposon end. Moreover, it preferred ssDNA and bound specifically and almost exclusively in a strand-specific way. These results have allowed us to propose alternative but related models of ISHp608 transposition and to relate this to other known families.
Results
Excision activity of ISHp608: the donor plasmid backbone
Previous results had shown that, in E. coli, insertion of ISHp608 into the conjugative plasmid pOX38 occurred in a sequence-specific manner, required tnpA but not tnpB and did not lead to fusion between the target and donor replicons (Kersulyte et al, 2002).
To explore ISHp608 transposition further, we examined the plasmid species present after overnight growth of E. coli carrying pBS:ISHp608wt (Kersulyte et al, 2002). As can be seen in the gel stained with Sybr green, Figure 2 (lane 1),this transposon donor plasmid (TDP, *) generated a smaller derivative. Restriction mapping and DNA sequencing (see below) demonstrated that this is a recircularised donor plasmid backbone (DPB, arrowhead).
Figure 2.
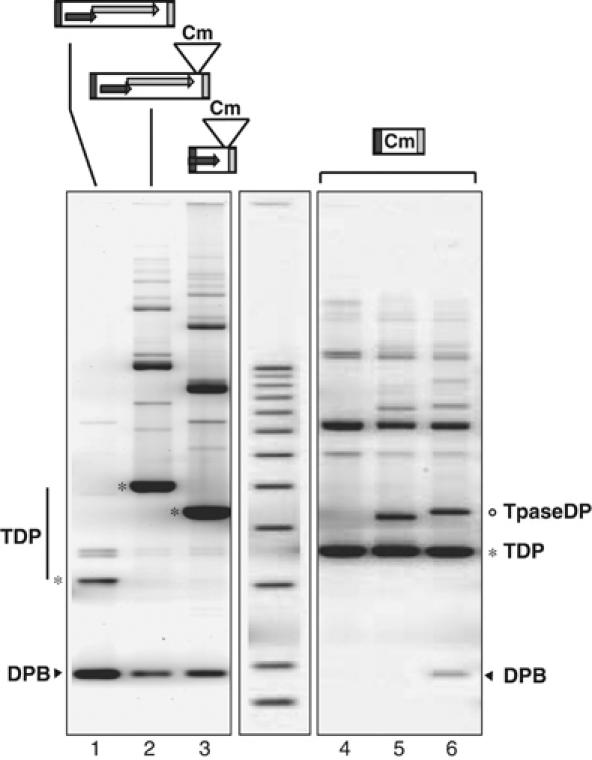
In vivo excision of ISHp608. Upper panel: Structure of the ISHp608 derivatives carried by pBS∷ISHp608 (WT), pBS101 (insertion of a Cm cassette of 4 kb between the end of tnpB and RE), pBS105 (substitution of the major part of tnpB by the Cm cassette) and pBS103 (entire tnpA and tnpB substituted by the Cm cassette). Symbols are as described in Figure 1A. Lower panel: Separation of plasmid DNA from alkaline lysates on 0.8% agarose gels. Lane 1: pBS:ISHp608 (wild-type TnpA-TnpB); lane 2: pBS101 (TnpA-TnpB-Cm); lane 3: pBS105 (TnpA-Cm); lane 4: pBS103 (LE-Cm-RE) alone; lane 5: pBS103+pAPT110, a vector plasmid without the tnpA gene; lane 6: pBS103+pBS107, the transposase donor plasmid (TpaseDP). The positions of TDP and TpaseDP are indicated by * and °, as is the DPB (arrowhead). The middle lane shows a standard 1 kb ladder.
Two additional plasmids were constructed in which a CmR gene was inserted downstream of tnpB leaving this frame intact (pBS101; lane 2) or was substituted for tnpB (pBS105; lane 3). In both cases, a smaller species of a similar structure and size to that found with the wild-type IS could be seen (DPB, arrowhead). We note that the relative quantity of DPB varied from experiment to experiment. This presumably reflects the time during growth of the culture at which the first excision events occurred to generate the replicative sealed donor backbone, and is in turn due to low levels of transposase produced stochastically from the resident ISHp608 promoter. This variation does not occur if TnpA is provided in trans under an inducible promoter (see below). These results imply that TnpA alone is responsible for the production of the DPB.
To define some of the requirements for production of the DPB and to determine whether TnpA is active in trans, derivative plasmids carrying the CmR gene were constructed in which both tnpA and tnpB had been removed leaving LE (143 base pairs (bp)) and RE (79 bp) of ISHp608 intact (pBS103). The behaviour of pBS103 was investigated in the presence or absence of TnpA supplied from a cloned gene under control of the inducible plac promoter on a compatible plasmid (pBS107). Growth was overnight in the presence of IPTG. Bands migrating high in the gel are presumably multimeric forms of donor transposon plasmids. The DPB species was clearly visible when TnpA was supplied in trans to pBS103 (arrowhead, lane 6). Similar results were obtained with a 6 h induction. It was not produced in the absence of the tnpA-carrying plasmid (lane 4) or with the transposase vector plasmid alone (lane 5). The results demonstrate that while TnpA is active in trans, the activity is rather poor as demonstrated by the weak DPB band.
Excision activity of ISHp608: the transposon
While the results shown in Figure 2 demonstrate TnpA-promoted production of the DPB (capable of autonomous replication), they do not provide information on the fate of the excised ISHp608 (which is presumably unable to replicate autonomously). To examine this, samples from cultures were collected after 5 h induction of TnpA production (pBS∷ISHp608, or pBS102 together with pBS107). pBS102 is similar to pBS103 but includes a longer CmR cassette (Materials and methods). DNA was extracted by the cleared lysate method and loaded onto an agarose gel at high concentration. Of the two plasmid configurations, the combination of pBS∷ISHp608 and pBS107 proved the most efficient in generating the DPB (data not shown).
Two different exposures of the same Sybr green stained gel with DNA samples from a strain carrying pBS∷ISHp608 and pBS107 are presented in Figure 3A (lanes 5 and 6, and 7 and 8, respectively). In addition to the intact TDP and the recircularised DPB (* and closed arrowhead, respectively, lanes 5 and 7), two faint bands migrating towards the bottom of the gel could be detected with the longer exposure (open arrowheads, lane 7). Following digestion with HincII, which cleaves once within the transposon and once within the DPB, the lower of the two bands disappeared, presumably converted into a linear form (lanes 6 and 8). This suggests that the excised IS exists in a double-stranded (ds) circular form although it does not rule out the presence of ss transposon species. Note that the HincII sites are separated by only 49 bp in the donor plasmid and that this second fragment is not detectable in this gel. We are not sure whether the upper faint band (lane 7) is a linear, open circular or ss form. In the latter case, this would be resistant to cleavage. While it has approximately the size expected for linear transposon, like the lower band, it gives a good PCR signal when tested for the presence of the junction. If it were a linear transposon form, the DNA must therefore be broken at a distance from the junction (nonspecific breakage would generate this pattern).
Figure 3.
