Abstract
Kar3, a kinesin-14 motor of Saccharomyces cerevisiae required for mitosis and karyogamy, reportedly interacts with Cik1, a nonmotor protein, via its central, predicted coiled coil. Despite this, neither Kar3 nor Cik1 homodimers have been observed in vivo. Here we show that Kar3 is a dimer in vitro by analytical ultracentrifugation. The motor domains appear as paired particles by rotary-shadow electron microscopy (EM) and circular dichroism (CD) spectroscopy of the nonmotor region shows characteristics of helical structure, typical of coiled coils. Remarkably, the Kar3/Cik1 nonmotor region shows greater helicity by CD analysis and rotary-shadow EM reveals a stalk joined to one large or two smaller particles. The highly helical Kar3/Cik1 nonmotor region and visible stalk indicate that dimerization with Cik1 causes structural changes in Kar3. The Cik1 and Kar3 stalk regions preferentially associate with one another rather than forming homodimers. Kar3/Cik1 moves on microtubules at 2–2.4 μm min−1, 2–5-fold faster than Kar3, and destabilizes microtubules at the lagging ends. Thus, structural changes in Kar3 upon dimerization with Cik1 alter the motor velocity and likely regulate Kar3 activity in vivo.
Keywords: Cik1 interactions, Kar3, kinesin-14 motor, microtubule motor, structure–function
Introduction
Saccharomyces cerevisiae encodes six ATP-driven kinesin motor proteins that potentially function in transport of vesicles and organelles along microtubules, spindle assembly and chromosome motility during cell division. Kar3, a kinesin-14 (formerly C-terminal motor) (Lawrence et al, 2004) family member and the only minus-end kinesin motor, is required for mitosis and karyogamy, or nuclear fusion following mating. Loss-of-function kar3 mutants show severe defects in karyogamy and exhibit unusually long cytoplasmic microtubules (Meluh and Rose, 1990). The Kar3 motor localizes to cytoplasmic microtubules, spindle pole bodies and the mitotic spindle (Meluh and Rose, 1990), where it produces force opposing that produced by two presumed plus-end kinesin-5 (formerly BimC) family members, Kip1 and Cin8 (Saunders and Hoyt, 1992; Saunders et al, 1997). Remarkably, recent evidence indicates that Kar3 promotes poleward transport of kinetochores along spindle microtubules (Tanaka et al, 2005). Kar3 destabilizes microtubules in gliding assays in vitro (Endow et al, 1994), and couples microtubule plus ends to the cortex of shmoo tips (Maddox et al, 2003), which may be important in regulating cytoplasmic microtubule assembly and mediating microtubule dynamics during vegetative growth and mating.
Strikingly, Kar3 has been reported to exist as a monomer in vivo, but is also found in the form of a dimer with a nonmotor protein, Cik1 (Barrett et al, 2000). A second interacting protein, Vik1, has been identified, which is thought to form a distinct functional complex with Kar3 (Manning et al, 1999). Of the two proteins, Kar3 interaction with Cik1 has been better studied (Barrett et al, 2000). Cik1 was originally identified in a genetic screen for mutants in spindle pole body and/or microtubule functions (Page and Snyder, 1992). Loss-of-function cik mutants cause chromosome instability and karyogamy defects, and exhibit abnormal mitotic spindles and prominent bundles of long microtubules. Like Kar3, Cik1 localizes to spindle pole bodies (Page and Snyder, 1992). Cik1 is tightly associated with Kar3 in vivo (Barrett et al, 2000), an interaction that has been inferred, but not demonstrated, to be due to the formation of a coiled coil by the two proteins. Both Kar3 and Cik1 contain a region with a high probability of coiled-coil formation by analysis with predictive computer algorithms. Although Barrett et al (2000) reported evidence that Kar3 and Cik1 interact tightly and probably exist in vivo as a heterodimer, they also reported that neither protein forms homodimers in vivo.
These observations raise several questions—can Kar3 dimerize with itself? What effects does dimerization with Cik1 have on Kar3 activity? We have addressed these questions by expressing Kar3 and Kar3/Cik1 in bacteria and analyzing the purified proteins. We find that both Kar3 and Kar3/Cik1 dimers form in vitro, but dimerization with Cik1 causes structural changes in Kar3 that alter its velocity. These findings strongly imply that Kar3 activity in vivo is regulated by heterodimer formation. This has important implications for kinesin-14 motor function.
Results
Kar3
A truncated Kar3 protein, Kar3 M278-K729, was expressed in bacteria and purified for biochemical and structural analysis. Kar3 M278-K729 is deleted for the N-terminal 277 residues of the full-length protein, but contains the second half of the central stalk domain (M278–V372), together with the neck (R373-R384) and conserved motor domain (N386-K729) (Figure 1A). By analysis with PAIRCOIL (Berger et al, 1995), residues Q335–V372 form coiled coil with a probability significantly higher than the cutoff of 0.500. This region consists of 38 residues and corresponds to 5.4 heptad repeats. The residues that follow, R373–R384, are immediately adjacent to the conserved motor domain and form coiled coil with a probability just below 0.500, representing the less stable neck. By analogy to the closely related kinesin-14 motor Ncd, G385 at the end of the Kar3 predicted coiled-coil domain is a pivot point for stalk/neck rotation (Yun et al, 2003).
Figure 1.
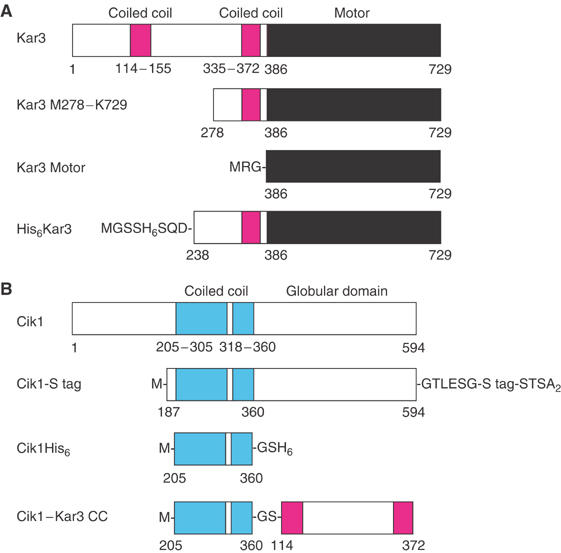
Kar3 and Cik1. (A) Full-length Kar3 is shown, with the two coiled-coil regions predicted by PAIRCOIL (Berger et al, 1995) (probability >0.5, magenta). The conserved motor domain (N386-K729, black) contains the highly conserved ATP- and microtubule-binding sites typical of the kinesin motor proteins. Kar3 M278–K729 consists of the second half of the central stalk region (M278–V372), neck (R373–R384), G385 pivot point residue at the stalk/neck–motor junction and conserved motor domain (N386–K729). The Kar3 motor domain protein, MRG–N386–K729, was expressed and purified for CD experiments. His6Kar3 has a His6 tag at the N-terminus of the stalk region, and was coexpressed with Cik1-S tag to give Kar3/Cik1. (B) Full-length Cik1 contains a region of predicted coiled coil (D205–E360, cyan) with a break or hinge region (V306–P317), followed by a putative C-terminal globular domain. Cik1-S tag with an S tag (KETA3KFERQHMDS) at the C-terminus was coexpressed with His6Kar3 to give Kar3/Cik1. Cik1His6 consists of the Cik1 central, predicted coiled-coil region. Cik1–Kar3 CC (coiled-coil) is a fusion of the predicted Cik1 coiled-coil region in tandem to that of Kar3.
