Abstract
Most of the transcription factors, RNA polymerases and enhancer binding factors are absent from condensed mitotic chromosomes. In contrast, epigenetic marks of active and inactive genes somehow survive mitosis, since the activity status from one cell generation to the next is maintained. For the zinc-finger protein CTCF, a role in interpreting and propagating epigenetic states and in separating expression domains has been documented. To test whether such a domain structure is preserved during mitosis, we examined whether CTCF is bound to mitotic chromatin. Here we show that in contrast to other zinc-finger proteins, CTCF indeed is bound to mitotic chromosomes. Mitotic binding is mediated by a portion of the zinc-finger DNA binding domain and involves sequence specific binding to target sites. Furthermore, the chromatin loop organized by the CTCF-bound, differentially methylated region at the Igf2/H19 locus can be detected in mitosis. In contrast, the enhancer/promoter loop of the same locus is lost in mitosis. This may provide a novel form of epigenetic memory during cell division.
Keywords: CTCF, cell cycle, chromosome conformation capture, mitotic chromosome
Introduction
During mitosis, chromatin fibers are folded and coiled, resulting in highly condensed chromosomes. Such a condensed structure is favorable for the movement and separation of genomic material. The condensation status of mitotic chromosomes coincides with the global silencing of gene transcription by all three RNA polymerases. Studies comparing the chromatin structure of specific genes in interphase and mitosis revealed that both the general nuclease sensitivity of active genes and presence of DNaseI-hypersensitive sites appear to be preserved in mitotic chromosomes (Gazit et al, 1982). However, the precise location of the hypersensitive sites upstream of some specific genes appears to differ between interphase and mitotic chromatin (Kuo et al, 1982), suggesting that there is a rearrangement of some of the proteins controlling these genes in mitosis. Indeed, dynamic changes of chromatin-associated proteins are observed when cells enter mitosis. Most of the basal transcription factors, RNA polymerases and enhancer binding factors are absent from the condensed, mitotic chromosomes (Martinez-Balbas et al, 1995; Gottesfeld and Forbes, 1997; Dovat et al, 2002; Zaidi et al, 2003), as well as enzymes that are involved in modification of histones, such as histone acetyltransferases (HATs) and deacetylases (HDACs), are excluded from the mitotic chromosomes (Kruhlak et al, 2001). In contrast, some types of proteins remain associated with mitotic chromosomes, including chromosome scaffold proteins (Hagstrom and Meyer, 2003), the chromosomal passenger proteins (Adams et al, 2001), components of basal transcription machineries (Michelotti et al, 1997; Chen et al, 2002; Christova and Oelgeschlager, 2002) and other proteins, such as the nuclear matrix protein (Berube et al, 2000), DNA glycosylase (Dantzer et al, 2002), poly (ADP-ribose) polymerase (PARP) (Kanai et al, 2000) and the BET bromodomain protein family (Dey et al, 2000). These proteins are thought to be either structural components of condensed chromosomes or gene markers for accurate propagation of gene expression throughout the cell cycle.
Eukaryotic genomes are organized into functional units containing individual genes or gene groups together with the corresponding regulatory elements. These functional units have to be insulated from each other in order to prevent illegitimate interactions with other transcriptional units. One particular aspect of insulation is enhancer blocking. In higher eukaryotes, several insulator sequences have been identified. A common model for the mechanism of enhancer blocking predicts association of looped chromatin domains with the nuclear matrix. Until now, only a single protein, CTCF, has been identified within vertebrates to mediate enhancer blocking activity (Ohlsson et al, 2001; Burgess-Beusse et al, 2002; Kuhn and Geyer, 2003). This 11 zinc-finger protein and the enhancer blocking function are conserved from Drosophila to man (Moon et al, 2005). In vertebrates, a role for CTCF in reading and propagating epigenetic marks has been shown (Pant et al, 2003; Schoenherr et al, 2003; Fedoriw et al, 2004; Mukhopadhyay et al, 2004; Rand et al, 2004). Furthermore, long-range chromatin interactions at the Igf2/H19 locus involving the differentially methylated regions (Murrell et al, 2004) are dependent on CTCF target sites (VK Tiwari, unpublished). The results presented here indicate that not only CTCF chromatin binding, but also ‘boundary type' long-range chromatin interaction can be detected in mitotic chromatin, whereas the enhancer/promoter interaction at the same locus is lost in mitosis.
Results
CTCF localizes to mitotic chromosomes
Previously, we have noticed that during mitosis, CTCF is found on centrosomes and midbody (Zhang et al, 2004). Subsequent tests with different fixation conditions revealed that after ethanol/acetic acid fixation, CTCF-specific antibodies stained centrosomes and midbody in addition to the chromosomes (Figure 1A). Confocal images of cells from individual stages of mitosis clearly demonstrate that CTCF antibody staining is associated with chromosomes in mitosis, from prophase, when the condensation of chromatin starts, up to telophase, when the decondensation of mitotic chromosomes initiates (Figure 1Aa–e). In addition, the mitotic centrosome staining by the anti-CTCF antibody was preserved in this fixation condition (Figure 1Ac and f). Indeed, when double immunofluorescent staining with antibodies against CTCF and γ-tubulin was carried out using ethanol/acetic acid fixation, a full pattern of the mitotic distribution of CTCF emerged, including the mitotic chromosome association, the spindle pole localization as identified by γ-tubulin (Figure 1Af), as well as the midbody association (Figure 1Ag). Specificity of the CTCF antibody was documented by blocking CTCF staining in the presence of the antigenic domain (Figure 1Ai).
Figure 1.
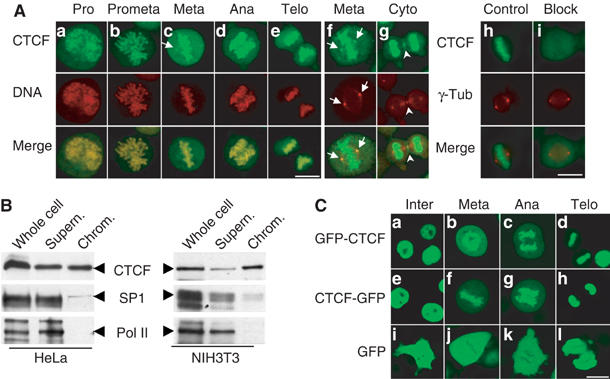
CTCF is associated with mitotic chromosomes. (A) Indirect immunofluorescence experiments were carried out on HeLa cells fixed with ethanol/acetic acid. The individual stages within mitosis, including prophase (a), prometaphase (b), metaphase (c), anaphase (d) and telophase (e), were selected according to the chromosomal morphology. CTCF was stained with the anti-CTCF antibody and a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (green), and DNA was stained with propidium iodide (red) (a–e). The overlay shows colocalization of CTCF and mitotic chromosomes. The arrows indicate the position of mitotic centrosomes. Indirect double immunofluorescence with antibodies against CTCF (green) and γ-tubulin (red) identifies CTCF in the mitotic centrosomes (f) and midbody (g). To test antibody specificity, the primary antibodies against CTCF and γ-tubulin were preincubated with bacterially expressed and GST-purified GST-CTCF (i) or GST (h). (B) Western blot with anti-CTCF, anti-Sp1 and anti-Pol II antibodies of HeLa and NIH3T3 whole cell extract, mitotic chromosome fraction (chrom.) and the corresponding supernatant. The respective proteins are indicated by arrows. (C) Living HeLa cells stably expressing GFP-CTCF (a–d), CTCF-GFP (e–h) and GFP (i–l) were analyzed by confocal microscopy and grouped according to cell cycle stage. Scale bar, 10 μm.