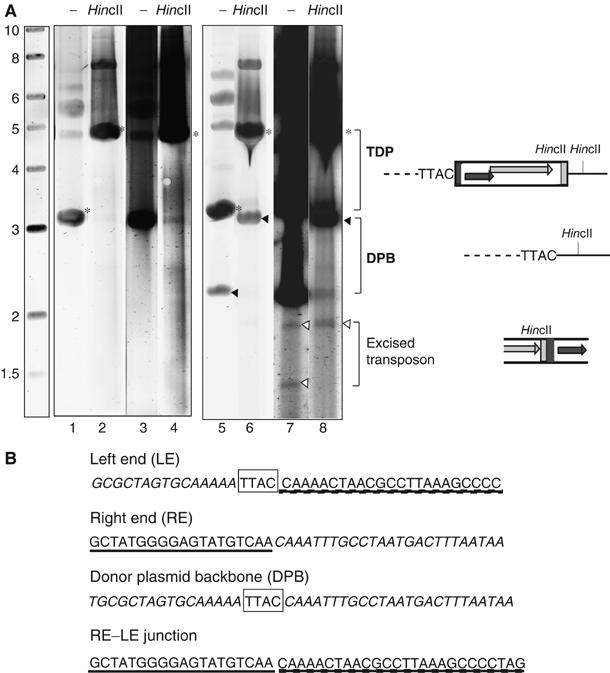
Excised transposon and donor plasmid backbone. (A) Separation of plasmid DNA from cleared lysates on a 0.8% agarose gel stained with Sybr green. Lanes 1–4: the host strain carried pBS107 (additional source of TnpA) and pBS109 (a derivative of pBS:Hp608 with a wild-type (WT) IS whose transposition has been inactivated by mutation of the TTAC tetranucleotide flanking LE). Lanes 1 and 2 show undigested and HincII-digested plasmid DNA, respectively. Lanes 3 and 4 are identical but overexposed. No products or transposition intermediates are observed. Lanes 5–8: the host strain carried pBS:Hp608 (WT IS) and pBS107. Lanes 5 and 7: native DNA; lanes 6 and 8: DNA digested with HincII. A schema of the different species is shown on the right. Symbols are as described in Figure 1A. The positions of TDP, DPB and excised transposon are indicated. Gel lanes 5 and 6 are from a normal exposure and lanes 6 and 8 are from a strong exposure. (B) Sequences of LE, RE, DPB and the RE–LE junction of the excised transposon. The TTAC target sequence is boxed. DNA sequences of LE and RE are underlined.
To clarify the assignment of bands we have made to the upper species on the gel, we have used a donor plasmid in which the transposon had been inactivated by mutation of the required TTAC (to GCTT) and therefore does not generate transposition products or intermediates (see below). The results (Figure 3A, lanes 1–4) show that this mutation eliminates the bands corresponding to the DPB and the excised transposon species (see also below). The circular nature of the transposon was confirmed using PCR analysis of isolated DNA bands to show the presence of a junction fragment carrying both left and right transposon ends (see below).
Nucleotide sequence of the donor backbone and transposon circle junctions
ISHp608 inserts site-specifically adjacent to the conserved tetranucleotide 5′TTAC (Kersulyte et al, 2002). To determine how ISHp608 might excise from its vector plasmid, the DNA sequence at the newly formed plasmid junction was determined. The results, shown in Figure 3B, demonstrate that the transposon had undergone a clean excision leaving the TTAC target sequence intact on the DPB. In order to obtain sufficient material of the potential junction of the excised ISHp608 molecule, it was first amplified by PCR on gel-purified products using oligonucleotides specific for each transposon end (Materials and methods). The sequence of the resulting fragment showed that LE and RE of the IS are abutted and carry no additional nucleotides (Figure 3B).
These results suggest that excision occurs by a mechanism involving sequence-specific recombination both in the DPB and in the IS element.
Requirement of flanking sequences for excision and transposition activity
To determine whether the 5′TTAC sequence and additional flanking nucleotides play a role in excision, a plasmid was constructed from pBS102 (carrying a CmR gene flanked by 143 bp of LE and 79 bp of RE of ISHp608) in which 5′TTAC was replaced by 5′GCTT while conserving the more distal flanking sequences (pBS115). TnpA was supplied in trans from pBS107. Both plasmids (p15A-based pBS107 and pBluscript-based pBS102 derivatives) migrate together under the conditions used here (Figure 4A, lanes 1 and 2, ○ and *). In contrast to pBS102 (Figure 4A, lane 2), no excised donor backbone species could be detected under these conditions with pBS115 (Figure 4A, lane 3).
Figure 4.
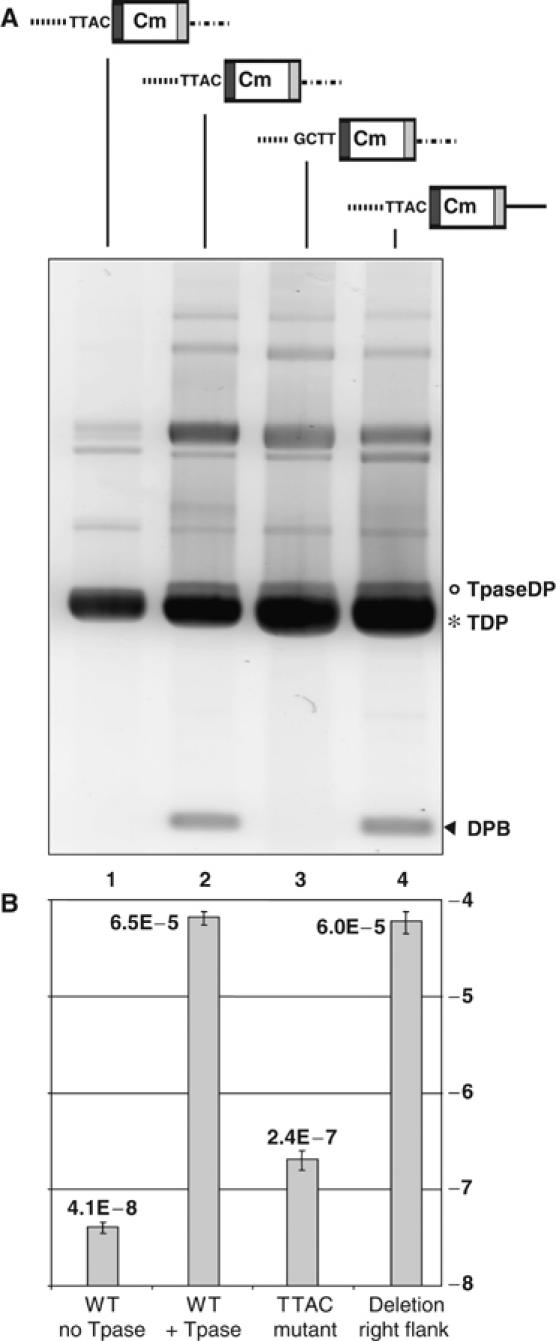
Requirement of flanking sequences. Upper panel: Structure of synthetic transposons: pBS102 (original flanking sequences); pBS115 (mutated for the target TTAC tetranucleotide); and pBS117 (retaining TTAC but with the right flanking sequences deleted). Symbols are as described in Figure 1A. Lower panel: (A) Separation of plasmid DNA from alkaline lysates on 0.8% agarose gel. Lane 1: pBS102 (WT)+vector plasmid pAPT110 with no TnpA; lanes 2–4: pBS102, pBS115 (-TTAC) and pBS117 (-flanking) with TnpA supplied from pBS107. The positions of pBS107 transposase donor plasmid TpaseDP (o), transposon donor TDP (*) and DPB (arrowhead) are indicated. (B) Frequency of transposition measured in a mating-out assay. Individual transposition frequencies are indicated on the figure on a log scale. Error bars are based on n⩾4 individual measurements. The columns correspond to the plasmid lanes of part A.