Kar3 stalk region is helical by circular dichroism (CD) spectroscopy
CD spectroscopy was performed to estimate the helical content of the Kar3 M278–K729 central stalk region. Scans of purified Kar3 M278–K729 protein from 300 to 190 nm were corrected for the buffer and contribution by the motor domain by scanning the buffer and motor domain (Kar3 MRG–N386–K729) alone. Spectra of the motor domain and Kar3 M278–K729 are shown in Figure 2A, together with the spectrum corresponding to the nonmotor, central stalk region, obtained by subtracting the motor domain CD signal from the Kar3 M278–K729 signal and calculating the mean molar ellipticity per residue. The difference profile, which corresponds to the 105 nonmotor stalk residues of Kar3 M278–K729, shows the characteristics of α-helical structure (Greenfield and Fasman, 1969). The mean molar ellipticity per residue for the difference spectrum is θ=−12 053 deg cm2 dmol−1 at 208 nm and −10 535 deg cm2 dmol−1 at 222 nm. The values at 208 and 222 nm indicate that the Kar3 M278–K729 stalk region is helical.
Figure 2.
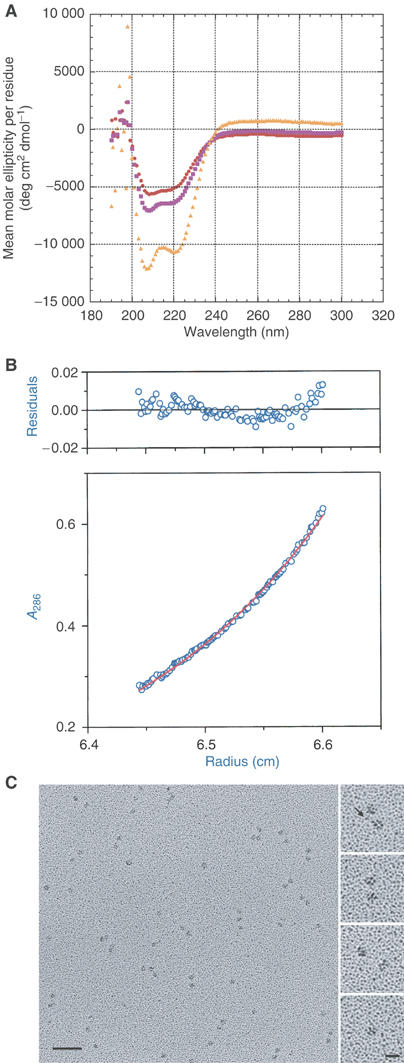
Kar3 M278–K729 analysis. (A) CD spectroscopy. Mean molar ellipticity per residue as a function of wavelength. The spectrum of residues corresponding to the nonmotor, stalk region of Kar3 M278–K729 was obtained by subtraction of the motor domain CD signal from the Kar3 M278–K729 signal (see Materials and methods). Spectra of Kar3 MRG–N386–K729 motor domain (19.2 μM) and Kar3 M278–K729 (9.69 μM) are shown for comparison. Motor domain,  ; Kar3 M278–K729,
; Kar3 M278–K729,  ; nonmotor stalk region,
; nonmotor stalk region,  . (B) Sedimentation equilibrium analysis. Absorbance of Kar3 M278–K729 (12.5 μM) at 286 nm as a function of distance from the center of rotation after reaching equilibrium at 8000 r.p.m. and 4°C. The scan was corrected for baseline. The curve fit shows a molecular weight of 99 200 Da for the protein. Residuals of the fit are shown above the scan. Absorbance, ○; curve fit, –. (C) Rotary-shadow EM. The field (left) shows single and paired globular particles of Kar3 M278–K729. Bar, 50 nm. The higher magnification images (right) show paired particles separated by variable distances (top, images 1–3) and a single particle (bottom image). The image at the top, taken from a different field, shows a putative stalk between the two particles (arrow). Bar, 12.5 nm.
. (B) Sedimentation equilibrium analysis. Absorbance of Kar3 M278–K729 (12.5 μM) at 286 nm as a function of distance from the center of rotation after reaching equilibrium at 8000 r.p.m. and 4°C. The scan was corrected for baseline. The curve fit shows a molecular weight of 99 200 Da for the protein. Residuals of the fit are shown above the scan. Absorbance, ○; curve fit, –. (C) Rotary-shadow EM. The field (left) shows single and paired globular particles of Kar3 M278–K729. Bar, 50 nm. The higher magnification images (right) show paired particles separated by variable distances (top, images 1–3) and a single particle (bottom image). The image at the top, taken from a different field, shows a putative stalk between the two particles (arrow). Bar, 12.5 nm.
The mean molar ellipticity per residue for the difference spectrum at 222 nm, θ=–10 535 deg cm2 dmol−1, is not as large as the value of –33 000 at 222 nm observed for a 100% coiled-coil protein (Carr and Kim, 1993); thus, only approximately a third of the 105 residues of the stalk region of Kar3 M278–K729 are helical. This is consistent with the prediction by PAIRCOIL that 36% of the 105 residues of the region (Q335–V372, 38 amino acids) show a significant probability (>0.500) of coiled-coil formation.
Kar3 is a dimer by analytical ultracentrifugation
Sedimentation equilibrium ultracentrifugation was performed to estimate the molecular weight of Kar3 M278–K729. Purified protein at 25 μM was serially diluted and three concentrations were brought to sedimentation equilibrium at 8000 r.p.m. at 4°C. Protein absorbance was measured at 268 and 286 nm as a function of distance from the center of rotation. The absorbance scans were corrected for baseline and curve fit to estimate the molecular weight of the major protein species. One such scan is shown in Figure 2B. The curve fits for all three protein concentrations showed a best-fit molecular weight close to that of the dimer as the major species. The residuals of the curve fits showed evidence for trace amounts of heterogeneity due to both higher and lower molecular weight species. The best-fit molecular weight average for the three concentrations is 96 500±3200 Da (mean±sem, n=6), close to the 102 822 Da predicted for dimeric Kar3 M278–K729. These data indicate that Kar3 M278–K729 exists in vitro primarily as a dimer. Together with the CD data, the analytical ultracentrifugation results are consistent with the interpretation that the nonmotor stalk region of Kar3 M278–K729 forms α-helical coiled coil, dimerizing the protein in vitro.
Kar3 is two-headed by rotary-shadow electron microscopy (EM)
Rotary-shadow EM was used to visualize Kar3 M278–K729 molecules. Purified protein from the same preparation used for the sedimentation equilibrium ultracentrifugation in Figure 2B was diluted 50- or 100-fold, shadowed with platinum and carbon, and examined in a Phillips 301 EM at 80 kV.
Micrographs showed single and paired particles (Figure 2C). Analysis showed that the distribution of paired particles was similar to that of single particles, but differed from the distribution of the second particle of each pair (see Supplementary data). The nonrandom distribution of the two particles of the pairs provides evidence for the interpretation that the paired particles represent the two heads of dimeric Kar3 M278–K729, rather than two adjacent monomers. The single particles were interpreted to represent monomer molecules.