An association of CTCF with mitotic chromosomes was quite unexpected since most transcription factors are not associated with condensed mitotic chromosomes. To further confirm this observation, biochemical purification of mitotic chromosomes was performed and the chromosome-enriched fractions were analyzed by Western blot using the anti-CTCF antibody. As shown in Figure 1B, CTCF is present in the mitotic chromosome fraction in both HeLa and NIH3T3 cells. In contrast, RNA polymerase II (Pol II) is nearly absent from the mitotic chromosome fraction, which is consistent with the silenced stage of DNA transcription during mitosis. In addition, binding of the Sp1 transcription factor to mitotic chromosomes is strongly reduced, which is in agreement with previous observations (Martinez-Balbas et al, 1995).
In order to verify the mitotic chromosome association of CTCF under living conditions, cell lines expressing GFP fusions with CTCF as a C-terminal or an N-terminal fusion with GFP were established. Since overexpression of CTCF leads to cell cycle arrest (Rasko et al, 2001), the resulting cell clones were checked for an expression level of the CTCF/GFP fusions below that of the endogenous CTCF protein (see Supplementary Figure 1) and the transgenes were sequenced. Living cells within different cell cycle stages were selected and analyzed by confocal microscopy (Figure 1C). In the control clones, as expected, GFP (Figure 1Ci–l) was distributed throughout the whole cell and mainly excluded from the condensed mitotic chromosomes during mitosis. In contrast, full-length CTCF was localized to the nucleus during interphase and both nucleolar exclusion and nucleolar accumulation of CTCF were observed (Figure 1Ca and e). During mitosis, association of CTCF with mitotic chromosomes was clearly shown by the bright green fluorescence on the condensed chromosomal arms. This association is seen in all mitotic stages (Figure 1Cb–d and f–h).
Taken together, using three different experimental approaches, we demonstrate that CTCF is associated with the mitotic chromosomes.
The C-terminal zinc-fingers are essential for mitotic chromosome targeting
After having shown that CTCF is a mitotic chromosome-associated protein, we wanted to know whether DNA binding, and therefore the DNA binding domain of CTCF, is responsible for the binding to mitotic chromosomes, or whether other domains and possibly other interactions are required. To this end, we generated GFP-CTCF deletion constructs (Figure 2). In order to simplify the experiments, we utilized transient DNA transfection into HeLa cells. All of the tested fusion proteins (except for GFP by itself) localized to the interphase nuclei (not shown). Similar to the stably transfected cell clones, transient expression of full-length CTCF is concentrated at the mitotic chromosomes (Figure 2Aa), whereas GFP alone is not concentrated on mitotic chromosomes (Figure 2Ai). When full-length CTCF is truncated in the middle, the C-terminal half of the protein (GFP-CTCF_ΔN) clearly shows an association with mitotic chromosomes (Figure 2Ab). This GFP fusion protein is localized to the chromosome arms in a similar manner as the full-length protein. The other half of CTCF, which encompasses the N-terminus and the N-terminal half of the zinc-fingers (GFP-CTCF_ΔC), does not paint the mitotic chromosomes, rather the whole cell is stained (Figure 2Ac). When the N-terminus, the C-terminus and the 11 zinc-finger domain were tested individually, only the zinc-finger domain showed a distinct association with the mitotic chromosomes (Figure 2Ad). In contrast, the C-terminus (GFP-CTCF_C.t.) and the N-terminus alone (GFP-CTCF_N.t.) showed no accumulation on the mitotic chromosomes (Figure 2Ag and h). Within the zinc-finger domain, the mitotic chromosome targeting region of CTCF was further mapped to the C-terminal half of the DNA binding domain (GFP-CTCF_ZnC) (Figure 2Ae). The N-terminal half of this DNA binding domain showed only a weak accumulation at the mitotic chromosome area (Figure 2Af), indicating that this part of the protein might have a minor contribution only to target CTCF to mitotic chromosomes. All of the localization data are summarized in Figure 2B, revealing that mitotic chromosome binding (ch) is found in all constructs expressing the C-terminal half of the 11 zinc-finger domain, whereas all other constructs showed GFP distribution throughout the mitotic cell (ce).
Figure 2.
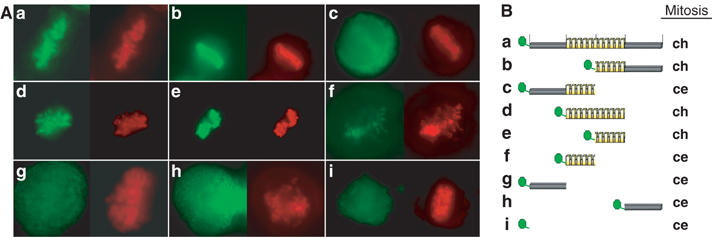
Delineation of the mitotic chromosome association domain of CTCF. (A) HeLa cells were transiently transfected with the GFP and GFP-CTCF constructs as schematically illustrated in (B). Except for the pEGFPC2 construct, all of the constructs resulted in interphase in a nuclear GFP signal (not shown). Living mitotic cells were analyzed by fluorescence microscopy after transfection of pEGFPC2-CTCF_f.l. (a), pEGFPC1-CTCF_ΔN (b), pEGFPC2-CTCF_ΔC (c), pEGFPC2-CTCF_Zn (d), pEGFPC2-CTCF_ZnC (e), pEGFPC2-CTCF_ZnN (f), pEGFPC2-CTCF_N.t. (g), pEGFPC2-CTCF_C.t. (h) and pEGFPC2 (i) (green) and stained with Hoechst 33342 (red). GFP signals are summarized in panel B. Mitotic GFP location is indicated by chromosomal (ch) or cellular distribution including the whole cellular space (ce). The N-terminal part of the zinc-finger domain (f) shows chromosomal localization as well, but at much reduced levels as compared to the full-length zinc-finger region or the C-terminal part of the zinc-finger domain.