To determine whether flanking sequences of the donor plasmid other than the 5′TTAC were involved in transposition, additional derivatives were constructed. In these plasmids, the TTAC was retained but 40 and 35 bp were deleted from the more distal flanking DNA on LE and RE sides of the transposon, respectively. Derivatives with deletions at either or both ends retained their ability to generate the excised backbone. Only pBS117 (with a deletion of RE flanking DNA) is shown (Figure 4A, lane 4).
To determine whether the sequence requirements for transposition of ISHp608 were similar to those for excision, the CmR-tagged ISHp608 derivative plasmids were used in a mating-out assay (Materials and methods). The conjugative plasmid pOX38Km was used as a target and pBS107 as a source of TnpA in the donor strain. In the absence of TnpA (vector plasmid pAPT110; Figure 4B, column 1), pBS102 carrying a transposon composed of wild-type ends and flanking sequences gave a background frequency of 4 × 10−8. However, in the presence of TnpA (pBS107), transfer of the transposon-based CmR occurred at a frequency of 6.6 × 10−5 (column 2). Mutation of the conserved tetranucleotide TTAC (pBS115, column 3) reduced this transposition frequency to a very low level (2.4 × 10−7) while plasmid pBS117 (column 4), which retains the 5′TTAC tetranucleotide but carries a deletion of the flanking 50 bp from RE of the transposon, exhibited a frequency of 6 × 10−5, identical to that of the wild-type pBS102. The fact that pBS115 shows a low transposition activity but does not generate detectable DPB simply reflects a lower sensitivity in the physical assay compared to the biological (mating) assay. Thus, there is a good correlation between the effect of 5′TTAC on excision (Figure 4A) and in transposition (Figure 4B).
Thus, the tetranucleotide TTAC is not only necessary for integration of ISHp608 (Kersulyte et al, 2002) but is also required for its excision leading to DPB formation while additional flanking DNA sequences are not.
Deletion analysis of LE and RE
Although elements of this IS family do not carry terminal inverted repeats, most carry sequences within LE and RE that are rich in repeats, imperfect palindromes and potential secondary structures (Beuzon and Casadesus, 1997; Odaert et al, 1998; Beuzon et al, 1999; see also Discussion). In the case of ISHp608, two nearly identical 22–23 bp palindromic sequences are present as direct repeats located 21 bp from LE and 10 bp from RE (Figure 5A and B; Kersulyte et al, 2002). Each end also includes two different smaller directly repeated sequences a-b (LE) and c-d (RE) (Figure 5A).
To determine which sequence features in LE and RE are necessary for excision and transposition activity, successive deletions from the interior were generated separately on each end (Figure 5A). The resulting ISHp608 derivatives were tested for their capacity to excise as a recircularised vector plasmid backbone. TnpA was supplied by pBS107.
The results (Figure 5C) show that RE1, which retains the repeated element ‘c', also retained activity similar to that of the ‘full-length' ends (WT). Removal of the ‘c' repeat (RE2) or the ‘c' repeat and part of the palindrome (RE3), however, eliminated detectable excision activity.
The situation was less clear for LE where successive deletion progressively reduced excision activity (Figure 5C). Thus, removal of the ‘b' repeat (LE2) reduced but did not eliminate activity. A deletion up to, but not including, the palindromic region (LE3) further reduced activity while partial deletion of the palindrome (LE4) eliminated detectable excision in this assay.
Mating-out experiments were undertaken with these ISHp608 deletion derivatives. The results (Figure 5D) largely confirmed those obtained from analysis of excision activity (Figure 5C). RE1, which retains the repeated element ‘c', was fully active (column 6, 5.0 × 10−5) compared to the ‘full-length' RE (column 1, 6.6 × 10−5), while removal of the ‘c' repeat (RE2) or the ‘c' repeat and part of the palindrome (RE3) greatly reduced activity (columns RE2 and RE3, 3.6 and 5.6 × 10−8, respectively) compared to the vector plasmid alone (4 × 10−8, data not shown).
Again, the situation is less clear for LE, where successive deletion progressively reduced activity (Figure 5D). Thus, removal of the ‘b' repeat (LE2) only slightly reduced activity (1.8 × 10−5) compared to 6.6 and 7.5 × 10−5 for the ‘full-length' LE and LE1, respectively. The deletion up to, but not including, the palindromic region (LE3) further reduced activity (2.3 × 10−6) while partial deletion of the palindrome (LE4) eliminated detectable transposition in this assay (4.5 × 10−8) compared to the vector alone (Figure 4B, column 1, 4.1 × 10−8). A possible problem in interpreting this type of deletion data is that the new neighbouring sequences may provide additional secondary structures that influence the reactions. However, analysis of these neighbouring sequences (Supplementary Figure 1) did not support this hypothesis.
Integration of the excised transposon
The mating assays above are measures of the overall transposition activity, which presumably includes both transposon excision and integration. The correlation between the transposition activity and the ability to produce transposon circles suggested that the potential ISHp608 circular forms with two abutted ISHp608 ends may represent transposition intermediates. If this were the case, a plasmid carrying abutted LE and RE might be expected to insert with high frequency.
To test this, plasmid pBS125 containing an ISHp608 junction (LE: 143 bp; RE: 79 bp) was assembled (Table I) and used in the mating-out assay. When TnpA was supplied in trans from pBS107, this plasmid underwent efficient transposition into the pOX38Km target plasmid with a frequency of 2 × 10−3 compared to that obtained with vector control (5 × 10−8) and to that obtained with ISHp608 derivatives in which the ends are in the normal configuration (6.5 × 10−5; Figure 4 and Table I). The results support the idea that ISHp608 transposes using a circular intermediate with abutted LE and RE.
Table 1.
Integration of excised transposon and transposition activity of Y127A mutant
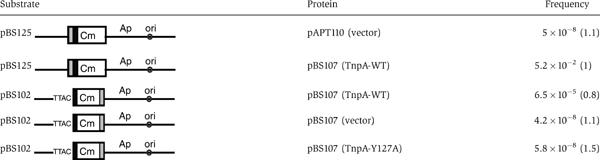 |
| Standard errors are shown in parentheses. |
Binding to LE and RE
To assess DNA binding and cleavage activities of TnpA, we developed several in vitro assays using TnpA purified on Ni-agarose resin as a TnpA-His6-tagged protein produced following induced expression from plac (pBS107; Materials and methods).