The ratio of single to paired particles in protein diluted to 500 nM before spreading was 1:1.6 (single particles=334, paired particles=520, total=854); thus ∼76% of the Kar3 M278–K729 molecules were dimers and ∼24% were monomers after diluting the protein for specimen preparation. The ratio of single to paired particles did not change significantly in protein at 250 nM, suggesting high-affinity binding for dimerization. The stalk of Kar3 M278–K729 is observed very infrequently in these preparations (Figure 2C, top right), probably because the coiled coil, predicted to be 5.4 heptad repeats by PAIRCOIL, is at the lower limit capable of being visualized by EM. Together with the CD results, the putative stalks that are observed indicate that Kar3 dimerizes by forming α-helical coiled coil.
Measurements using the line tool of NIH Image v1.63 gave an estimate of 57.2±0.7 Å (mean±s.e.m., n=70) for the diameter of the single or paired particles after correction for the platinum shell. The diameter fits well with the dimensions of approximately 61 × 56 × 42 Å for the Kar3 motor domain in a crystal structure (PDB 3Kar) (Gulick et al, 1998), estimated by assuming an elliptical shape of the motor domain. The paired particles or heads are positioned at variable distances from one another in the micrographs (Figure 2C). The distances between heads of the paired particles showed a broad range of 47–193 Å (n=112). The large distance between the paired Kar3 heads differs strikingly from closely related Ncd, where the two heads are positioned very close to one another in crystal structures and both heads interact extensively with the end of the coiled-coil stalk (Sablin et al, 1998; Kozielski et al, 1999; Yun et al, 2003).
Kar3/Cik1 expression and purification
To determine if Kar3 interaction with Cik1 causes detectable structural changes in Kar3, we expressed Kar3/Cik1 and analyzed the purified protein. Truncated His6Kar3 and Cik1-S tag proteins (Figure 1) were expressed from the same plasmid, pETDuet-1, each gene regulated by its own promoter. His6Kar3, MGSSH6SQD P238–K729, contains the second half of the central stalk (P238–V372), including the 38-residue region of predicted coiled coil (Q335–V372), together with the neck and conserved motor domain. Cik1-S tag consists of M-N187–D594–GTLESG-S tag-STSA2, and includes the Cik1 central region, D205–E360, predicted to form coiled coil by PAIRCOIL, together with the C-terminal, presumed globular domain. The His6Kar3 and Cik1-S tag proteins were identified by Western blot analysis in tests of induced cells for protein expression.
His6Kar3/Cik1-S tag protein was purified by Ni-affinity chromatography, followed by FPLC on Sephacryl S-200 gel filtration and Hi-Trap SP-Sepharose columns. SDS–PAGE showed a peak from the gel-filtration and SP-Sepharose columns that contained both the His6Kar3 and Cik1-S tag proteins in approximately equal amounts (see Supplementary data), consistent with dimer formation. The two protein bands after SP-Sepharose chromatography were confirmed as Kar3 and Cik1 by mass spectroscopy.
Sedimentation equilibrium analysis of the purified His6Kar3/Cik1-S tag protein gave a best-fit molecular weight of 115 000 Da at 2.58 μM with evidence for higher molecular weight species at higher protein concentrations. The theoretical molecular weight of 108 401 Da for the dimer is very close to the best-fit molecular weight, providing evidence that the purified protein exists primarily as a dimer at low concentration.
Kar3/Cik1 nonmotor region is highly helical by CD spectroscopy
Analysis of purified His6Kar3/Cik1-S tag protein by CD spectroscopy gave a mean molar ellipticity per residue of θ=−16 881 deg cm2 dmol−1 at 208 nm and −17 511 deg cm2 dmol−1 at 222 nm after subtraction of one Kar3 motor domain from the His6Kar3/Cik1-S tag signal (Figure 3A). These values indicate a strikingly high helical content of ∼53% (assuming θ=−33 000 deg cm2 dmol−1 at 222 nm for a 100% coiled-coil protein) (Carr and Kim, 1993) for the difference protein, which includes the Cik1 C-terminal domain. As the mean molar ellipticity is given per residue and the Cik1 C-terminal domain is predicted to be a mixture of α-helices and β-strands by nnPREDICT (Kneller et al, 1990) and other secondary structure prediction algorithms, this difference value is most likely an underestimate of the helicity of the Kar3/Cik1 central stalk. Thus, we would predict the helical content of the stalk region alone to be even greater, ∼70%, assuming ∼35% helicity of the Cik1 C-terminal domain. Despite this underestimate, the value of θ=−17 511 deg cm2 dmol−1 at 222 nm for the Kar3/Cik1 nonmotor region is significantly larger than the θ=−10 535 deg cm2 dmol−1 at 222 nm that we observe for the Kar3 M278–K729 nonmotor stalk region. The additional 40 amino acids of the Kar3 stalk region in the His6Kar3/Cik1-S tag protein compared to Kar3 M278–K729 probably contributes to the signal, but the CD signal has been averaged over these residues and the others remaining in the difference protein; thus, their contribution alone does not account for the increased helical content of the difference protein. The higher helical content of the Kar3/Cik1 nonmotor region compared to dimeric Kar3 suggests that interaction of Kar3 with Cik1 stabilizes the formation of α-helical coiled coil by the central stalk region.
Figure 3.
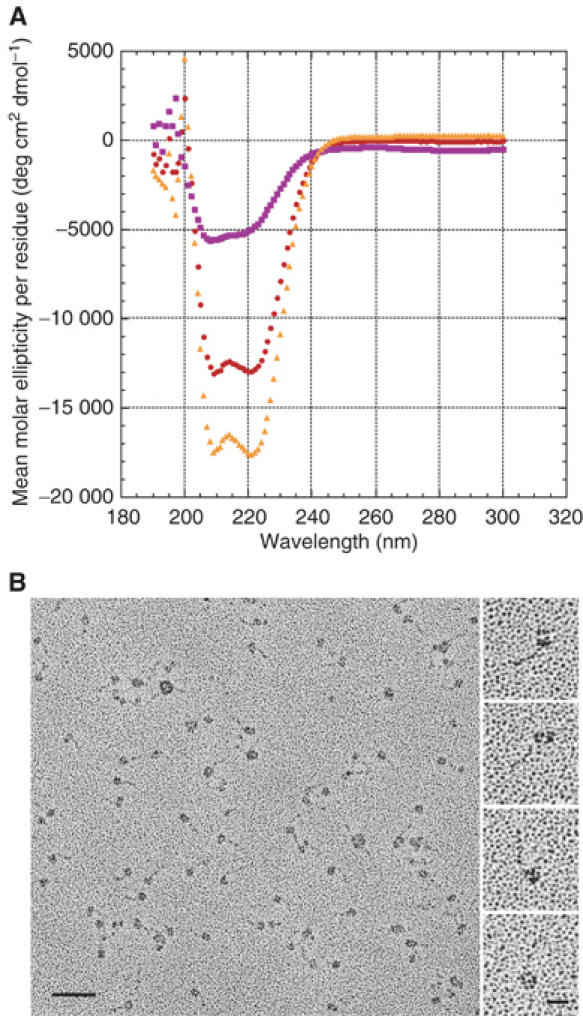
Kar3/Cik1 analysis. (A) CD spectroscopy. Mean molar ellipticity per residue as a function of wavelength. The spectrum of residues corresponding to the nonmotor region of the His6Kar3/Cik1-S tag protein was obtained by subtraction of the Kar3 motor domain CD signal from the His6Kar3/Cik1-S tag signal. Spectra of Kar3 MRG–N386–K729 motor domain (read at 19.2 μM and adjusted to 8.58 μM) and His6Kar3/Cik1-S tag (8.58 μM) are shown for comparison. Kar3 motor domain,  ; His6Kar3/Cik1-S tag,
; His6Kar3/Cik1-S tag,  ; nonmotor region,
; nonmotor region,  . (B) Rotary-shadow EM. The field (left) shows the His6Kar3/Cik1-S tag, which appears as single and double globular particles joined to a stalk. Bar, 50 nm. The higher magnification images (right), taken from different fields, show double particles joined to a stalk (top two images) and single particles with a stalk (bottom two images). Bar, 12.5 nm.