CTCF and HP1α have distinct distribution patterns on chromosomes
Chromatin is generally divided into euchromatin and heterochromatin. Euchromatin consists of much less condensed chromatin fibers that have a relatively dispersed appearance in the nucleus and occupy most of the nuclear region. Heterochromatin comprises very densely packed chromatin fibers, which are typically found at centromeres and appear as compact regions clustered near the nucleolus and nuclear membrane. We wanted to know whether CTCF binds to euchromatin and/or to heterochromatin and if CTCF is preferentially distributed on chromatin during mitosis. To address this question, HP1α was used as a marker for heterochromatin (Minc et al, 1999) and double immunofluorescence was carried out in HeLa cells. In order to visualize the complete subcellular localization of both CTCF and HP1α throughout the cell cycle, cells were fixed with ethanol/acetic acid (Figure 3A). In addition, paraformaldehyde fixation was also tested in order to check the consistency of HP1α staining (data not shown). Both proteins were exclusively localized to the nuclear interior during interphase (Figure 3Aa–c). However, HP1α showed a different subnuclear distribution as compared to CTCF, irrespective of the fixation condition (Figure 3A and data not shown). HP1α was distributed as intensely labeled, discrete nuclear foci against a diffuse nucleoplasmic background. The foci stained by HP1α were mostly concentrated at the nuclear and nucleolar periphery (Figure 3Ab). CTCF was distributed throughout the nucleus and usually excluded from the nucleolus (Figure 3Aa). Furthermore, CTCF appeared to be mainly excluded from the nuclear foci that are characterized by HP1α (Figure 3Ac). Similar patterns were also observed at cytokinesis (Figure 3Ag–i). The HP1α distribution pattern is in agreement with a previous report where those foci were identified as centromeric heterochromatin (Cheutin et al, 2003). Therefore, the predominantly mutually exclusive distribution of CTCF and HP1α in nuclei suggests that CTCF may be preferentially excluded from heterochromatin during interphase.
Figure 3.
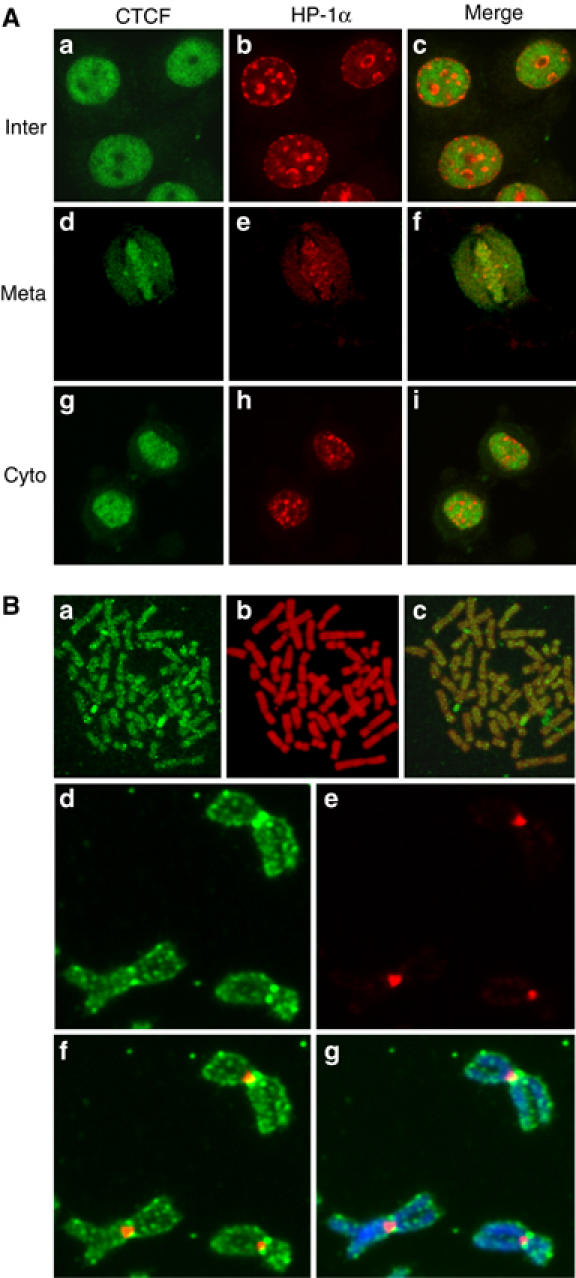
The cell cycle-dependent subcellular distribution of CTCF and HP1α. (A) Double indirect immunofluorescence experiments were carried out in HeLa cells fixed with ethanol/acetic acid. Cells were stained with primary antibodies against CTCF and HP1α followed by FITC-conjugated (green) or Texas red dye-conjugated (red) secondary antibodies, respectively. Interphase (a–c), metaphase (d–f) and early cytokinesis (g–i) are shown. The merged panel allows the comparison of the localization of CTCF and HP1α. (B) Mitotic chromosome spreads were prepared with synchronized human primary amnion cells (46,XX) (a–c). Indirect immunofluorescence was carried out with the anti-CTCF antibody and an FITC-conjugated secondary antibody (green). DNA was stained with propidium iodide (red). The overlay shows colocalization of CTCF and DNA. Unfixed HeLa cell mitotic chromosome spreads were stained with Hoechst and antibodies against CTCF and HP1α (d–g). The slide was visualized for CTCF (d), HP1α (e), CTCF+HP1α (f) and CTCF+HP1α+Hoechst (g).
It has been shown previously that HP1α stays bound to the chromosomes during mitosis and is primarily concentrated at heterochromatic sites like centromeres and pericentromeres (Minc et al, 2001). Since CTCF is bound to the mitotic chromosomes as well, the maintenance of this distribution of CTCF and HP1α during interphase was investigated during mitosis. Punctuated HP1α staining patterns at the centromeric regions of mitotic chromosomes were observed (Figure 3Ae and data not shown). In contrast, the staining pattern of CTCF, which labeled the entire chromosome arms, is clearly different from that of HP1α (Figure 3Ad–f). In order to obtain higher resolution images and to test the mitotic distribution of CTCF in primary cells, mitotic chromosomes from primary amnion cells were spread before staining. As shown in Figure 3B, CTCF is distributed along the whole chromosome arms. HP1α binding to mitotic chromosomes has been carefully analyzed on unfixed spread chromosomes (Minc et al, 2001). This we repeated for a double staining to visualize CTCF as well as HP1α on HeLa spread chromosomes (Figure 3Ad–g). The staining pattern of CTCF suggests that, unlike the selective centromeric association of HP1α, CTCF is present at multiple regions along the chromosomes with a slightly higher concentration at the centromeres. Such a distribution over the chromosome arms would be predicted from the estimation of the number of CTCF binding sites in the genome, which is in the order of 104 (Mukhopadhyay et al, 2004). Such a high number of binding sites cannot be resolved on the compacted mitotic chromosomes, rather a staining pattern with irregular intensity along the entire chromosomes would be predicted and is seen.