To determine whether TnpA-His6 was capable of binding the ISHp608 ends, its ability to retard electrophoretic migration of a 70 bp dsDNA fragment containing a complete copy of RE was first tested. This DNA fragment is specifically retarded by purified TnpA (Figure 6A, lanes 1–6). Incubation with increasing quantities of TnpA and an excess of nonspecific competitor DNA gave rise to one retarded species, in contrast to a vector control preparation (data not shown) (see Materials and methods). In view of the potential secondary structures (Figure 5) and their similarity to single-strand bacteriophage replication origins (see Discussion), the capacity of TnpA to bind ssDNA was determined. Single-strand oligonucleotides carrying 70 bases of the top or bottom strand of the same sequence of RE used above (Figure 6A) were tested in the gel retardation assay. The results demonstrate not only that TnpA binds ssRE (lanes 7–12) but that binding is strand specific (lanes 13–18). Only the top strand of RE can be efficiently retarded giving rise to two bands, one major and a minor, less retarded band. We estimate that it exhibits at least 102-fold higher affinity than the dsRE since the TnpA-His6 concentration in lane 6 was 1.2 × 10−6 M while that in lane 12 was only 4 × 10−8 M. In contrast, little or no binding to the bottom strand was detected (Figure 6A, lanes 13–18; final TnpA concentration, lane 18, 1.2 × 10−6 M). A preference for top strand binding was also observed with LE (Figure 6B). The patterns were almost identical for dsLE (Figure 6B, lanes 1–4) and for ‘top' strand LE (Figure 6B, lanes 5–8). Interestingly, some binding was observed with the ‘bottom' LE strand (Figure 6B, lane 12) at the highest TnpA concentration used. The detailed binding activities of TnpA are under investigation.
Figure 6.
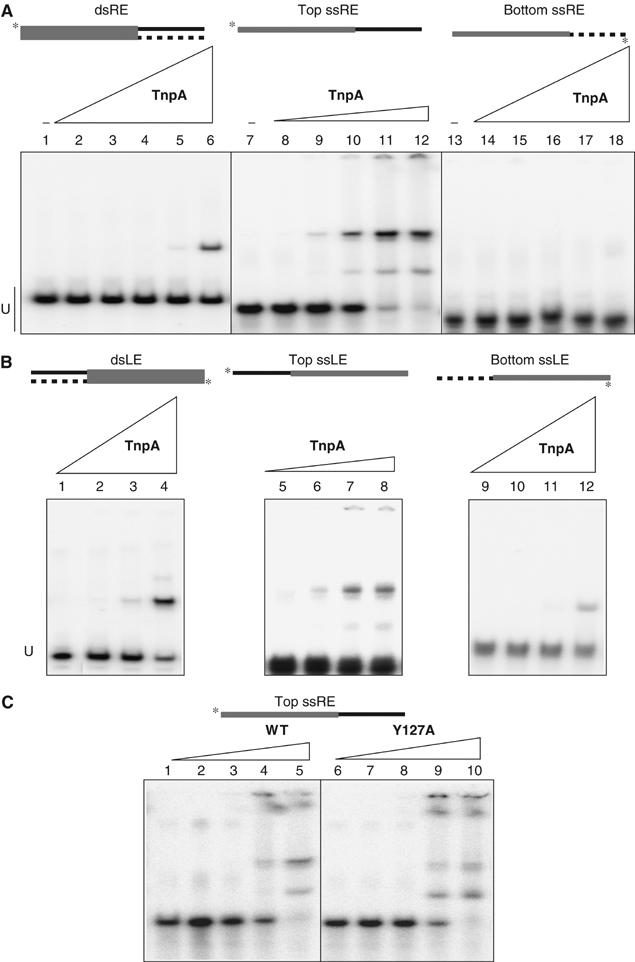
LE and RE binding activity of TnpA-His6 protein by electrophoretic mobility shift assay (EMSA). The LE- or RE-carrying DNA fragments are schematised at the top of the figure. (A) Left panel: EMSA of a 70 bp RE-carrying DNA fragment performed with 0, 0.2 × 10−8, 0.1 × 10−7, 0.5 × 10−7, 2.4 × 10−7 and 1.2 × 10−6 M of TnpA-His6 (lanes 1–6, respectively). Unbound DNA (U) and complexes are indicated. Middle panel: Binding of 70 bases ss top strand RE oligonucleotide with 0, 0.6 × 10−10, 0.3 × 10−9, 1.6 × 10−9, 8 × 10−9 and 4 × 10−8 M TnpA-His6 (lanes 7–12, respectively). Right panel: Binding of 70 bases ss bottom strand RE oligonucleotide with 0, 0.2 × 10−8, 0.1 × 10−7, 0.5 × 10−7, 2.4 × 10−7 and 1.2 × 10−6 M of TnpA-His6 (lanes 13–18). (B) Left panel: EMSA of an 80 bp LE-carrying DNA fragment performed with 0.1 × 10−7, 0.5 × 10−7, 2.4 × 10−7 and 1.2 × 10−6 M of TnpA-His6 (lanes 1–4, respectively). Unbound DNA (U) and complexes are indicated. Middle panel: Binding of 80 bases ss top strand LE oligonucleotide with 0.3 × 10−9, 1.6 × 10−9, 8 × 10−9 and 4 × 10−8 M TnpA-His6 (lanes 5–8, respectively). Right panel: Binding of 80 bases ss bottom strand LE oligonucleotide with 0.1 × 10−7, 0.5 × 10−7, 2.4 × 10−7 and 1.2 × 10−6 M of TnpA-His6 (lanes 9–12, respectively). (C) Binding activity of Y127A mutant: EMSA of a 70 bases ss top strand RE oligonucleotide performed with 0, 0.6 × 10−10, 0.3 × 10−9, 1.6 × 10−9 and 8 × 10−9 M TnpA-His6 (lanes 1–5 and 6–10 for wild-type and Y127A mutant protein, respectively).
These results demonstrate that TnpA-His6 specifically binds LE and RE, in particular with high affinity for ssDNA corresponding to the top strand.
Cleavage at LE and RE
To relate TnpA binding in vitro to cleavage, nicking assays were performed. dsDNA fragments, 5′ labelled with [γ-32P]ATP on one strand, were incubated with TnpA in reaction buffer containing Mg2+ and products were analysed on a denaturing sequencing gel. A discrete band corresponding to a cleavage product at LE was observed (Figure 7, lane 2). As judged by the size of the labelled single-strand fragment (60 nt), the point of cleavage occurred 3′ to the target 5′TTAC on the top strand. No additional species were observed when the bottom strand was labelled (data not shown). A similar result was obtained using a ds transposon junction fragment (lane 8). Surprisingly, no cleavage could be observed on dsRE DNA (lane 4). However, when presented with an ss oligonucleotide representing the top strand of RE, TnpA catalysed efficient cleavage. This occurred 3′ to the 5′TCAA, which defines the end of RE (lane 6).
Figure 7.