. (B) Rotary-shadow EM. The field (left) shows the His6Kar3/Cik1-S tag, which appears as single and double globular particles joined to a stalk. Bar, 50 nm. The higher magnification images (right), taken from different fields, show double particles joined to a stalk (top two images) and single particles with a stalk (bottom two images). Bar, 12.5 nm.
Kar3/Cik1 forms a stalk by rotary-shadow EM
We also examined the His6Kar3/Cik1-S tag protein by EM after rotary shadowing the purified protein with platinum and carbon. Electron micrographs showed predominantly single particles that were, strikingly, attached to a short stalk (Figure 3B). The single particles are shown at higher magnification in the images at the right in Figure 3B (bottom two images). Measurements of the particles gave a mean diameter of 78.5±1.8 Å (n=70), larger by ∼11.3 Å than the 57.2±0.7 Å diameter we observed for single Kar3 heads. Thus, we interpret the single particles to represent the Kar3 motor domain together with the Cik1 C-terminal domain. We also observed double particles, smaller than the single particles, attached to a stalk (Figure 3B, top two images at the right), which we interpret to be the Kar3 head and Cik1 C-terminal domain.
The mean stalk length of the His6Kar3/Cik1-S tag was 21.7±0.3 nm (n=70), consistent with ∼149 residues of coiled coil, based on the length of the Ncd coiled-coil stalk in a crystal structure (M292–R346, 55 residues≈8 nm, PDB 1N6M) (Yun et al, 2003). This accounts for the predicted Cik1-S tag coiled-coil region (D205–E360, 156 residues), together with the central, nonmotor region of His6Kar3 (MGSSH6SQD P238–V372, 148 residues). A stalk was associated with 89% of the single or double particles (n=55, total=62) in protein diluted to 515 nM before spreading. Protein spread at 257 nM showed 64% single or double particles associated with a stalk (n=42, total=66). These values are significantly different from one another (χ2=10.94, 1 d.f., P<0.005), indicating that Kar3/Cik1 stalk stability is dependent on protein concentration.
Cik1 central region is helical
We also expressed a protein consisting only of the Cik1 central, predicted coiled-coil region, Cik1His6 (M-D205–E360–GSH6) (Figure 1B). Purification of the Cik1His6 protein by Ni-affinity chromatography gave a single band by SDS–PAGE. The protein was further size-purified by FPLC on a Superose 12 gel-filtration column prior to CD spectroscopy. The CD spectrum showed values of θ=−25 703 deg cm2 dmol−1 at 208 nm and −22 453 deg cm2 dmol−1 at 222 nm, indicating an extremely high helical content, ∼68% (assuming θ=−33 000 deg cm2 dmol−1 at 222 nm for a 100% coiled-coil protein). The value of θ=−22 453 deg cm2 dmol−1 at 222 nm for Cik1H6 is significantly greater than the values of θ=−10 535 deg and −17 511 deg cm2 dmol−1 at 222 nm that we observe for the nonmotor region of Kar3 M278–K729 and His6Kar/Cik1-S tag, respectively (Table I).
Table 1.
Helical content of stalk or nonmotor region
| Protein | [θ]208 (deg cm2 dmol−1) | [θ]222 (deg cm2 dmol−1) | % Helixa |
|---|---|---|---|
| Kar3 M278–K729 stalk | −12 053 | −10 535 | 32 |
| Kar3/Cik1 nonmotor regionb | −16 881 | −17 511 | 53 |
| CikHis6 stalk |
−25 703 |
−22 453 |
68 |
| aBased on θ=−33 000 deg cm2 dmol−1 at 222 nm for a 100% coiled-coil protein (Carr and Kim, 1993). | |||
| bIncludes the Kar3/Cik1 stalk and Cik1 C-terminal domain. | |||
Cik1 and Kar3 stalk regions form heterodimers
To determine the relative tendency for the Cik1 and Kar3 central stalk regions to form homodimers compared to Kar3/Cik1 heterodimers, we expressed the Cik1 predicted coiled-coil region in tandem to that of Kar3 as a fusion protein, Cik1–Kar3 CC, consisting of Cik1 M-D205–E360–GS-Kar3 E114–V372 (Figure 1B). The Cik1 and Kar3 regions of the fusion protein are expected to form coiled-coil homodimers of ∼34.5 nm in length (Figure 4A), based on ∼8 nm/55 residues for the Ncd stalk (see above). Alternatively, formation of Kar3/Cik1 coiled-coil heterodimers would result in protein filaments of multiples of ∼23 nm, the expected length of the Kar3/Cik1 stalk (based on 156 residues of the Cik1 predicted coiled-coil region and ∼8 nm/55 residues for the Ncd stalk), formed by overlapping molecules of the fusion protein (Figure 4A). The expected Kar3/Cik1 heterodimer stalk length of ∼23 nm is close to the length of 21.7±0.3 nm that we observe for the stalk of His6Kar3/Cik1-S tag.
Figure 4.
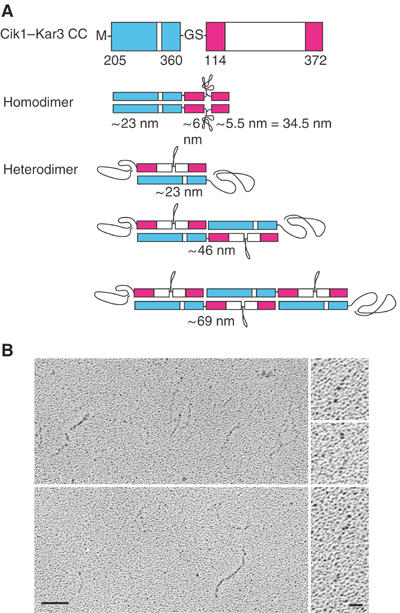
Coiled-coil formation by Cik1 and Kar3. (A) Cik1-Kar3 CC (coiled-coil) protein consists of the Cik1 predicted coiled-coil region fused in tandem to that of Kar3. Formation of homodimers is expected to result in a protein of ∼34.5 nm in length, based on the length of the Ncd coiled coil in a crystal structure (PDB 1N6M) (Yun et al, 2003). Formation of heterodimers should result in proteins of multiples of ∼23 nm in length, the length of the Kar3/Cik1 coiled coil, formed by overlapping molecules. Regions that do not form coiled coil are assumed to form random coil. (B) Rotary-shadow EM of Cik1–Kar3 CC shows proteins of varying lengths (left). Bar, 50 nm. The higher magnification images show molecules of 31.4, 50.0 and 98.1 nm. Bar, 12.5 nm.