CTCF is bound in mitosis at known target sites
Microscopic images and cellular fractionation document the overall distribution of CTCF within the nucleus and on mitotic chromosomes. But these results cannot distinguish between CTCF binding at specific sequences, which are also bound by CTCF during interphase, and some other kind of binding to mitotic chromatin, possibly irrespective of the presence or absence of specific target sequences. Therefore, we wanted to analyze well-characterized CTCF binding sites for occupancy during mitosis by chromatin immunoprecipitation (ChIP) with an antibody against CTCF. We chose the β-globin insulator (Tanimoto et al, 2003), the H19 locus (Bell and Felsenfeld, 2000), the DM-1 locus (Filippova et al, 2001) and the myc-N site (Lutz et al, 2003). For all of these sites, CTCF function and/or occupancy in human chromatin has been documented. Since these chromatin samples have been prepared from nonsynchronized cells, they primarily consist of interphase chromatin. ChIP assays (Figure 4) demonstrated specific CTCF binding with asynchronous chromatin preparations on all four binding sites, in contrast to a non-CTCF binding site, which remains negative (H19neg). Chromatin prepared from synchronized mitotic cells was similarly used for ChIP experiments and revealed specific CTCF binding in all cases (Figure 4). There is never an exact match in the amount of precipitated material in asynchronous versus mitotic chromatin. This is somehow expected since interphase and mitotic chromatin have quite different compaction and biophysical properties. As a positive control, we precipitated mitotic and asynchronous chromatin with an antibody against histone H3. The DM1 promoter from both chromatin samples was similarly recovered (Figure 4) with a standard error similar to the CTCF ChIP experiments. The negative control H19neg was negative in mitosis as well as in asynchronous cells, demonstrating that sequence specific DNA binding of CTCF is observed, rather than a nonspecific ‘sticking' to mitotic chromosomes. Furthermore, using an antibody against Pol II, we observed that mitotic chromosomes lost Pol II association with the promoter of the H2B gene (Figure 4; Pol II). For this gene, loss of Pol II binding during mitosis has been found previously (Christova and Oelgeschlager, 2002), supporting the hypothesis that CTCF binding to mitotic chromosomes is specific for CTCF as well as for specific CTCF target sites. We wanted to extend these results generated from human HeLa cell chromatin to another cell line and another species. The best studied species for CTCF function is mouse, since target site deletions have clearly documented the dependence of enhancer blocking on CTCF binding (for review, see Schoenherr et al, 2003). We used nonsynchronized chromatin and mitotic chromatin from a mouse cell line (NIH3T3) and carried out ChIP analyses with a CTCF-specific antibody. A conserved CTCF binding site exists in the gene coding for the amyloid β-protein precursor (APP) with a functional CTCF site in the promoter region in man (Vostrov and Quitschke, 1997) as well as in mouse (unpublished). Furthermore, the H19 ICR at the mouse Igf2/H19 locus and two additional CTCF target sites (Igf2R and #396) were analyzed. The CTCF site at the Igf2R gene resides within a differentially methylated region (R Ohlsson, unpublished) and the #396 site was identified in a genome-wide screen (Mukhopadhyay et al, 2004). When analyzed by ChIP, all sequences clearly show CTCF occupancy in mitotic chromatin (Figure 5) with target sequences being detected in the precipitates of both asynchronous and mitotic chromatin. Again, differences in the amount of target sequences that are precipitated in interphase or mitotic cells may have technical reasons (see above) or may reflect heterogeneity in mitotic cells with respect to CTCF binding. Sequence specificity of the binding to mitotic chromatin is demonstrated by the analysis of a nonbinding site in the myc gene, which cannot be detected in either of the chromatin preparations (Figure 5). Therefore, we can conclude that all of the eight different CTCF sites tested show binding in interphase as well as in mitotic chromatin.
Figure 4.
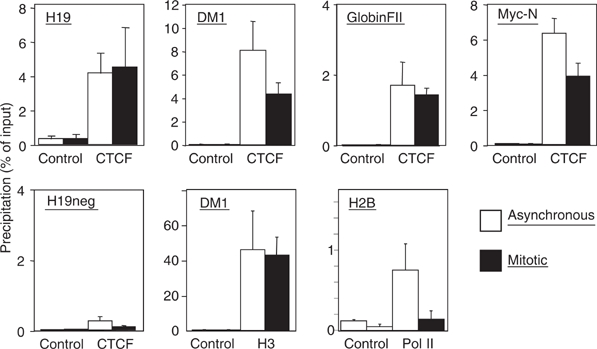
Human CTCF target sites are occupied in interphase and mitosis. Chromatin prepared from interphase HeLa cells (asynchronous) and mitotic HeLa cells was precipitated with antibodies against CTCF or RNA Pol II or histone H3 (H3). To control unspecific precipitation preimmune serum in the case of CTCF and H3 or an unrelated antibody (IgG) in the case of Pol II (control) was used. Precipitates were tested by real-time PCR for the presence of DNA sequences of known CTCF target sites at the myc-N site (Lutz et al, 2003), the DM-1 locus (Filippova et al, 2001), the H19 locus (Bell and Felsenfeld, 2000) and the β-globin insulator FII (Tanimoto et al, 2003). A non-CTCF binding site (H19neg) could not be detected in the precipitates. The values are expressed as percentage of precipitated input chromatin. Error bars indicate the standard error of 2–3 independent chromatin preparations.
Figure 5.
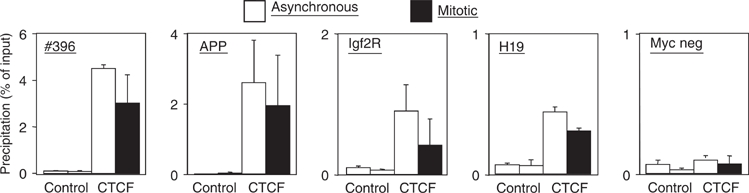
Mouse CTCF target sites are occupied in interphase and mitosis. Chromatin prepared from interphase NIH3T3 cells (asynchronous) and mitotic NIH3T3 cells was precipitated with antibodies against CTCF or with preimmune serum (control). Precipitates were tested by real-time PCR as in Figure 4 for the presence of DNA sequences of known CTCF target sites at the mouse APP promoter (unpublished), the CTCF site at the Igf2R gene (unpublished), the #396 site (Mukhopadhyay et al, 2004) and the H19 ICR (Bell and Felsenfeld, 2000; Hark et al, 2000; Kanduri et al, 2000; Szabo et al, 2000). The non-CTCF binding site (Myc neg) could not be detected in the precipitates.