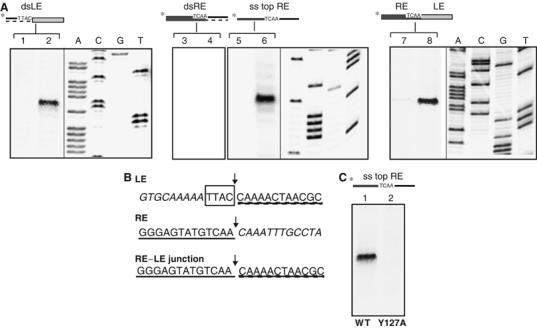
TnpA-His6 cleavage. (A) 141 bp LE-carrying fragment (lanes 1 and 2), 80 bp ds or 80 bases top strand RE-carrying fragment (lanes 3 and 4, and 5 and 6) and 177 bp RE–LE junction-carrying fragment (lanes 7 and 8) fragment were 5′ 32P-radiolabelled at each end separately. The DNA was incubated with 0 or 6 × 10−6 M TnpA-His6 in reaction buffer in the presence of Mg2+ as divalent cation (Materials and methods). The reactions were stopped with SDS and separated on an 8% sequencing polyacrylamide gel. The gel was then analysed by phosphorimaging. The uncleaved substrates are not shown on this gel. (B) Cleavage products on LE (64 bases), RE (53 bases) or RE–LE junction (79 bases) are indicated by arrowheads. (C) Cleavage of RE single-strand ‘top' strand performed with 1.2 × 10−6 M of wild-type and Y127A mutant TnpA-His6 (lanes 1 and 2, respectively).
Thus, TnpA cleaved 3′ to the tetranucleotide 5′TTAC target sequence on dsLE DNA (i.e. at the 5′ end of the transposon) introducing an ss break. Similarly, it cleaved 3′ to the RE tip 5′TCAA on a ds junction fragment. Since signals within RE are not sufficient for cleavage on a dsRE fragment, it seems probable that signals in LE are responsible for cleavage of the circle junction.
Is Y127 part of the catalytic site?
The alignment in Figure 1B shows that several residues are completely conserved throughout the family. These do not include motifs characteristic of classical DDE, Y-, S- or Y2 transposases. They do, however, include Y127, and H64 and H66 separated by a hydrophobic residue. These are similar to a subset observed in the Y2 (or RCR) enzymes, although significantly, TnpA does not carry a second Y residue. This implies that while the initial chemistry of both ISHp608 and the RCR transposons is the same using a tyrosine residue as the nucleophile, the enzymology of subsequent steps might be different.
To determine whether Y127 was involved in catalysis, a Y127A mutant was constructed and tested for transposition in vivo and for binding and cleavage in vitro. Transposition was reduced to background levels (Table I). While binding to the ends was similar to that obtained with the wild type (Figure 6C), the Y127A mutant did not support detectable cleavage (Figure 7C). This does not prove, but strongly suggests that Y127 is involved in catalysis.
Discussion
The results presented here show that the H. pylori insertion sequence, ISHp608, transposes by a mechanism different from that adopted by other known transposons. A plasmid carrying ISHp608 gave rise to two novel DNA species in overnight cultures of E. coli. These were identified as a circular copy of the plasmid backbone and, at very low levels, a copy of the transposon that also appears to be an excised circle (Figures 2 and 3). Species with the expected mobility and which generate a junction fragment with appropriate oligonucleotides in a PCR test were observed using several alternative plasmid configurations and transposon sizes (Figure 3 and data not shown).
Comparison of the DNA sequence at the plasmid–IS junctions of the original ISHp608-carrying plasmid with the newly formed junctions of the circular vector backbone and the excised ISHp608 confirmed that the vector backbone had been precisely resealed. Moreover, the excised transposon carried abutted LE and RE with no deletion or addition of base pairs (Figure 3B).
Inactivation of the second ISHp608 orf, tnpB, had no effect on excision activity, whereas removal of tnpA eliminated all activity (Figure 2). Moreover, the presence of an additional plasmid expressing TnpA in trans from a plac promoter restored excision activity to an ISHp608 copy ablated for both TnpA and TnpB (Figure 2). These results were recapitulated in a mating-out assay using the conjugative plasmid pOX38Km as a target, an assay that measures overall transposition activity (data not shown). The results of the mating assay fully confirmed those of Kersulyte et al (2002), showing that this short polypeptide of 155 aa is the only ISHp608 protein required for transposition in E. coli.
To explore the DNA sequences required for ISHp608 transposition activity, various deletion derivatives were constructed and their activity was examined using TnpA supplied in trans. ISHp608 inserts exclusively into the tetranucleotide TTAC (Kersulyte et al, 2002). Mutation of this sequence also eliminated detectable excision and drastically reduced transposition frequency in the mating-out assay. On the other hand, deletion of left or right flanking sequences while retaining the TTAC tetranucleotide had no effect (Figure 4).
Relation of ISHp608 to other transposons
TnpA encoded by ISHp608 and other members of this family is clearly different from all other known transposases. TnpA is very closely related to that of the S. typhimurium element IS200. orfs related to tnpA are widely distributed among eubacteria and archae and show high conservation. Comparison of TnpA, a predicted protein of 155 aa, with other members of the group is shown in Figure 1B (a full alignment can be found at www-is.biotoul.fr). They share the majority of conserved residues typical of the entire family (Figure 1B). The level of identity is high despite the large range of bacterial hosts, and extends over almost the entire length of the proteins. Several blocks of highly conserved amino acids are apparent. These include HxuxxxKyR, HuH and YuxxQ (marked * in Figure 1B). No DDE motif typical of many transposases is evident and no obvious relationship with the Y- or S-recombinases or the Y2 RC transposases could be detected although the HuH block is one of three motifs present in Y2 enzymes (see Ilyina and Koonin, 1992; Mahillon and Chandler, 1998; Curcio and Derbyshire, 2003).
The conserved Y residues in the protein initially suggested that this family may transpose using a reaction similar to that catalysed by Y-transposases. Indeed, mutation of the conserved Y127 resulted in abolition of both transposition (Table I) and cleavage (Figure 7C) but had no effect on TnpA binding to the ends (Figure 6C). This suggests that Y127 may be part of the catalytic site. However, the absence of an effect of flanking sequences on transposition and particularly the requirement of a conserved target tetranucleotide (5′TTAC) for integration and excision (Figure 4) is more reminiscent of IS91, which inserts at specific target tetranucleotides, CTTG or GTTC (Garcillan-Barcia et al, 2002). Examination of the target nucleotides of other members of the IS200/IS605 group reveals that several exhibit conserved blocks adjacent at LE (IS200: TTTT/TA, GT or AT; IS1541: TTTT or TXTT where X is A, G or C (Odaert et al, 1998; Devalckenaere et al, 1999); IS605: TTTAA or TTAAC (Kersulyte et al, 1998); IS606: TTAT (Kersulyte et al, 1998)). The sequence of additional members identified in the public databases is also consistent with the presence of a short conserved target sequence although, in the absence of observed transposition events, this is difficult to establish with certainty.