The Cik1-Kar3 CC fusion protein was expressed in bacteria and purified by SP-Sepharose chromatography. Analysis by rotary-shadow EM revealed protein filaments that varied in length (Figure 4B) with a mean of 60.8±3.0 nm (n=90) and range from 23.7 to 153.6 nm (see Supplementary data). The proteins of ∼23 nm length can be attributed to formation of Kar3/Cik1 coiled-coil heterodimers by two overlapping Cik1–Kar3 CC molecules (Figure 4A). Those of ∼29–35 nm in length probably arise by formation of coiled-coil homodimers by two protein molecules and visualization of the Cik1 region (∼23 nm), together with one or both of the Kar3 regions (∼6 nm, ∼5.5 nm). The proteins longer than ∼35 nm, the theoretically predicted maximal length of Cik1–Kar3 CC homodimers, we attribute to Kar3/Cik1 coiled-coil heterodimers formed by several overlapping molecules. Approximately 80% of the proteins were >35 nm in length (n=72, total=90), with a mean of 68.8±3.2 nm. These presumably represent Kar3/Cik1 coiled-coil heterodimers formed by an average of four overlapping molecules (Figure 4A). This means that ∼94% of the protein molecules are present as heterodimers. This is probably an underestimate, since it excludes heterodimers of <35 nm in length formed by two protein molecules. Thus, Kar3/Cik1 molecules form heterodimers at least 16 times more frequently than Cik1 or Kar3 homodimers, presumably due to greater heterodimer stability.
Kar3/Cik1 is a microtubule motor
Microtubules were reported to glide on Kar3 motor-coated coverslips with a velocity of 1–2 μm min−1 in in vitro assays (Endow et al, 1994). Unexpectedly, slow depolymerization of microtubules gliding on the Kar3 motor was observed, mostly from the lagging or minus ends. Kar3/Cik1 behavior in gliding assays has not yet been reported. We examined microtubule gliding using lysates of cells expressing the His6Kar3 and Cik1-S tag proteins. Based on our analysis indicating that the Cik1 and Kar3 stalk regions tend to form heterodimeric coiled coil, we expected that the predominant form of His6Kar3 in the lysates would be His6Kar3/Cik1-S tag heterodimers, although His6Kar3 homodimers may also be present.
The gliding assays showed good binding of microtubules to the motor attached to the coverslip surface by anti-His6 antibodies. Microtubule gliding was observed only when the lysates were diluted with 1–2 parts of buffer—microtubules were bound to the glass, but showed little or no movement at higher or lower lysate concentrations, indicating a critical dependence of gliding on motor density, a characteristic of nonprocessive motors (Howard, 2001). Microtubules were shorter at the end of gliding sequences compared to the beginning, indicating that they disassemble slowly at the ends. Tracking gave a mean velocity of 1.96±0.05 μm min−1 (n=9) for microtubule leading ends. The lagging end velocity was significantly faster, 2.34±0.05 μm min−1 (n=10). The mean difference between lagging and leading end velocities is 0.34±0.02 μm min−1 (n=7), which we attribute to microtubule disassembly at the lagging end.
We also tested purified His6Kar3/Cik1-S tag protein in microtubule gliding assays. Microtubules bound to the motor attached to the glass surface and glided with a velocity of 2.39±0.49 μm min−1 (n=9) measured from the leading ends, similar to the velocity of 1.96±0.05 μm min−1 observed for lysates (Table II). Purified His6Kar3/Cik1-S tag showed a lagging end velocity of 2.56±0.39 μm min−1 (n=10), close to the 2.34±0.05 μm min−1 obtained for lysates. The difference between the lagging and leading end velocities is 0.35±0.08 μm min−1 (n=6). Length measurements showed that microtubules shorten during the videotaped sequences by 0.55±0.10 μm min−1 (n=6). The length difference, together with the difference in lagging and leading end velocities, indicates that the Kar3/Cik1 motor destabilizes microtubules at the lagging end in gliding assays.
Table 2.
Microtubule gliding velocity
| Protein | Velocity (μm min−1) |
||
|---|---|---|---|
| Leading end | Lagging end | Lagging–leading end | |
| His6Kar3+Cik1-S tag lysate | 1.96±0.05 (n=9) | 2.34±0.05 (n=10) | 0.34±0.02 (n=7) |
| His6Kar3/Cik1-S tag purified protein | 2.39±0.49 (n=9) | 2.56±0.39 (n=10) | 0.35±0.08 (n=6) |
| His6Kar3 lysate | 0.49±0.09 (n=7) | ND | ND |
| GST/Kar lysatea |
1.19±0.16 (n=10) |
1.80±0.11 (n=10) |
0.61±0.12 (n=10) |
| aData from Endow et al (1994) for GST/Kar3 H277–K729. ND, not determined. | |||
Lysates of cells expressing only His6Kar3 from the pETDuet-1 vector were tested in motility assays using the same methods as for His6Kar3/Cik1-S tag protein. The lysates showed good binding of microtubules to the coverslip, but most of the microtubules were immobile and shortened over time from one or both ends. The few microtubules that moved were tracked from the leading end, giving a velocity of 0.49±0.09 μm min−1 (n=7) (Table II), slower than the 1–2 μm min−1 we reported previously for GST/Kar3 (H277–K729) (Endow et al, 1994). The slower velocity is most likely due to the smaller nonspecific protein region provided by the His6 than the GST for attachment of the motor to the glass. The GST may help to prevent the motor from being flattened onto the glass surface. Re-analysis of our previous microtubule gliding data for GST/Kar3 gives 1.19±0.16 μm min−1 (n=10) and 1.80±0.11 μm min−1 (n=10) for leading and lagging ends, respectively. The difference between leading and lagging end velocities is 0.61±0.12 μm min−1 (n=10) (Table II). The His6Kar3 M278–K729 and GST/Kar3 leading end velocities are 2–5-fold slower than His6Kar3/Cik1-S tag lysates or purified protein. Thus, the gliding assays show that the Kar3/Cik1 motor moves on microtubules significantly faster than Kar3 and, like Kar3, destabilizes microtubules at the lagging ends (Endow et al, 1994).
Assays of purified His6Kar3/Cik1-S tag protein for directionality showed that it is a minus-end microtubule motor, like GST/Kar3 (Endow et al, 1994) and GST/Ncd (GST/MC1) (Chandra et al, 1993), which was used for a control in the assays. Axoneme–microtubule complexes glided with the microtubule leading on coverslips coated with either His6Kar3/Cik1-S tag (n=7) or GST/MC1 (n=19). The faster velocity of His6Kar3/Cik1-S tag protein in the gliding assays leaves open the possibility that its motility properties differ from those of Kar3.
Discussion
Kinesin protein dimerization by coiled coils
A predominant structural theme in the kinesin family is dimerization by α-helical coiled-coil formation. Formation of kinesin-1 dimers occurs by a coiled coil and is believed to underlie the ability of single motors to take many successive steps along the microtubule before detaching—single kinesin-1 motors walk processively by alternate binding to the microtubule and hydrolysis of ATP by the two heads (Asbury et al, 2003; Kaseda et al, 2003; Higuchi et al, 2004; Yildiz et al, 2004). Recently, dimerization was reported to convert nonprocessive monomeric Unc104, a kinesin-3 (formerly Unc104/Kif1) family member, into a processive motor (Tomishige et al, 2002). Unc104 dimerization may occur on membranes, as the motor bound to lipid substrates shows diffusional movement in vitro that could allow molecules to move towards one another and interact in regions where their local concentration is high (Klopfenstein et al, 2002). The proposed ability of Unc104 to convert from a monomer to a dimer is of considerable interest, as this structural change may regulate vesicle transport by the motor along microtubules in vivo.