Igf2-H19 long-range interaction in mitosis
Despite a high compaction ratio, mitotic chromosomes are in general accessible to transcription factors and some structural proteins (Chen et al, 2005). It has been speculated that the dynamic exchange of transcription factors observed during mitosis might help prepare the genome for its reactivation by maintaining a relatively open chromosome configuration. If this is the case, long-range chromosome interactions exemplified by the β-globin locus (Tolhuis et al, 2002; Palstra et al, 2003) or the Igf2/H19 locus (Murrell et al, 2004) have to be re-established after mitosis to physically associate active genes with multiple cis-regulatory elements in the nuclear space. Alternatively, some of these long-range interactions might be maintained during mitosis. To distinguish between these two possibilities, we analyzed the Igf2/H19 locus. Previously, it has been demonstrated that the H19 ICR interacts with a differentially methylated region in the 5′-flank of the Igf2 gene separated by more than 80 kb (Murrell et al, 2004). Given that this interaction depends on CTCF target sites at the H19 ICR (Kurukuti et al, submitted) and that CTCF remains associated with the H19 ICR in mitotic chromosomes, as documented here, there was a distinct possibility that such long-range interactions would actually survive chromosome compaction. To this end, we carried out a ‘chromosome conformation capture' (3C) assay (Dekker et al, 2002) in interphase and mitotic cells. The 3C technology involves quantitative PCR analysis of crosslinking frequencies between two given DNA restriction fragments, which gives a measure of their proximity in the nuclear space. Local chromatin configuration has no effect on digestion efficiency, implying that the assay is not biased owing to preferential restriction enzyme digestion of one site over the other (Dekker et al, 2002). Analysis of five independent samples by 3C revealed that a short-range interaction within the Calreticulin gene, which is used to normalize crosslinking frequencies, is maintained in mitotic chromosomes (Figure 6B). PCR analysis using one primer from Igf2 DMR1 and one of the primers from Calreticulin gene (Tolhuis et al, 2002) did not generate any ligation product in any of the mitotic and interphase chromosome samples (Figure 6B), demonstrating that random intramolecular ligation is not a problem in this assay. Nevertheless, the strong overall compaction of mitotic chromosomes might impose a problem by unspecific chromatin contacts generating positive signals in the 3C assay. To test for such a potential problem, we used the ubiquitously transcribed gene region of the Ercc3 locus for which a spatial conformation in interphase has been described, causing a positive 3C signal (Palstra et al, 2003). We envisaged that the shut down of transcription during mitosis should erase the 3C signal generated by primers within the transcribed region separated by 14 kb. Indeed, the specific Ercc3 signal in asynchronous chromatin is lost during mitosis (Figure 6C), thus showing that the mitotic compaction does not cause unspecific 3C signals. Therefore, we tested the CTCF-mediated 3C signal between the Igf2 DMR1 and H19 ICR elements, separated by more than 80 kb. The 3C analysis revealed that the long-range interaction between Igf2 DMR1 and H19 ICR is maintained in both mitotic and interphase chromosomes. We reached this conclusion by analyzing this interaction using two different primer combinations, each of which produced nearly identical results. The primer set DF and IF gave one nonspecific band below the specific one (shown as asterisk), which was not observed after Southern hybridization using a specific DMR1 probe (data not shown). In contrast, the interaction of the enhancer with the Igf2 promoter, separated by 100 kb, resulted in a 3C signal specific for asynchronous cells and not seen in mitotic chromatin (Figure 6C). Thus our results indicate that the epigenetically controlled long-range interaction within the Igf2/H19 expression domain is maintained during mitosis, whereas an interaction of similarly separated enhancer and promoter fragments and short-range interactions within the Ercc3 gene are lost.
Figure 6.
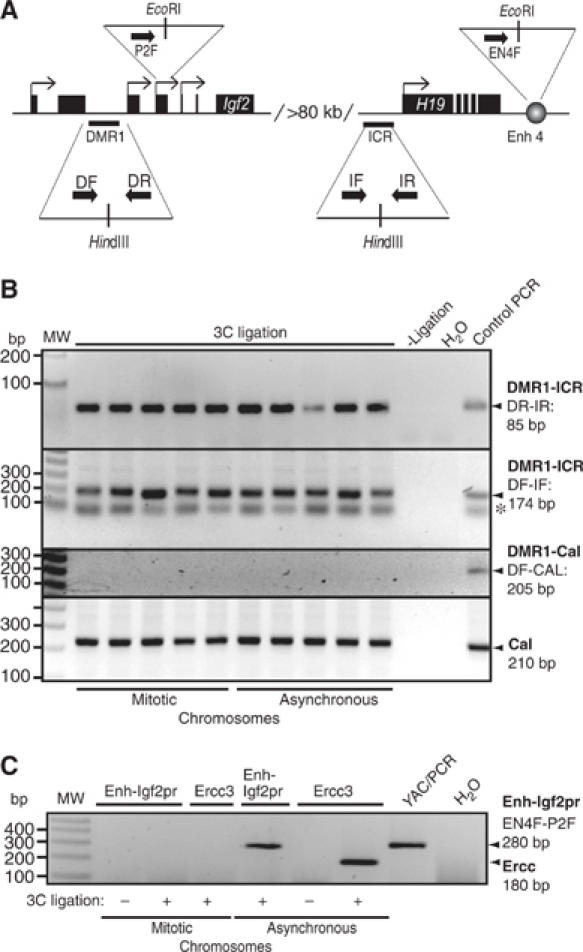
The Igf2 DMR1–H19 ICR interaction is an epigenetic mark. (A) Schematic representation of the primers used. The Igf2 DMR1 and H19 ICR as well as Igf2 promoter 2 and H19 enhancers, which are separated by more than 80 kb of intervening sequences, are indicated in the image. The primers used are DF and DR for Igf2 DMR1, IF and IR for H19 ICR, P2F for the Igf2 promoter 2 and EN4F for the enhancer 4 downstream of H19. (B) Long-distance ICR/DMR1 interaction is maintained in mitotic chromosomes. Chromatin interaction between the H19 ICR and Igf2 DMR1 was studied by 3C analysis using chromatin from NIH3T3 cells and two different primer combinations. PCR products from primer combinations DR plus IR, and DF plus IF were run on 2.5 and 1.5% agarose gel, respectively, with ethidium bromide staining. The last lane in both the gels shows the PCR products with template obtained after digestion and ligation of YAC (for Igf2 DMR1 and H19 ICR) or a control plasmid (containing the DMR1 and Calreticulin sequences, see below). The band in all the lanes labeled with an asterisk is a nonspecific band, which does not hybridize to a specific DMR1 probe (not shown). All individual PCR analyses of five independent 3C samples were performed at least twice. To account for random background interactions, we performed PCR using one primer each from Calreticulin (CalR) and Igf2 DMR1 as indicated in the panel. The control template for this analysis was generated by mixing, digesting and ligating CalR and Igf2 DMR1 HindIII amplicons in equimolar amounts. The CalR products were used for normalizing signals to account for crosslinking efficiency of mitotic/asynchronous chromosomes with previously used primers (Tolhuis et al, 2002). (C) Long-distance enhancer/promoter interaction is not maintained in mitosis. One of the samples (#3 in both mitotic and asynchronous chromosome preparations) presented in panel B was subjected to additional 3C analysis to test for unspecific interaction. The interactions within the Ercc3 locus (encompassing 14 kb) (Palstra et al, 2003) and between the H19 enhancer and Igf2 promoter 2 (encompassing 101 kb) (Tiwari et al, unpublished observation) were examined in mitotic and asynchronous chromosomes. In both cases, the interaction seen in asynchronous cells (Ercc and Enh-Igf2pr) is lost in mitotic chromatin. Signals were always dependent on the ligation step (3C ligation).