Although it remains to be formally demonstrated, IS91 probably transposes by an RC transposition mechanism. Members of this family encode a large transposase with sequence similarity to Rep proteins of the pC194/pUB110 plasmid family and to gpA proteins of φX174 family of ssDNA phages, which all undergo RC replication (Mendiola and de la Cruz, 1992; Garcillan-Barcia et al, 2002). These proteins display some sequence motifs also conserved in Rep proteins. They include a motif containing a pair of tyrosine residues that form the catalytic pocket, as well as a triad of histidines (Garcillan-Barcia et al, 2002). These two motifs are also carried, although in a different order, by the relaxase family, proteins involved in conjugative DNA transfer (Byrd and Matson, 1997; Llosa et al, 2002).
Like ISHp608, IS91 produces circular dsDNA containing transposon sequences covalently joined at its exact termini. However, ssIS91 circles, one of the hallmarks of RC replication, were also observed (del Pilar Garcillan-Barcia et al, 2001). As yet, we have no evidence for production of ISHp608 ss circles.
In contrast to ISHp608, no recircularised IS91 DPB could be identified. Moreover, TnpA (155 aa) is less than half the size of the IS91 transposase and other RC enzymes (Mendiola et al, 1992; Khan, 1997). This suggests that the transposition mechanisms of IS91 and ISHp608 differ. However, like those of IS91, the ISHp608 termini contain extensive potential secondary structures. ISHp608 carries a pair of subterminal directly repeated sequences, as well as repeated sequences specific for each end (Figure 5A). Secondary structure prediction using ‘MFolds' (www.bioinfo.rpi.edu) revealed potential structures in LE and RE (Figure 5B) that could correspond to a replication origin and terminator, characteristic for RC plasmids and members of the IS91 family. Systematic analysis of many other members of the family revealed similar potential structures (data not shown). The distances from the nick site to the first potential structures can vary between 5 and 19 pb: LE alternatively 19 or 9/RE 9 (ISHp608), 11/8 (IS606), 10/8 (IS200), 11/8 (IS1541), 8/8 (IS605), 5/8 (IS8301), 10/11 (ISH1–8). In the case of IS91 and other members of this family, these distances are 5–7/7 (Garcillan-Barcia et al, 2002).
For ISHp608, successive deletions in LE and RE (Figure 5) progressively remove the potential structures. In both ends, deletions retaining the central palindromic sequences (LE3 and RE2) exhibited seriously reduced transposition frequencies. Thus, these sequences are not sufficient for transposition and supplementary sequences, presumably those carrying other repeated elements are important for activity. In the case of IS91, deletion of LE does not eliminate transposition activity but leads to ‘one-ended' transposition where a sequence resident in the donor plasmid is recognised as a second end (Mendiola et al, 1994; Bernales et al, 1999). This is not observed for ISHp608. The role of these regions is being analysed in detail using the in vitro binding and cleavage assays described above.
For IS200, the nucleic acid secondary structures at the ends were thought to play a role in assuring low gene expression levels (Beuzon et al, 1999): one located within 20 bp of LE can adopt a perfect hairpin that may act as transcriptional terminator, preventing activation of IS200 by impinging transcription (Lam and Roth, 1983); another may sequester the ribosome binding site of the transposase gene (Beuzon and Casadesus, 1997). While this may also be true for ISHp608, it is clear that the LE and RE regions including the potential secondary structures play a mechanistic role in transposition. These structures may therefore play dual roles. It will be interesting to compare IS200, IS608 and IS1541 (85% identity to IS200), which is found in numerous copies in Yersinia genomes (Odaert et al, 1998; Devalckenaere et al, 1999), to determine the reason for the relative inactivity of the widely spread IS200 (Haack, 1995).
Model of ISHp608 transposition
It is difficult to reconcile ISHp608 transposition with any of the presently understood mechanisms of transposition. Moreover, as described above, TnpA does not carry the complete sequence signatures of any of the other known transposases. In the absence of a convincing DDE motif, and since there are no conserved serine residues in TnpA, we assumed that the attacking nucleophile would be the single conserved tyrosine in TnpA (Y127). We have also provided data to support this idea (Table I and Figures 6 and 7). Cleavage would therefore probably involve formation of a transitory covalent enzyme–DNA intermediate.
Based on the above observations, we propose a two-step working model involving transposon excision (Figure 8A–C) followed by insertion (Figure 8D). Model (A) is perhaps the simplest. It involves cleavage of LE and RE (either simultaneously or consecutively), reciprocal strand transfer using, for example, a 3′OH generated on the flanking DNA at LE and on the transposon end at RE to generate a Holliday junction (HJ) and resolution. Although we observe no cleavage activity on linear dsRE, supercoiling of the donor plasmid could provide the energy necessary for generating a cleavable ssRE simply by extrusion of the potential secondary structure (Mizuuchi et al, 1982; Furlong et al, 1989; Bowater et al, 1991). The model generates both DPB and excised transposon.
Figure 8.
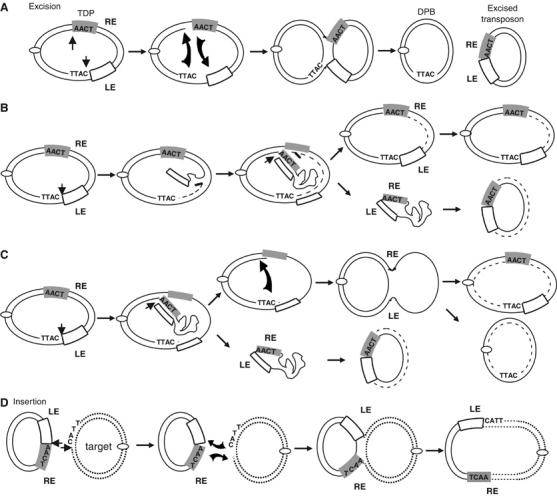
ISHp608 transposition model. Donor plasmid and transposon DNAs are shown as thick black lines. Left (LE) and right (RE) transposon ends are indicated. Replicated DNA is shown as a dashed line. Target DNA is shown as fine dotted lines. The open oval represents the plasmid origin of replication. Three alternative schemes are proposed for excision. In (A), cleavage at LE and RE (simultaneously or consecutively) and reciprocal strand transfer would generate an HJ, which could be resolved into double-strand circular copies of the transposon and the DPB. Pathway (B) involves a cleavage at LE and displacement of this strand by replication primed by 3′OH of flanking donor plasmid DNA. This could assist formation of a single-strand region for cleavage of RE and generate a single-strand circle that could then undergo replication to a double-strand form. In the third pathway (C), cleavage again occurs at LE but the transposon strand is displaced but without replication. Cleavage and transfer at RE could generate a single-strand circle and a sealed donor backbone with a single-strand loop transposon. Vegetative replication from plasmid would give rise to the original donor plasmid and DPB. Insertion (D) could occur by the reverse process: formation of a synaptic complex between TnpA, the abutted transposon ends in the circle junction and the target 5′TTAC motif; cleavage both 3′ to the TTCA on the RE–LE junction and to the target TTAC; reciprocal strand transfer using the free 3′OH to generate an HJ, and resolution.