Kar3 dimer formation
The kinesin-14 proteins have a C-terminal motor domain, and are thought to all be minus-end motors. Ncd, a member of this group, dimerizes when a region of its central α-helical coiled coil is expressed with the conserved motor domain (Chandra et al, 1993). Kar3 may be an exception to other kinesin-14 family members—the motor contains a central domain predicted to form α-helical coiled coil by computer algorithms, but Barrett et al (2000) reported that Kar3 does not form homodimers in vivo. Instead, Kar3 was reported to form complexes in vivo with a nonmotor protein, Cik1 or Vik1 (Manning et al, 1999).
We show here by analytical ultracentrifugation that a truncated protein, Kar3 M278–K729, containing less than half of the central nonmotor region, exists in solution as a dimer. Sedimentation equilibrium ultracentrifugation indicates the presence of an equilibrium mixture of monomer and dimer, with an estimated molecular weight of 96 500±3200 Da, close to the predicted dimer molecular weight of 102 822 Da. At the concentrations used in this study, the dimer is by far the major species. CD spectroscopy shows helical characteristics, consistent with coiled-coil formation. Finally, rotary-shadow EM shows paired particles interpreted to represent the two motor domains or heads, infrequently attached to a putative stalk. Based on these findings, we conclude that Kar3 exists in solution as a dimer and formation of dimers occurs via an α-helical coiled coil.
The absence of Kar3 homodimers in vivo in the study by Barrett et al (2000) was based on hydrodynamic analysis of proteins in yeast cell lysates using antibodies to detect the proteins. Calculation of protein molecular mass from the Stoke's radius and sedimentation coefficient, determined by antibody staining of proteins in column or sucrose gradient fractions, led to the conclusion that no Kar3 or Cik1 homodimers were present in cells. This was substantiated by the failure to coimmunoprecipitate proteins corresponding to homodimers from cells expressing Kar3 or Cik1 proteins of somewhat different molecular mass. However, the gel-filtration and sucrose gradient analyses could be misleading, as the molecular mass of the Kar3 and Cik1 homodimers would be close to that of the heterodimer and not resolvable using these methods. Thus, the failure to detect homodimers in these previous experiments could have been due to limitations of the methods that were used.
Alternatively, the failure to detect homodimers may have been because Kar3 dimers exist, but are present in low abundance in cells because interaction of Kar3 with Cik1 is more stable. We present evidence here that Kar3 dimer formation is concentration dependent and that homodimers of Kar3 or Cik1 form less readily than heterodimers when the central, nonmotor region of both proteins is present. Thus, interaction with Cik1 or Vik1 may prevent or greatly reduce Kar3 dimer formation in vivo, and a low concentration of Kar3 dimers might not be detectable with the methods that were used by Barrett et al (2000).
Recently, Mackey et al (2004) reported analytical ultracentrifugation data that they interpreted as showing that GST/Kar3 277–729 is a monomer in vitro. This conclusion was based on the determination of a sedimentation coefficient of 5.36 s for the protein. However, a value of 5.36 s is consistent with either a globular monomer or an elongated dimer. The s value alone is not sufficient to calculate the protein molecular mass and determine its association state—the Stokes radius is also required and this was not determined by Mackey et al (2004). Instead, the conclusion that GST/Kar3 277–729 is a monomer was based only on the s value. Mackey et al (2004) also performed sedimentation equilibrium studies with GST/Kar3 277–729. However, they interpreted their data to show that the protein was a monomer ‘at the early equilibrium times… but was prone to irreversible aggregation to dimmers' during the centrifugation runs, even at the lowest protein concentration (2 μM), where they reported that the aggregation was the worst. Although the meaning of these statements is not entirely clear, taken at face value, these statements are consistent with the finding we report here that Kar3 278–729 exists in solution as a dimer.
Kar3/Cik1 dimer formation
We further demonstrate that Kar3/Cik1 heterodimers form in vitro by several criteria. CD spectra show characteristics of a high helical content for the nonmotor region of the protein and rotary-shadow EM shows a single particle, larger than single Kar3 heads, or two smaller particles, that we interpret to represent the Kar3 head together with the Cik1 C-terminal domain, joined to a stalk. Unexpectedly, the Kar3/Cik1 stalk is visible by rotary-shadow EM, in contrast to the absence of a dimeric Kar3 stalk, which is presumably too short to be visualized by EM. The visible stalk, together with the CD results, indicate that the Kar3/Cik1 protein dimerizes by forming a coiled coil.
Cik1 central region is helical
We also expressed and purified a protein consisting of the Cik1 central, predicted coiled-coil region, and analyzed it by CD. The CD spectra show characteristics of strikingly high helical content with a signal greater than that of the nonmotor region of dimeric Kar3 or Kar3/Cik1. This indicates that Cik1 probably exists in vitro as a dimer.
We examined the relative tendency of Cik1 and Kar3 to form homodimers rather than heterodimers by expressing their central, predicted coiled-coil regions in tandem to one another and examining the length of the proteins by rotary-shadow EM. The mean protein length longer than that expected for the homodimer is consistent with formation of coiled-coil heterodimers by an average of four molecules. Our analysis indicates that heterodimers are at least 16 times more frequent than homodimers. This implies that Kar3/Cik1 is the predominant form of Kar3 in the cell where Cik1 is also present.
Kar3/Cik1 is a microtubule motor
We further asked whether the Kar3/Cik1 protein is a microtubule motor and, if so, whether its motility properties differ from those of Kar3. Using in vitro assays, we show here that Kar3/Cik1 is a motor that moves on microtubules with a velocity of 2–2.4 μm min−1, 4–5-fold faster than His6Kar (0.49 μm min−1), the same form of Kar3 in the His6Kar3/Cik1-S tag protein that we tested in motility assays, and ∼2-fold faster than GST/Kar3 (1.2 μm min−1) (Endow et al, 1994). The Kar3/Cik1 protein depolymerizes taxol-stabilized microtubules at the lagging end, like GST/Kar3 (Endow et al, 1994), and shows dependence on motor density for gliding, characteristic of nonprocessive motors. Purified His6Kar3/Cik1-S tag protein exhibits a mean velocity similar to that of lysates expressing Kar3 and Cik1, but a wider range of velocities. This suggests that the His6Kar3/Cik1-S tag motor is partially inactivated during purification and its true velocity may be greater than reported here. Despite this, our data show that the Kar3/Cik1 velocity of movement on microtubules is significantly faster than Kar3. This raises the possibility that Kar3/Cik1 heterodimer formation in cells regulates Kar3 by altering motor activity.
The faster velocity of the Kar3/Cik1 motor may be due to a more rigid or stable stalk/neck, indicated by the higher helicity of the nonmotor stalk region by CD spectroscopy and visible stalk by rotary-shadow EM. The stalk of Ncd, the kinesin-14 motor most closely related to Kar3 (Dagenbach and Endow, 2004), has been proposed to function like a lever arm to produce force and direct motor movement to the microtubule minus end (Yun et al, 2003). A similar mechanism may operate for the Kar3 motor. The unexpected findings we report here are that Kar3 requires an interacting protein to convert it into a faster motor and single-headed Kar3/Cik1 is faster than two-headed Kar3 in gliding assays in vitro.