Discussion
Nuclear transcription is repressed during mitosis, a phenomenon that has been known for decades (reviewed by Gottesfeld and Forbes, 1997). Mechanisms of transcriptional repression have been attributed to phosphorylation of RNA Pol II and of the general transcription factors TFIIH and TFIID (Segil et al, 1996; Bellier et al, 1997; Long et al, 1998; Akoulitchev and Reinberg, 1998). Epigenetic marks of active and inactive genes have to be maintained throughout mitosis in order to preserve the activity status of every gene. Such ‘bookmarks' have been identified on the level of chromatin structure (Gazit et al, 1982; Michelotti et al, 1997; John and Workman, 1998), as well as for the factors TFIID and TFIIB, which remain associated with active gene promoters during mitotic inactivation (Christova and Oelgeschlager, 2002). In contrast, enhancer or promoter factors are displaced from the condensed chromosomes during mitosis (Martinez-Balbas et al, 1995; Gottesfeld and Forbes, 1997; Parsons and Spencer, 1997). Displacement may be mediated by an active phosphorylation event as exemplified with the zinc-finger proteins Ikaros and Sp1 (Dovat et al, 2002). After mitosis, enhancer and promoter factors have to resume their activity on the active genes. Guiding factors, such as the enhancer blocking factor CTCF, should be active and bound at the CTCF target sites in order to direct enhancer function to the appropriate promoters (Ohlsson et al, 2001). Since CTCF is a zinc-finger factor like Ikaros or Sp1, which are displaced from mitotic chromatin, we expected that CTCF would also be lost from mitotic chromatin. Nevertheless, we demonstrate that CTCF remains bound. This was established by several in vivo methods, including indirect immunofluorescence staining, localization of GFP-CTCF in living cells and biochemical fractionation.
Further analysis mapped the major chromosome targeting domain of CTCF to the C-terminal part of the 11 zinc-finger region, whereas the N-terminal half of the zinc-finger contributes to binding without mediating strong binding on its own. The DNA binding property of the zinc-finger domain might mediate the direct attachment of CTCF to the DNA of mitotic chromosomes. Similarly, the mitotic chromosome targeting domain of HMGB1/2 is overlapping with the DNA binding domain (Pallier et al, 2003). ChIP analysis revealed that CTCF binding on mitotic chromosomes involves sequence-specific binding to the same target sites relevant for interphase CTCF function, whereas nonbinding sites remain unoccupied in interphase and mitosis. Besides direct binding to DNA, interaction with other mitotic chromosome proteins might facilitate the association of CTCF to mitotic chromosomes.
These observations support a model where CTCF mediates a cellular memory from one cell generation to the next. This may occur within a substructure such as an active chromatin hub (ACH) since it has been shown that all DNA elements involved in ACH formation in the β-globin locus contain CTCF binding sites (Palstra et al, 2003). Alternatively, the preservation of CTCF-dependent higher order chromatin conformation, such as those within the Igf2/H19 domain (Kurukuti et al, submitted), might ensure the stability of expression domains at all parts of the cell cycle, most notably from M to G1. Indeed, here we demonstrated that the epigenetically controlled long-range interaction within the Igf2/H19 locus in interphase (Murrell et al, 2004) is maintained during mitosis.
In trying to include these results together with previous publications about CTCF function into a single picture, one might envisage CTCF as a factor involved in the organization of a higher order chromatin structure. Such a structure would allow active transcription units to be separated from inactive ones. This separation has been attributed to insulator elements intensively studied in Drosophila (for review, see Labrador and Corces, 2002). One of the functions of insulators is to block enhancers from activating the ‘wrong' promoters. It has been speculated that the loop model of insulated chromatin domains in Drosophila may similarly be true for vertebrates with CTCF and insulator sequences bound to the nuclear matrix (Dunn et al, 2003; Yusufzai and Felsenfeld, 2004) and to nucleophosmin. This interaction functionally tethers insulator sequences to the nucleolus (Yusufzai et al, 2004). Although the nucleolus is degraded during mitosis, possibly the CTCF self-interaction when bound to two separated sequences (Pant et al, 2004) and other CTCF interaction partners may help to stabilize these insulator loops during mitosis. Since the enhancer/promoter interaction is lost during mitosis, as exemplified at the Igf2/H19 locus in this manuscript, these loops can easily be re-established after mitosis within the framework of the maintained insulator loops. These results offer a mechanism that higher order interactions, involved in maintaining proper expression domains, are retained in mitosis to provide a novel form of epigenetic memory during cell division.
Materials and methods
Cell culture, synchronization and DNA transfection
Cells were cultured in appropriate media supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml) and streptomycin (100 μg/ml) in an atmosphere containing 5% CO2 at 37°C. Primary amnion cells (46,XX; kindly provided by Karl-Heinz Grzeschik, Institute for Human Genetics, Marburg University) were maintained in AMF medium (WSK-Labordiagnostik, Frankfurt). HeLa cells or NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium. For mitotic synchronization, cells were incubated in fresh medium containing 50 ng/ml nocodazole for 11–12 h. Enriched mitotic cells were collected by manually shaking the dishes. Microscopic inspection and FACS analysis of the mitotic cell fraction revealed no detection of interphase cells (below 1%). This material was used for ChIP and for 3C analysis.