In a variation of this scheme, which reconciles the requirement of ssRE DNA (Figure 8B), the transposon is excised as an ss circle by cleavage first at LE and one strand of the liberated ssLE is displaced by replication. This uses the 3′OH of the flanking donor plasmid DNA and regenerates the ds transposon in situ. Cleavage of LE and strand displacement could generate an ssRE with the secondary structure necessary for recognition and cleavage. In the example shown, a 3′OH RE generated by cleavage of the distal, RE, strand is used as a nucleophile to attack the 5′ transposon end and would generate an ss circle. This model is attractive since the energy for displacement can be generated by the replication machinery and a simple mechanism is provided to explain why cleavage of LE ‘top' strand can occur in both ds and ss forms while RE is only cleaved in a single ‘top' strand form. The liberated ss transposon circle could then undergo simple replication to a ds form (or integration as an ss form as proposed by Tavakoli et al, 2000). However, it is difficult to explain how this model could generate a recircularised DPB.
A third mechanism is shown in Figure 8C. Cleavage again occurs at LE but the transposon ‘top' strand is in some way displaced without replication. Formation and liberation of the ss circle could be coupled to strand transfer between the free ss ends of the donor plasmid to generate a sealed donor backbone with a loop of ss transposon ‘bottom' strand. Vegetative replication of the plasmid would presumably resolve these structures into a copy of the original donor plasmid with the transposon and one resealed DPB. These alternative models are at present being addressed experimentally.
Insertion could occur by the reverse process: formation of a synaptic complex between TnpA, the abutted transposon ends in the circle junction and the target 5′TTAC motif (Figure 8D); cleavage both 3′ to the 5′TCAA on the RE–LE junction and to the target 5′TTAC; reciprocal strand transfer using the free 3′OH to generate an HJ, and resolution. Integration could also be imagined to occur with a single-strand transposon circle in a reversal of the excision pathway (Figure 8C).
These results provide some intriguing hints revealing a new class of transposase. However, they do not identify its catalytic activities, its relationship to transposases of known catalytic activity or the way it interacts with the transposon ends. Neither do they explain how a transposase of only 155 aa, less than half the length of most transposases, deals with the multiple functions of DNA structure/sequence recognition, cleavage and strand transfer reactions. Answers to these important questions must await further biochemical and structural analysis, complementary approaches that are being pursued at present.
Materials and methods
Bacterial strains and media
JS219 (MC1061, recA1, lacIq; Cam et al, 1988) was used as a donor strain in mating-out experiments and in all other transposition-related assays. The recipient strain for mating-out experiments was MC240 (XA103, F−, ara, del(lac pro), gyrA (nalR), metB, argEam, rpoB, thi, supF). Cell extracts for TnpA-His6 were prepared from MC1061 endA. Cultures were grown in LB supplemented, where necessary, with ampicillin (Ap, 100 μg/ml), kanamycin (Km, 25 μg/ml) or chloramphenicol (Cm, 30 μg/ml). Selection was on L plates supplemented with appropriate antibiotics.
Plasmids for in vivo assays and TnpA-His6 production. The TnpA reading frame was cloned into pAPT110 (Polard and Chandler, 1995) with a C-terminal His6 tag under the control of plac to generate plasmid pBS107. The plasmid also carried the lacIq gene to regulate TnpA-His6 expression. TnpA was amplified with oligonucleotides IS608NdeI and B52HisSpHI on pBS∷ISHp608.
![]()
pBS∷ISHp608 (Kersulyte et al, 2002), pBS101, pBS105, pBS102 and pBS103 are pBluscript-derived and carry respectively wild-type ISHp608, wild-type ISHp608 with an insertion of a Cm cassette, ISHp608 without TnpB with Cm cassette and a synthetic transposon in which the 143 bp LE and 79 bp RE flank the Cm cassette. The Cm cassette (approximately 3 kb) was derived from pAPT99 (Polard et al, 1992) by digestion with HindIII and filling with Klenow fragment. pBS103 is the same as pBS102 but carries a shorter cassette.
pBS125 was constructed using Jonc1 and Jonc2:
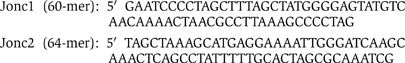
All plasmids with modified flanking sequences and LE and RE deletions were constructed from pBS102 using the following oligonucleotides:

DNA procedures
Standard techniques were used for DNA manipulation and cloning. Restriction and DNA modifying enzymes were purchased from New England Biolabs. DNA was isolated from agarose gels using the QIAquick gel extraction kit (Qiagen). Plasmid DNA was extracted using miniprep or maxiprep kits (Qiagen). PCR products were purified using the QIAquick PCR purification kit (Qiagen). Mutagenesis was performed by inverse PCR (Weiner and Costa, 1994). PCR reactions were carried out with Pfu polymerase (Promega; eight cycles: 1 min 94°C, 1 min 55°C, 5 min 72°C). The final PCR product was digested with DpnI (New England Biolabs) and self-ligated.
In vivo excision assay
E. coli strain MC1061recA was transformed with both the compatible plasmids pBS102 (Apr, Cmr, carrying the transposon substrate) and pBS107 (Kmr specifying TnpA) and plated at 37°C with appropriate antibiotic selection. Plasmid DNA was extracted by the alkaline lysis method from overnight cultures of single transformant colonies grown at 37°C with or without 0.5 mM IPTG and analysed on 0.8% TAE agarose gel. Cleared lysates were prepared as described (Turlan and Chandler, 1995) after 5 h induction with 0.5 mM IPTG.
TnpA-His6 purification
A preculture of MC1061 endA carrying TnpA-His6 expression plasmid pBS107 and grown at 37°C in L broth containing Km was diluted 50-fold into the same medium at 30°C. Protein expression was induced at A600=0.5–0.6 by adding 0.5 mM IPTG. After 2½ − 3h, the bacteria were centrifuged and the pellet was resuspended in buffer A (50 mM phosphate buffer pH 8, 450 mM NaCl, 1 mM β-mercaptobetanol), 10 mM imidazole supplemented with 1 mg/ml lysozyme and EDTA-free protease inhibitor cocktail (Roche). The bacteria were sonicated and the lysate was cleared by centrifugation. The supernatant was then mixed with Ni-agarose resin (Qiagen). After washes in buffer A and 50 mM imidazole, TnpA-His6 was eluted with 200 mM imidazole. The preparation was then dialysed in 25 mM HEPES pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT and 20% glycerol and stored at −80°C.
Electrophoretic mobility shift assay
All RE (B85, B85c) and LE (B81up and B81com) carrying oligonucleotides were 5′-end-labelled with 32P.

In a standard gel retardation assay, 4 nM of the DNA fragment or oligonucleotide (2500 c.p.m. Cerenkov) was incubated with TnpA-His6 in a final volume of 8 μl of binding mixture (20 mM HEPES pH 7.5, 100 mM NaCl, 2 mM CaCl2, 1 mM DTT, 2 mg/ml BSA and 15% glycerol) with 0.5 μg of poly-dIdC competitor. Complexes were separated in an 8% native polyacrylamide gel in TGE buffer.