Kar3 provides an example of a kinesin motor that exists in different forms in vitro and undergoes structural changes upon association with its partner protein, Cik1, that alter the motor velocity. These forms probably also exist in cells, where they may differ in motor activity and stability. The ability of the Kar3 motor to exist in several forms, including an alternative form with Vik1 (Manning et al, 1999), implies a higher level of regulation involving structural changes that affect motor function.
Materials and methods
Protein expression and purification
Plasmids encoding truncated Kar3 and Kar3/Cik1 proteins were constructed in pMW172 (Way et al, 1990) or pETDuet-1 (Novagen) and transformed into BL21(DE3)pLysS host cells for expression in bacteria using the T7 RNA polymerase system (Studier et al, 1990). The Kar3 proteins were Kar3 M278–K729 (452 residues, 51 411 Da), His6Kar3 MGSSH6SQD P238–K729 (505 residues, 57 876 Da) and a monomeric Kar3 motor domain protein, MRG–N386–K729 (347 amino acids, 38 870 Da) (Gulick et al, 1998), for use in CD experiments. The Kar3/Cik1 protein was His6Kar3 expressed with Cik1 M–N187–D594–GTLESGKETA3KFERQHMDSSTSA2 (S tag underlined) (435 residues, 50 525 Da). Plasmids to express the Cik1 predicted coiled-coil region by itself, or fused in tandem to that of Kar3 encoded Cik1His6 M–D205–E360–GSH6 (165 residues, 19 692 Da) or Cik1 M–D205–E360–GS-Kar3 E114–V372 (418 residues, 49 907 Da), respectively.
Cells were grown to mid-log phase (OD550∼0.4–0.8), induced by adding 0.4 mM IPTG for 4–6 h at 22°C, harvested and stored at −80°C until use. Kar3 and Cik1–Kar3 CC proteins were purified by SP-Sepharose chromatography in HEM (10 mM HEPES, pH 7.2, 1 mM EGTA, 1 mM MgCl2) with elution in HEM200 (HEM+200 mM NaCl), followed by FPLC on Superose 12 in HEM200 (Song and Endow, 1997), and Cik1His6 by Ni-affinity and Superose 12 chromatography. His6Kar3/Cik1-S tag protein was purified by Ni-affinity, followed by Sephacryl S-200 gel-filtration and Hi-Trap SP-Sepharose (Amersham Biosciences) chromatography. The identity of the Kar3 and Cik1 subunits of the purified protein was confirmed by matrix-assisted laser-desorption ionization-time of flight (MALDI-TOF) mass spectroscopy.
Protein concentration was estimated by reading a spectrum from 220 to 350 nm and converting the OD280 value to μM using an extinction coefficient calculated from the protein amino-acid composition (Gill and von Hippel, 1989) with the assumptions that the proteins were isolated under reducing conditions, thus, Cys–Cys disulfide bridges did not contribute to the absorbance, and each Kar3 motor domain was bound to 1 ADP (λex=2310 M−1 cm−1).
CD spectroscopy
CD spectroscopy was performed on an Aviv CD Spectrometer Model 202 using 1 mm and 1 cm path-length cuvettes. Purified Kar3 M278–K729 protein at 9.69, 9.98 or 11.4 μM in HEM200 was scanned from 300 to 190 nm at 1-nm intervals with 10 s averaging time at 4°C in a 1 mm path-length cuvette, then diluted 10- or 12-fold with buffer and scanned in a 1-cm cuvette. A baseline for buffer alone was determined and subtracted from the spectra. To subtract the contribution by the motor domain, purified monomeric Kar3 motor domain protein (MRG–N386–K729) (Gulick et al, 1998) at 19.2, 19.8 or 22.3 μM was scanned and the CD signal in millidegrees for the motor domain was subtracted from the signal for Kar3 M278–K729 to obtain the net signal for the nonmotor region of the protein. This signal was converted to degrees and divided by the protein concentration in g cm−3, the cuvette path length, and 105 × 2, the number of residues in the nonmotor region of Kar3 M278–K729, and multiplied by the dimer molecular weight of the region (12 541 Da × 2=25 082 Da) to obtain the mean molar ellipticity per residue for the difference spectrum.
CD scans for the His6Kar3/Cik1-S tag protein at 8.58 μM in HEM200 were performed in the same manner as for Kar3 M278–K729, except that the signal for His6Kar3/Cik1-S tag was corrected by subtracting the signal for the monomeric Kar3 motor domain protein read at 19.2 μM and adjusted to 8.58 μM. The difference spectrum for His6Kar3/Cik1-S tag corresponds to the nonmotor region of the His6Kar3 and Cik1-S tag proteins, including the Cik1 C-terminal domain (total nonmotor region=593 residues, 69 600 Da). CD spectra of the Cik1H6 protein at 8.51 μM (dimer concentration) were also read.
Analytical ultracentrifugation
Kar3 M278–K729 was analyzed by short-column (80 μl samples) sedimentation equilibrium ultracentrifugation to determine the molecular weight of the protein. Purified Kar3 M278–K729 at 25, 12.5 and 6.25 μM in HEM200 was brought to sedimentation equilibrium in a Beckman XL-A analytical ultracentrifuge at 4°C by centrifuging at 15 000 r.p.m. for 2 h and then lowering the speed to 8000 r.p.m. Absorbance scans were made at 268 and 286 nm at the time of the speed change, and at 6, 12 and 18 h at 8000 r.p.m. Equilibrium was established at 12–18 h. This rapid approach procedure was adopted because a run at 4°C for 24 h at 8000 r.p.m., followed by 24 h at 12 000 r.p.m., showed a small amount of protein degradation during the run. At 18 h, the speed was raised to 50 000 r.p.m. for 2.5 h and the cells were scanned to establish a baseline against which the absorbance scans were corrected. The data were fit to the Ideal-1 model (Beckman Instruments, Inc.) using 0.728 for the partial specific volume (calculated from the amino-acid composition and corrected for 4°C) and ρ=1.0064 for solvent density at 4°C. Best-fit molecular weights for scans of the three protein concentrations at 268 and 286 nm were averaged to give an estimated molecular weight for the Kar3 M278–K729 protein.
The His6Kar3/Cik1-S tag protein at 10.3, 5.15 and 2.58 μM was analyzed by sedimentation equilibrium ultracentrifugation using methods similar to those used for the Kar3 M278–K729 protein.
Electron microscopy (EM)
Purified protein was diluted on ice in 0.2 M NH4HCO3 pH ∼8.2+30% glycerol to a final concentration of 250 or 500 nM Kar3 M278–K729, 257 or 515 nM Kar3/Cik1, and 293 nM, 366 or 488 nM Cik1–Kar3 CC protein. Rotary shadowing was performed as described (Fowler and Erickson, 1979) using a Balzers BAE 120 vacuum evaporator. Briefly, 10 μl of the diluted sample was drawn into a glass capillary tube at room temperature, the tube was inserted into a nozzle attached to a can of compressed air, and the protein solution was sprayed onto a small piece of freshly cleaved mica. The mica was mounted on the rotary-shadowing platform using double-sided tape, and the samples were placed in the vacuum evaporator and dried under vacuum. Shadowing with platinum and carbon was performed after the sample buffer was fully evaporated.