Calcium phosphate precipitation was used for transient and stable transfection in HeLa cells essentially as described (Chen and Okayama, 1987). For stable selection, cells were passaged at 1:50 into selection medium containing 400 μg/ml G-418 48 h after transfection. Positive clones were expanded individually.
Indirect immunofluorescence and Western blot
Double immunofluorescence was carried out on HeLa cells as described previously (Zhang et al, 2004) except that cold ethanol containing 5% (v/v) acetic acid was used for the fixation of cells. After 10 min fixation, cells were rehydrated in cold PBS containing 0.5% Triton X-100 for 5 min. Cells were then treated with blocking buffer followed by incubation with primary antibodies, either monoclonal antibodies against HP1α (1:2000 in PBS; EUROMADEX), or γ-tubulin (1:200 in PBS; Sigma), or a polyclonal antibody against the C-terminus of CTCF (1:200 in PBS; Klenova et al, 1993) for 1 h at room temperature. Secondary antibodies were either Texas red dye-conjugated affinity pure goat anti-mouse IgG (1:200; Dianova) or FITC-conjugated affinity pure goat anti-rabbit IgG (1:200; Dianova). DNA was stained with a red DNA dye, propidium iodide. Slides were mounted and analyzed by confocal laser scanning microscopy (Leica TCS 4D or API DeltaVision microscope). To show specificity of the signals, primary antibodies were preincubated with bacterially expressed and GST-purified GST fusion proteins representing the C-terminus of CTCF or GST alone as control. For the living cell imaging, cells expressing EGFP fusions with CTCF were directly subjected to the confocal analysis in the culture medium, or counterstained with Hoechst 33342 (1 μg/ml) and analyzed by fluorescence microscopy (Zeiss Axioplan 2 with FluoARC fluorescence source). Pictures were taken with AxioCam MRm camera.
Western blot was performed as described previously (Zhang et al, 2004). Primary antibodies used in the Western blot were a rabbit anti-Pol II antibody (1:200 in PBST containing 0.5% milk; Santa Cruz N-20), a rabbit anti-Sp1 antibody (1:400 in PBST; kindly provided by Dr Guntram Suske, Marburg) and a rabbit anti-CTCF antibody.
Mitotic chromosome spreads and isolation
To prepare mitotic chromosome spreads, primary amnion cells were incubated in medium containing 1 μg/ml colcemid for 3–4 h, collected by shake-off, recovered by centrifugation and resuspended in hypotonic buffer (0.25% (w/v) sodium citrate, 37.5 mM KCl). After incubation for 20 min at 37°C, cells were pelleted at 900 g for 10 min, resuspended in 0.5 ml hypotonic buffer and 5 ml ME buffer (methanol/acetic acid (4:1)) was added and mixed gently. Cells were repelleted and resuspended in fresh ME buffer. After 1 h incubation at room temperature, cells were spun onto a coverslip by centrifugation at 1000 g and air-dried. The spreads were then kept in blocking buffer and subjected to indirect immunofluorescence. Alternatively, HeLa mitotic chromosomes were spread under native conditions as described (Minc et al, 2001) to allow colocalization studies with CTCF and HP1α antibodies.
Mitotic chromosomes were isolated from cells harvested by mitotic shake-off after nocodazole treatment as described above. Cells were collected by centrifugation at 500 g for 10 min at 4°C, washed with cold PBS and resuspended in 75 mM KCl. After 20 min on ice, cells were pelleted at 3000 r.p.m. for 5 min at 4°C, and homogenized in disruption buffer (10 mM Tris–HCl, pH 7.4, 120 mM KCl, 20 mM NaCl, 0.1% Triton X-100, 2 mM CaCl2, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 0.5 μg/ml pepstatin A and 0.1 mM PMSF) by drawing through a 26-gauge needle five times. Cell lysates were pelleted by centrifugation at 6000 r.p.m. for 5 min at 4°C and the supernatant retained. Pellets were washed once with disruption buffer and the chromosome-enriched pellets were dissolved in SDS sample buffer.
3C assay
Cells growing in culture were arrested in M phase using nocodazole, and were crosslinked using formaldehyde. The reaction was quenched by the addition of glycine to 0.125 M followed by harvesting. Typically, per experiment, around 107 cells were used. Nuclei were harvested after lysis of the cells in ice-cold lysis buffer (10 mM Tris, 10 mM NaCl, 0.2% NP-40) containing protease inhibitors. Nuclei were resuspended in the appropriate restriction buffer containing 0.3% SDS and incubated for 1 h at 37°C while shaking. Triton X-100 was added to 1.8%, and the nuclei were further incubated for 1 h at 37°C to sequester SDS. The crosslinked DNA samples were digested overnight with the restriction enzyme of choice (HindIII or EcoRI). The restriction enzyme was inactivated by the addition of SDS to 1.6% and incubation at 65°C for 20 min. The reaction was diluted (to 2.5 ng/μl of genomic DNA) with ligase buffer (30 mM Tris–HCl, 10 mM MgCl2, 10 mM DTT, 1 mM ATP (pH 7.8)), and Triton X-100 was added to 1% and incubated for 1 h at 37°C. The DNA was ligated using T4 ligase for 4.5 h at 16°C followed by 30 min at room temperature. Proteinase K was added, and samples were incubated overnight at 65°C to reverse the crosslinks. The following day, samples were incubated for 30 min at 37°C with RNase, and the DNA was purified by phenol extraction and ethanol precipitation.
The PCR analysis was optimized by digesting and ligating a YAC clone, which encompassed the entire Igf2-H19 locus. The correctly ligated fragments were singled out among all possible ligation products by amplification using the specific primers. Typically, 30 ng of ligated material from the above processed samples from mitotically arrested as well as asynchronized cells was used.
The primer sequences are shown in Supplementary data. The Calreticulin primers have been used previously (Tolhuis et al, 2002). The conditions used for PCR were 94°C for 5 min, 94°C for 30 s, 55°C for 1 min, 72°C for 30 s, PCR following repeat step 2–36 times, final elongation at 72°C for 10 min and 4°C forever. Control samples without ligation (3C ligation: −; Figure 6C) were always negative in the PCR.