In vitro cleavage
RE, LE or RE–LE junction-carrying fragments 5′ 32P-radiolabelled at each end separately or 5′ 32P-radiolabelled oligonucleotide B85 (RE top strand) were used. The reaction was performed in a final volume of 15 μl containing 28 nM substrate, 6 μM TnpA in 20 mM HEPES (pH 7.5), 0.1 M NaCl, 1 mM DTT, 20 ng/ml BSA, 0.5 μg of poly-dIdC and 14% glycerol. After 20 min of incubation at 37°C, MgCl2 was added to a final concentration of 5 mM and incubation was continued for 30 min. Reactions were terminated by addition of 1.6 μl of 1% SDS followed by 15 min of incubation at 37°C and finally by addition of 16 μl of 80% formamide loading buffer. Nicking products were analysed on an 8% denaturing sequencing gel.
Supplementary Material
Supplementary Figure 1
Table 1
Acknowledgments
We thank members of the Mobile Genetic Elements group (E Gueguen, N Pouget, P Rousseau, P Siguier, G Duval-Valentin, J Filé) and AB Hickman, P Rousseau, G Duval-Valentin and B Hallet for discussions and for critical reading of the manuscript. We are also grateful to D Berg for his advice in the choice of the transposon derivative and for generously providing us with the initial clone of ISHp608. This work was supported by grants from the CNRS (UMR5100), the ‘Programme microbiologie' (MENRST) and the Groupement de Recherche (GDR 2157).
References
- Bernales I, Mendiola MV, de la Cruz F (1999) Intramolecular transposition of insertion sequence IS91 results in second-site simple insertions. Mol Microbiol 33: 223–234 [DOI] [PubMed] [Google Scholar]
- Beuzon CR, Casadesus J (1997) Conserved structure of IS200 elements in Salmonella. Nucleic Acids Res 25: 1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzon CR, Chessa D, Casadesus J (2004) IS200: an old and still bacterial transposon. Int Microbiol 7: 3–12 [PubMed] [Google Scholar]
- Beuzon CR, Marques S, Casadesus J (1999) Repression of IS200 transposase synthesis by RNA secondary structures. Nucleic Acids Res 27: 3690–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater R, Aboul-ela F, Lilley DM (1991) Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A+T)-rich regions that promote low-salt cruciform extrusion. Biochemistry 30: 11495–11506 [DOI] [PubMed] [Google Scholar]
- Byrd DR, Matson SW (1997) Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol 25: 1011–1022 [DOI] [PubMed] [Google Scholar]
- Cam K, Bejar S, Gil D, Bouche JP (1988) Identification and sequence of gene dicB: translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res 16: 6327–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M, Mahillon J (2002) Insertion sequences revisited. In Mobile DNA, Craig NL, Craigie R, Gellert M, Lambowitz A (eds), Vol. II, pp 305–366. Washington, DC: ASM Press [Google Scholar]
- Cornet F, Chandler M (2004) Non-homologous recombination. In Evolution: Gene Establishment, Survival, and Exchange, Miller CJ, Day M (eds), pp 36–66. Washington, DC: ASM Press [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM (2003) The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol 4: 865–877 [DOI] [PubMed] [Google Scholar]
- del Pilar Garcillan-Barcia M, Bernales I, Mendiola MV, de la Cruz F (2001) Single-stranded DNA intermediates in IS91 rolling-circle transposition. Mol Microbiol 39: 494–501 [DOI] [PubMed] [Google Scholar]
- Devalckenaere A, Odaert M, Trieu-Cuot P, Simonet M (1999) Characterization of IS1541-like elements in Yersinia enterocolitica and Yersinia pseudotuberculosis. FEMS Microbiol Lett 176: 229–233 [DOI] [PubMed] [Google Scholar]
- Espinosa M, del Solar G, Rojo F, Alonso JC (1995) Plasmid rolling circle replication and its control. FEMS Microbiol Lett 130: 111–120 [DOI] [PubMed] [Google Scholar]
- Furlong JC, Sullivan KM, Murchie AI, Gough GW, Lilley DM (1989) Localized chemical hyperreactivity in supercoiled DNA: evidence for base unpairing in sequences that induce low-salt cruciform extrusion. Biochemistry 28: 2009–2017 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia MP, Bernales I, Mendiola MV, De la Cruz F (2002) IS91 rolling circle transposition. In Mobile DNA, Craig NL, Craigie R, Gellert M, Lambowitz A (eds) pp 891–904. Washington, DC: ASM press [Google Scholar]
- Haack KR (1995) The activity of IS200 in Salmonella typhimurium. PhD thesis, University of Utah
- Ilyina TV, Koonin EV (1992) Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20: 3279–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Akopyants NS, Clifton SW, Roe BA, Berg DE (1998) Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223: 175–186 [DOI] [PubMed] [Google Scholar]
- Kersulyte D, Mukhopadhyay AK, Shirai M, Nakazawa T, Berg DE (2000) Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J Bacteriol 182: 5300–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Velapatino B, Dailide G, Mukhopadhyay AK, Ito Y, Cahuayme L, Parkinson AJ, Gilman RH, Berg DE (2002) Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J Bacteriol 184: 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA (1997) Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev 61: 442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S, Roth JR (1983) IS200: a Salmonella-specific insertion sequence. Cell 34: 951–960 [DOI] [PubMed] [Google Scholar]
- Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F (2002) Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45: 1–8 [DOI] [PubMed] [Google Scholar]
- Mahillon J, Chandler M (1998) Insertion sequences. Microbiol Mol Biol Rev 62: 725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola MV, Bernales I, de la Cruz F (1994) Differential roles of the transposon termini in IS91 transposition. Proc Natl Acad Sci USA 91: 1922–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola MV, de la Cruz F (1992) IS91 transposase is related to the rolling-circle-type replication proteins of the pUB110 family of plasmids. Nucleic Acids Res 20: 3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola MV, Jubete Y, de la Cruz F (1992) DNA sequence of IS91 and identification of the transposase gene. J Bacteriol 174: 1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K, Mizuuchi M, Gellert M (1982) Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol 156: 229–243 [DOI] [PubMed] [Google Scholar]
- Odaert M, Devalckenaere A, Trieu-Cuot P, Simonet M (1998) Molecular characterization of IS1541 insertions in the genome of Yersinia pestis. J Bacteriol 180: 178–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P, Chandler M (1995) An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev 9: 2846–2858 [DOI] [PubMed] [Google Scholar]
- Polard P, Prere MF, Fayet O, Chandler M (1992) Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J 11: 5079–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley TL, Ellermeier CD, Slauch JM (2000) Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar typhimurium survival in Peyer's patches. J Bacteriol 182: 4406–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli N, Comanducci A, Dodd HM, Lett MC, Albiger B, Bennett P (2000) IS1294, a DNA element that transposes by RC transposition. Plasmid 44: 66–84 [DOI] [PubMed] [Google Scholar]
- Turlan C, Chandler M (1995) IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J 14: 5410–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MP, Costa GL (1994) Rapid PCR site-directed mutagenesis. PCR Methods Appl 4: S131–S136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Table 1