After shadowing, the Pt/C replica was floated off the mica in a small tray of deionized water and mounted on a 400-mesh copper EM grid. Specimens were examined in a Phillips 301 EM and images were recorded at ∼ × 49 500 magnification. Negatives were scanned at actual magnification and 600 dpi using an Epson Model 4870 scanner, and black/white levels were adjusted in Adobe Photoshop v8.0. Particles were measured using the line tool of NIH Image v1.63 calibrated with a 100 nm scale bar in each image. Measurements were corrected for the platinum shell by subtracting 2 × 1 nm from the measured head diameter or length of the Cik1-Kar3 CC protein, or by adding 2 × 1 nm to the measured Kar3 inter-head distance (HP Erickson, personal communication). Measurements for analysis of particle distribution were made using ImageJ v1.33u calibrated with a 100 nm scale bar in each image from the center of the paired or monomer particle to the center of the nearest and second nearest particles, and were not corrected for the platinum shell.
Motility assays
Lysates for motility assays were prepared in PEM100 (10 mM NaPO4, pH 7.4, 1 mM EGTA, 1 mM MgCl2, 100 mM NaCl) using 2 ml g−1 induced cells, as described (Song et al, 1997). Clean glass coverslips were coated with 2–3 μl of 1 mg ml−1 anti-His6 antibodies (ICL, Inc.)+3–6 μl PEM for 1 min, followed by 2 × 30 μl PEM washes. The cleared lysate (1–6 μl)+PEM to make 6 μl volume was placed on the coverslip with 0.6–0.9 μl of 2 μM microtubules ([tubulin])+40 μM taxol and 0.9 μl of 50 mM Mg·ATP, pH ∼7. Coverslips were inverted onto clean slides, the edges were sealed with VALAP (1:1:1 vasoline/lanolin/paraffin, heated until clear) and microtubule gliding was observed using VEDIC microscopy (Walker et al, 1990) and recorded onto an S-VHS videotape. Gliding velocities were determined by tracking microtubules from a videotape using custom software (a gift of N Gliksman and T Salmon). Directionality assays were performed using axoneme–microtubule complexes, assembled as described (Song et al, 1997).
Supplementary Material
Supplementary Data
Supplementary Data
Supplementary Data
Acknowledgments
We thank T Oas and P Chugha for valuable discussions and help with CD spectroscopy, the Duke University Protein Facility for the use of an analytical ultracentrifuge, and KC Lounes and HP Erickson for helpful comments on the manuscript. This work was supported by grants from the NIH and HFSP to SAE and St Jude Children's Research Hospital Cancer Center and American Lebanese Syrian Associated Charities to HWP.
References
- Asbury CL, Fehr AN, Block SM (2003) Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302: 2130–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JG, Manning BD, Snyder M (2000) The Kar3p kinesin-related protein forms a novel heterodimeric structure with its associated protein Cik1p. Mol Biol Cell 11: 2373–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Wilson D, Wolf E, Tonchev T, Milla M, Kim P (1995) Predicting coiled coils by use of pairwise residue correlation. Proc Natl Acad Sci USA 92: 8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Kim PS (1993) A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73: 823–832 [DOI] [PubMed] [Google Scholar]
- Chandra R, Salmon ED, Erickson HP, Lockhart A, Endow SA (1993) Structural and functional domains of the Drosophila ncd microtubule motor protein. J Biol Chem 268: 9005–9013 [PubMed] [Google Scholar]
- Dagenbach EM, Endow SA (2004) A new kinesin tree. J Cell Sci 117: 3–7 [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED (1994) Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J 13: 2708–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler WE, Erickson HP (1979) Trinodular structure of fibrinogen: confirmation by both shadowing and negative stain electron microscopy. J Mol Biol 134: 241–249 [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326 [DOI] [PubMed] [Google Scholar]
- Greenfield N, Fasman GD (1969) Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8: 4108–4116 [DOI] [PubMed] [Google Scholar]
- Gulick AM, Song H, Endow SA, Rayment I (1998) X-ray crystal structure of the yeast Kar3 motor domain complexed with Mg·ADP to 2.3 Å resolution. Biochemistry 37: 1769–1776 [DOI] [PubMed] [Google Scholar]
- Higuchi H, Bronner CE, Park HW, Endow SA (2004) Rapid double 8-nm steps by a kinesin mutant. EMBO J 23: 2993–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J (2001) Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Kaseda K, Higuchi H, Hirose K (2003) Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol 5: 1079–1082 [DOI] [PubMed] [Google Scholar]
- Klopfenstein DR, Tomishige M, Stuurman N, Vale RD (2002) Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell 109: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller DG, Cohen FE, Langridge R (1990) Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol 214: 171–182 [DOI] [PubMed] [Google Scholar]
- Kozielski F, De Bonis S, Burmeister WP, Cohen-Addad C, Wade RH (1999) The crystal structure of the minus-end-directed microtubule motor protein ncd reveals variable dimer conformations. Structure 7: 1407–1416 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LSB, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy ASN, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L (2004) A standardized kinesin nomenclature. J Cell Biol 167: 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey AT, Sproul LR, Sontag CA, Satterwhite LL, Correia JJ, Gilbert SP (2004) Mechanistic analysis of the Saccharomyces cerevisiae kinesin Kar3. J Biol Chem 279: 51354–51361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Stemple JK, Satterwhite L, Salmon ED, Bloom K (2003) The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr Biol 13: 1423–1428 [DOI] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M (1999) Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol 144: 1219–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD (1990) KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 60: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Page BD, Snyder M (1992) CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev 6: 1414–1429 [DOI] [PubMed] [Google Scholar]
- Sablin EP, Case RB, Dai SC, Hart CL, Ruby A, Vale RD, Fletterick RJ (1998) Direction determination in the minus-end-directed kinesin motor ncd. Nature 395: 813–816 [DOI] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA (1997) Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell 8: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA (1992) Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70: 451–458 [DOI] [PubMed] [Google Scholar]
- Song H, Endow SA (1997) Rapid purification of microtubule motor domain proteins expressed in bacteria. BioTechniques 22: 82–85 [DOI] [PubMed] [Google Scholar]
- Song H, Golovkin M, Reddy ASN, Endow SA (1997) In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc Natl Acad Sci USA 94: 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185: 60–89 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU (2005) Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434: 987–994 [DOI] [PubMed] [Google Scholar]
- Tomishige M, Klopfenstein DR, Vale RD (2002) Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297: 2263–2267 [DOI] [PubMed] [Google Scholar]
- Walker RA, Gliksman NR, Salmon ED (1990) Using video-enhanced differential interference contrast microscopy to analyze the assembly dynamics of individual microtubules in real time. In Optical Microscopy for Biology, Herman B and Jacobson K (eds) pp 395–407. New York: Wiley-Liss [Google Scholar]
- Way M, Pope P, Gooch J, Hawkins M, Weeds AG (1990) Identification of a region in segment 1 of gelsolin critical for actin binding. EMBO J 9: 4103–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR (2004) Kinesin walks hand-over-hand. Science 303: 676–678 [DOI] [PubMed] [Google Scholar]
- Yun M, Bronner CE, Park C-G, Cha S-S, Park H-W, Endow SA (2003) Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd. EMBO J 22: 5382–5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Data
Supplementary Data