To test whether the PCR products obtained above are not a result of random ligation among various digested samples but are due to specific intramolecular ligation because of close proximity in the nucleus, we performed PCR using one primer each from Calreticulin and Igf2 DMR1, which will give amplification if the ligation observed here is random and intermolecular. The control template to check the PCR efficiency of these primers was prepared by amplifying regions spanning HindIII sites in Calreticulin and Igf2 DMR1 used in the 3C assays, mixing them in an equimolar amount, followed by digestion overnight at 37°C with HindIII and subsequent ligation. Typically, 5 ng of this ligation mix was used for amplification using the same conditions stated above.
Supplementary Material
Supplementary Figure 1
Supplementary information: Materials and Methods
Acknowledgments
We thank Leni Schäfer-Pfeiffer for technical assistance, Karl-Heinz Grzeschik for primary cells and Guntram Suske for antibodies. This work was supported in part by grants from the Swedish Science Research Council, and the Swedish Cancer Research Foundation (to RO), the Netherlands Organisation of Scientific Research NWO-ALW (to NG) and the Deutsche Forschungsgemeinschaft DFG (to RR). NWO-ALW and DFG grants have been evaluated by the European Science Foundation (EuroDYNA).
References
- Adams RR, Carmena M, Earnshaw WC (2001) Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol 11: 49–54 [DOI] [PubMed] [Google Scholar]
- Akoulitchev S, Reinberg D (1998) The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev 12: 3541–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Bellier S, Dubois MF, Nishida E, Almouzni G, Bensaude O (1997) Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol Cell Biol 17: 1434–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube NG, Smeenk CA, Picketts DJ (2000) Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet 9: 539–547 [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G (2002) The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci USA 99 (Suppl 4): 16433–16437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Okayama H (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7: 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S (2005) Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol 168: 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Hinkley CS, Henry RW, Huang S (2002) TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell 13: 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 [DOI] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T (2002) Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol 4: 79–82 [DOI] [PubMed] [Google Scholar]
- Dantzer F, Luna L, Bjoras M, Seeberg E (2002) Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res 30: 2349–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K (2000) A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol 20: 6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovat S, Ronni T, Russell D, Ferrini R, Cobb BS, Smale ST (2002) A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev 16: 2985–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KL, Zhao H, Davie JR (2003) The insulator binding protein CTCF associates with the nuclear matrix. Exp Cell Res 288: 218–223 [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS (2004) Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303: 238–240 [DOI] [PubMed] [Google Scholar]
- Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ (2001) CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet 28: 335–343 [DOI] [PubMed] [Google Scholar]
- Gazit B, Cedar H, Lerer I, Voss R (1982) Active genes are sensitive to deoxyribonuclease I during metaphase. Science 217: 648–650 [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ (1997) Mitotic repression of the transcriptional machinery. Trends Biochem Sci 22: 197–202 [DOI] [PubMed] [Google Scholar]
- Hagstrom KA, Meyer BJ (2003) Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet 4: 520–534 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- John S, Workman JL (1998) Bookmarking genes for activation in condensed mitotic chromosomes. BioEssays 20: 275–279 [DOI] [PubMed] [Google Scholar]
- Kanai M, Uchida M, Hanai S, Uematsu N, Uchida K, Miwa M (2000) Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem Biophys Res Commun 278: 385–389 [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856 [DOI] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV (1993) CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol 13: 7612–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, Verdin E, Bazett-Jones DP (2001) Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem 276: 38307–38319 [DOI] [PubMed] [Google Scholar]
- Kuhn EJ, Geyer PK (2003) Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol 15: 259–265 [DOI] [PubMed] [Google Scholar]
- Kuo MT, Iyer B, Schwarz RJ (1982) Condensation of chromatin into chromosomes preserves an open configuration but alters the DNase I hypersensitive cleavage sites of the transcribed gene. Nucleic Acids Res 10: 4565–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Corces VG (2002) Setting the boundaries of chromatin domains and nuclear organization. Cell 111: 151–154 [DOI] [PubMed] [Google Scholar]
- Long JJ, Leresche A, Kriwacki RW, Gottesfeld JM (1998) Repression of TFIIH transcriptional activity and TFIIH-associated cdk7 kinase activity at mitosis. Mol Cell Biol 18: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz M, Burke LJ, LeFevre P, Myers FA, Thorne AW, Crane-Robinson C, Bonifer C, Filippova GN, Lobanenkov V, Renkawitz R (2003) Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. EMBO J 22: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C (1995) Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83: 29–38 [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D (1997) Marking of active genes on mitotic chromosomes. Nature 388: 895–899 [DOI] [PubMed] [Google Scholar]
- Minc E, Allory Y, Courvalin JC, Buendia B (2001) Immunolocalization of HP1 proteins in metaphasic mammalian chromosomes. Methods Cell Sci 23: 171–174 [DOI] [PubMed] [Google Scholar]
- Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B (1999) Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108: 220–234 [DOI] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke LJ, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V (2005) CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, Quitschke W, Chernukhin I, Klenova E, Lobanenkov V, Ohlsson R (2004) The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res 14: 1594–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W (2004) Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893 [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Pallier C, Scaffidi P, Chopineau-Proust S, Agresti A, Nordmann P, Bianchi ME, Marechal V (2003) Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol Biol Cell 14: 3414–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R (2004) Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol 24: 3497–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, Ohlsson R (2003) The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev 17: 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons GG, Spencer CA (1997) Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol Cell Biol 17: 5791–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand E, Ben-Porath I, Keshet I, Cedar H (2004) CTCF elements direct allele-specific undermethylation at the imprinted H19 locus. Curr Biol 14: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Rasko JE, Klenova EM, Leon J, Filippova GN, Loukinov DI, Vatolin S, Robinson AF, Hu YJ, Ulmer J, Ward MD, Pugacheva EM, Neiman PE, Morse HC III, Collins SJ, Lobanenkov VV (2001) Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res 61: 6002–6007 [PubMed] [Google Scholar]
- Schoenherr CJ, Levorse JM, Tilghman SM (2003) CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet 33: 66–69 [DOI] [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N (1996) Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev 10: 2389–2400 [DOI] [PubMed] [Google Scholar]
- Szabo P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR (2000) Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10: 607–610 [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Sugiura A, Omori A, Felsenfeld G, Engel JD, Fukamizu A (2003) Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol Cell Biol 23: 8946–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Quitschke WW (1997) The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem 272: 33353–33359 [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Felsenfeld G (2004) The 5′-HS4 chicken β-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc Natl Acad Sci USA 101: 8620–8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SM, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS (2003) Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc Natl Acad Sci USA 100: 14852–14857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Burke LJ, Rasko JE, Lobanenkov V, Renkawitz R (2004) Dynamic association of the mammalian insulator protein CTCF with centrosomes and the midbody. Exp Cell Res 294: 86–93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary information: Materials and Methods


